Abstract
In this work, multi-functional bio-synthetic hybrid nanostructures were prepared and studied for their potential utility in the recognition and inhibition of mRNA sequences for inducible nitric oxide synthase (iNOS), which are overexpressed at sites of inflammation, such as in cases of acute lung injury. Shell crosslinked knedel-like polymer nanoparticles (SCKs) that present peptide nucleic acids, for binding to complementary mRNAs, and cell penetrating peptides (CPPs), to gain cell entry, along with fluorescent labels and sites for radiolabeling, were prepared by a series of robust, efficient and versatile synthetic steps that proceeded from monomers to polymers to functional nanoparticles. Amphiphilic block graft copolymers having combinations of methoxy- and thioacetyl-terminated poly(ethylene glycol) (PEG) and DOTA-lysine units grafted from the backbone of poly(acrylic acid) (PAA) and extending with a backbone segment of poly(octadecyl acrylate-co-decyl acrylate) (P(ODA-co-DA)) were prepared by a combination of reversible addition-fragmentation chain transfer (RAFT) polymerization and chemical modification reactions, which were then used as the building blocks for the formation of well-defined SCKs decorated with reactive thiols accessible to the surface. Fluorescent labeling with Alexa Fluor 633 hydrazide was then accomplished by amidation with residual acrylic acid residues within the SCK shells. Finally, the PNAs and CPP units were covalently conjugated to the SCKs via Michael addition of thiols on the SCKs to maleimide units on the termini of PNAs and CPPs. Confirmation of the ability of the PNAs to bind selectively to the target iNOS mRNAs when tethered to the SCK nanoparticles was determined by in vitro competition experiments. When attached to the SCKs having a hydrodynamic diameter of 60 ± 16 nm, the Kd values of the PNAs were ca. an order of magnitude greater than the free PNAs, while the mismatched PNA showed no significant binding.
INTRODUCTION
The mortality associated with acute lung injury (ALI) and acute respiratory distress syndrome (ARDS) remains high in the U.S. and around the world 1, establishing the need for improved treatment and prevention strategies. ALI is an inflammatory process in the airspaces and lung parenchyma that is characterized by fluid accumulation (pulmonary edema) and it can progress to an acute stage that presents necrosis of the lung tissue 2. One of the major indications of ALI is the overexpression of inducible nitric oxide synthase (iNOS), an enzyme that is found predominantly in the cells of epithelial origin. The production of iNOS is caused by the activation of proinflammatory cytokines such as interleukin-lβ (IL-lβ) and tumor necrosis factor-α (TNF-α), which are signaling molecules that are released as a part of the immune response due to the presence of bacterial products (e.g., lipopolysaccharides (LPS)) 3. The upregulation of iNOS results in the production of nitric oxide (NO), which when present in regulated amount can act as a mediator of vital biological functions in the lung 4. However, the overexpression of this enzyme leads to the production of high concentrations of NO in an unregulated fashion, causing harm to cells and resulting in cell death 5. Studies have shown that the inhibition of iNOS prevents the accumulation of NO and improves the symptoms of ALI 6, 7.
Because inducible nitric oxide synthase is both a biomarker and an exacerbating agent for ALI, there is great interest in the development of diagnostic agents and inhibitors for iNOS. Most of the work in this area has focused on imaging agents 8–10 and inhibitors for the iNOS protein 11 as well as imaging agents for detecting the formation of the nitric oxide that iNOS catalyzes 12–14. An alternative strategy would be to target the mRNA encoding the iNOS protein with antisense agents that could have a dual function of both detecting iNOS mRNA expression level and inhibiting translation of the mRNA to the protein. One ideal class of antisense agents for this purpose are peptide nucleic acids (PNAs), which are synthetic analogs of DNA, in which the sugar phosphate backbone is replaced with an uncharged pseudopeptide backbone composed of N-(2-amino-ethyl) glycine units to which the nucleobases are linked through methylene carbonyl linkers 15, 16. Their attractiveness stems from their stability to degradation in vivo, high affinity for mRNA, and ability to block translation 17–19. Additionally, PNAs are able to invade regions of secondary structure in the target mRNA, and the resulting PNA·RNA complexes are resistant to the action of RNase H which would otherwise degrade the mRNA and result in a loss of signal. While there are several reports on the use of PNAs to both inhibit 19 and image gene expression 20–24, application of PNAs as regulators and imaging agents of gene expression in vivo has been hampered by their poor cellular uptake and rapid clearance 15, 25, 26. To overcome the permeability problems, PNAs have been conjugated to a variety of cell penetrating peptides and other ligands, which have been shown to improve their uptake in cell culture through endocytosis though they often remain trapped in endosomes 27, 28. In addition, simple cell penetrating PNA conjugates suffer from rapid clearance in vivo due to their small size. One way to circumvent these problems is to conjoin PNAs with nanoparticles having optimal cell penetrating, endosomal escape, pharmacokinetic, and biodistribution properties.
In our group, we have had long standing interest in developing novel polymeric systems and chemistries for the preparation of shell crosslinked knedel-like (SCK) nanoparticles 29–33 and fine tuning of existing nanostructures 34–39 for a diverse set of applications in biomedical research 40–48. SCKs are composed of block copolymers with hydrophilic and hydrophobic segments, making them amphiphilic and prone to self assembly in selective solvents. Due to crosslinking between adjacent groups present in the shell, SCKs possess structural stability. Moreover, their tunable size, shape and ability to perform chemical modifications within selective sub-particle regions 49, 50, allows for the conjugation of biologically active ligands 41, 43, 51, 52, such as PNAs 53, to their surfaces, making them a versatile agent for gene delivery and transfection 52, 54–57. In the past, we have employed PNAs conjugated to SCKs to direct their assembly into higher order structures via selective and tunable binding interactions 53, and for intracellular delivery of PNA through a bioreductively-cleavable linker 54. The versatility of SCKs also allows for fine tuning of the core properties, resulting in shape adaptable nanoparticles 58–61 that might enhance multivalent binding recognition events. Herein, we report the design and synthesis of hierarchically-assembled, multi-functional SCKs that are conjugated with PNAs specific for iNOS mRNA to promote recognition and inhibit iNOS expression. These SCKs were also conjugated to the protein transduction domain of human immunodeficiency virus type 1 TAT protein to facilitate cellular entry, and to a fluorescent probe or DOTA for labeling with the PET imaging radionuclide 64Cu for imaging purposes. The PNA-SCK conjugates were shown to sequence-specifically bind to iNOS mRNA with high affinity by comparing the binding of matched and mismatched PNA-SCK conjugates.
EXPERIMENTAL SECTION
Materials and Methods
Polymerizations were performed on a double manifold with glassware and syringes that were dried in an oven (100 °C) for at least 1 h, and with syringes that were washed with N2 (3 ×), prior to use. tert-Butyl acrylate (t-BA, 99%) was received from Sigma-Aldrich Company (St. Louis, MO), distilled over calcium hydride, and stored under N2 prior to use. Octadecyl acrylate, decyl alcohol, trifluoroacetic acid (TFA, 95%; Aldrich), 2,2′-(ethylenedioxy)-bis(ethylamine) (97%, Aldrich), 1-(3′-dimethylaminopropyl)-3-ethylcarbodiimide methiodide (EDCI, 98%, Aldrich), hydroxybenzotriazole (HOBT, Aldrich), 2–(1H–benzotriazol–1–yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU, Aldrich), Boc-NH-PEG3kDa-NH2 (Rapp Polymere, Tübingen, Germany), MeO-PEG2kDa-NH2 (Rapp Polymere, Tübingen, Germany), hydroxyl amine hydrochloride (98%, Aldrich), Sephadex G50 (Medium, GE Healthcare, Piscataway, NJ), N-succinimidyl-S-acetylthiopropionate (SATP) (Pierce, Rockford, IL), and 4-maleimido butyric acid (Aldrich), N,N-diisopropylethylamine (DIPEA) (Aldrich), diethyl ether (anhydrous) (Aldrich), Alexa Fluor 633 hydrazide (Invitrogen) and O-(7-Azabenzo-triazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HATU) were used as received. Fmoc-XAL PEG-PS resin was purchased from PerSeptive Biosystems (Ramsey, MN). GGYGRKKRRQRRR-maleimide was purchased from Chengdu Kaijie Bio-pharmaceuticals Co., Ltd (Chengdu, China). PNA monomers (Fmoc-A-(Bhoc)-OH, Fmoc-C-(Bhoc)-OH, Fmoc-G-(Bhoc)-OH and Fmoc-T-OH) were obtained from Panagene (Daejeon, Korea). (S-1-dodecyl-S′-(α,α′-dimethyl-α″-acetic acid)trithiocarbonate) (DDMAT) and Boc-protected DOTA-lysine (a lysine derivative of 1, 4, 7, 10-tetraazocyclododecane-N, N′, N″, N‴-tetraacetic acid (DOTA)) were synthesized as reported previously 44–46, 62. All other reagents were obtained from Sigma-Aldrich and used as received. Ellman’s reagent kit (Pierce) was used for Ellman’s assay, with the calibration curve constructed using cysteine monohydrate in aqueous buffer (100 mM PBS, 0.1 M NaCl, 10 mM EDTA, pH 7.4) at 412 nm. Spectra/Por membrane tubes were purchased from Spectrum Medical Industries, Inc., and were used for dialysis. Nanopure water (18 MΩ•cm) was acquired by means of a Barnstead Nanopure ultrapure water purification system (Thermo Scientific, Asheville, NC).
1H NMR and 13C NMR spectra were recorded on an Inova 300 or Mercury 300 spectrometer interfaced to a UNIX computer using VnmrJ software. Samples were prepared as solutions in CDCl3, CD2Cl2, or DMF-d7 and solvent protons were used as internal standard. IR spectra were recorded on an IR Prestige 21 system (Shimadzu Corp., Japan). A small amount of sample was placed to cover the ATR crystal for IR measurements. Data were analyzed using IRsolution software. UV-vis absorption measurements were made using a UV-2550 system (Shimadzu Corp., Japan) using PMMA cuvettes. Spectra were analyzed with UV-Probe 2.33 software.
Gel permeation chromatography was conducted on a system equipped with a Waters Chromatography, Inc. (Milford, MA) model 1515 isocratic pump and a model 2414 differential refractometer with a three-column set of Polymer Laboratories, Inc. (Amherst, MA) Styragel columns (PLgel 5μm Mixed C, 500 Å, and 104 Å, 300 × 7.5 mm columns) and a guard column (PLgel 5μm, 50 × 7.5 mm). The system was equilibrated at 40 °C in tetrahydrofuran (THF), which served as the polymer solvent and eluent (flow rate set to 1.00 mL/min). The differential refractometer was calibrated with Polymer Laboratories, Inc. polystyrene standards (300 to 467,000 Da). Polymer solutions were prepared at a concentration of ca. 3 mg/mL with 0.05% vol toluene as flow rate marker and an injection volume of 200 μL was used. Data were analyzed using Empower Pro software from Waters Chromatography Inc.
Differential scanning calorimetry studies were performed on a Mettler Toledo DSC822 calibrated according to the standard procedures using Indium. The heating rates were 10 °C min−1 and cooling rates were 5 °C min−1 with a temperature range of −120–150 °C. The crystalline melting point (Tm) and crystallization temperature (Tc) were determined during the second heating or cooling cycles, respectively. Thermogravimetric analysis was performed under Ar atmosphere using a Mettler Toledo model TGA/DSC1 with a heating rate of 10 °C/min. Measurements were analyzed using Mettler Toledo STARe software v.10.00.
Samples for TEM were prepared by depositing 5 μL of sample to glow discharged carbon coated copper grids. Excess sample was wicked off using filter paper and the grids were allowed to dry in air for 1 minute. The grids were then stained with 5 μL of 2% uranyl acetate and excess stain was wicked off using filter paper. Specimens were observed on a JEOL 1200EX transmission electron microscope operating at 100 kV and micrographs were recorded at calibrated magnifications using an SIA-15C CCD camera. The final pixel size was 0.42 nm/pixel. The number-average particle diameters (Dav) and standard deviations were generated from the analysis of particles from at least two different micrographs.
Dynamic light scattering (DLS) measurements were conducted using Delsa Nano C from Beckman Coulter, Inc. (Fullerton, CA) equipped with a laser diode operating at 658 nm. Size measurements were made in water (n = 1.3329, η = 0.890 cP at 25 ± 1 °C; n = 1.3293, η = 0.547 cP at 50 ± 1 °C; n = 1.3255, η = 0.404 cP at 70 ± 1 °C). Scattered light was detected at 165° angle and analyzed using a log correlator over 70 accumulations for a 0.5 mL of sample in a glass sizing cell (0.9 mL capacity). The photomultiplier aperture and the attenuator were automatically adjusted to obtain a photon counting rate of ca. 10 kcps. The calculations of the particle size distribution and distribution averages were performed using CONTIN particle size distribution analysis routines. Prior to analysis, the samples were filtered through a 0.45 μm Whatman Nylon membrane filter (Whatman, Inc.). The samples in the glass sizing cell were equilibrated at the desired temperature for 5 min before measurements were made. The peak average of histograms from intensity, volume or number distributions out of 70 accumulations was reported as the average diameter of the particles.
PNA synthesis was carried out on an automated ABI 8909 synthesizer (Applied Biosystems, Carlsbad, CA) with a PNA option. High-pressure liquid chromatography (HPLC) of PNAs was carried out on Beckman Coulter System Gold 126 HPLC system. Matrix-assisted laser desorption ionization (MALDI) mass spectra of PNA conjugates were measured on a PerSeptive Voyager RP MALDI-time of flight (TOF) mass spectrometer using CHCA as a matrix and calibrated versus insulin (average [M + H] = 5734.5) which was present as an internal standard.
DNA was 5′-labeled by [γ-32P]-ATP with T4 polynucleotide kinase. DNA, T4 polynucleotide kinase buffer, H2O, [γ-32P]-ATP and T4 polynucleotide kinase were mixed together and incubated for 1 h at 37 °C, followed by boiling for 20 min to inactivate the kinase. Radiolabeled DNA bands with corrected sizes were purified from polyacrylamide gel and taken forward for further experiments.
Synthesis of poly(tert-butyl acrylate) (PtBA140) (1)
To a 100 mL Schlenk flask equipped with a stir bar, DDMAT (284 mg, 0.779 mmol) was added as a chain transfer agent followed by AIBN (6.40 mg, 39.0 μmol) and 2-butanone (11.2 g, 156 mmol). Monomer tBA (20.0 g, 156 mmol) was added and the Schlenk flask was then sealed with a rubber septum. The reaction mixture was allowed to stir at room temperature to ensure homogeneity. After performing four freeze-pump-thaw cycles, the reaction mixture was allowed to equilibrate to room temperature before being inserted into a preheated oil bath at 60 °C. The kinetics of polymerization were monitored by analyzing aliquots at predetermined times via 1H NMR spectroscopy. After 2 h, the desired conversion was achieved and the reaction was quenched by immersing the Schlenk flask into liquid N2. A minimum amount of THF was used to dissolve the reaction mixture which was precipitated three times into a mixture of methanol and ice to afford the homopolymer of PtBA. The polymer was dissolved in dichloromethane and dried over magnesium sulfate. Finally, the solvent was removed under vacuum to yield (13.0 g, 68 % yield, based upon 80% conversion) of poly(tert-butyl acrylate) as a bright yellow powder. (Mn)NMR = 18.3 kDa. (Mn)GPC = 23.6 kDa. (Mw)GPC = 26.4 kDa. PDI = 1.12. IR: 2920, 2830, 1716, 1443, 1364, 1285, 1125, 813, 745 cm−1. 1H NMR (CD2Cl2) δ: 0.85 (t, J = 7.0 Hz, CH3CH2-), 1.20–1.85 (br, -CHCH2- of the polymer backbone, alkyl chain initiator and HOOCC(CH3)2-), 2.20 (br, -CHCH2- of the polymer backbone), 3.20 (t, J = 7.0 Hz, -SCSCH2-), 4.6–4.7 (br, -CH2CHS-) ppm. 13C NMR (CD2Cl2) δ: 12.5, 22.7, 27.9, 29.5, 36.1, 37.3, 41.9–42.4, 80.2, 173.9–174.1 ppm. DSC: (Tg) = 48 °C. TGA: Tonset = 186 °C, Tdecomposition: (186–228 °C) 37.5 % mass loss; (228–307 °C) 7.5% mass loss; (307–472 °C) 27% mass loss; 28% mass remaining.
Synthesis of poly(tert-butyl acrylate)140-b-poly(octadecyl acrylate16-co-decyl acrylate43) (PtBA140-b-P(ODA16-co-DA43) (2)
To a 50 mL flame–dried Schlenk flask equipped with a stir bar, ODA (1.67 g, 5.14 mmol) was added followed by DA (2.95 g, 13.9 mmol), PtBA (3.50 g, 0.190 mmol) and a stock solution of AIBN (1.56 mg, 9.50 μmol) prepared in toluene. The monomers and macrochain transfer agent were allowed to dissolve in toluene (3.50 g, 38.0 mmol). The reaction mixture was degassed by three freeze pump thaw cycles and allowed to warm to room temperature. Polymerization was facilitated by immersing the Schlenk flask into a preheated oil bath at 70 °C. Progress of polymerization was monitored by 1H NMR spectroscopy of aliquots at pre-determined times. Upon attaining the desired degree of polymerization, after 1.5 h, the reaction was quenched by immersing the reaction flask into liquid N2. The reaction mixture was dissolved in a minimum amount of THF (6.00 mL) and three consecutive precipitations were performed in methanol in an ice bath. After the final precipitation, a yellow powder (3.35 g, 70% yield, based on 60% conversion) was obtained after removal of solvent and subsequent drying in vacuo. (Mn)NMR = 32.6 kDa. (Mn)GPC = 26.8 kDa. (Mw)GPC =29.7 kDa. PDI = 1.10. IR: 2920, 2830, 1716, 1443, 1364, 1285, 1125, 813, 745 cm−1. 1H NMR (CD2Cl2) δ: 0.88 (t, J = 6.9 Hz, -CH3CH2- of ODA, DA and chain transfer agent), 1.35–1.76 and 1.70–1.98 (br, -CH2CH-backbone of PODA, PDA, CTA and HOOCC(CH3)2-), (2.05–2.40 (br, -CH2CH-), 4.00 (br, -OCH2CH2-, PODA, PDA) ppm. 13C NMR (CD2Cl2) δ: 13.8, 22.7, 25.9, 27.7–27.8, 28.7, 29.4, 29.6, 31.9, 34.5–37.5, 41.4–41.9, 64.6, 80.2, 173.8–174.1 ppm. DSC: (Tg) = 41 °C. (Tm) = −2.2 and 9.1 °C. (Tc) = −6.8 and 3.4 °C. TGA: Tonset = 209 °C, Tdecomposition: (209–250 °C) 11 % mass loss; (250–485 °C) 70% mass loss; 20% mass remaining.
Synthesis of poly(acrylic acid)140-b-poly(octadecyl acrylate16-co-decyl acrylate43) (PAA140-b-P(ODA16-co-DA43)) (3)
A round bottom flask equipped with stir bar was charged with 2 (1.00 g, 30.7 μmol). Dichloromethane (10 mL) was added followed by dropwise addition of TFA (10 mL) and the reaction mixture was allowed to stir for 24 h at RT. The solvent was removed under vacuum and the polymer was allowed to dissolve in a minimum amount of THF. Upon extensive dialysis against nanopure water and lyophilization, a pale white fluffy solid of PAA140-b-P(ODA16-co-DA43) (3) (0.74 g, 98%) was obtained. (Mn)NMR = 24.7 kDa. IR: 3600-2350, 1716, 1443, 1250, 1159, 830, 750 cm−1. 1H NMR (DMF-d7) δ: 0.86 (t, J = 6.6 Hz, -CH3CH2- of ODA, DA and chain transfer agent), 1.15–1.4 (br, -CH2-alkyl chain of PODA, PDA, CTA and HOOCC(CH3)2-), 1.95–2.1 (br, -CH2CH- backbone of PODA, PDA and CTA), 2.05–2.20 (br, -CH2CH- backbone of PODA, PDA and CTA), 4.00 (br, -OCH2CH2-, PODA, PDA) ppm. 13C NMR (DMF-d7) δ: 14.5, 23.5, 27.0, 29.5, 30.5, 31.0, 33.0, 35.1–37.5, 41.5–43.0, 65.0, 174.1–175.1, 175.9 ppm. DSC: (Tg) = 129.5 °C. (Tm) = 6.3 °C, (Tc) = 3.1 °C. TGA: Tonset = 190°C, Tdecomposition: (190–294 °C) 18 % mass loss; (294–495 °C) 68% mass loss; 14% mass remaining.
Synthesis of poly(acrylic acid137)-g-DOTA3-b-poly(octadecyl acrylate16-co-decyl acrylate43) (PAA137-g-(CONH-lysine-DOTA-Boc)3-b-P(ODA16-co-DA43)) (4)
General procedure for amidation, involving the attachment of DOTA
A 50 mL flame-dried RB flask equipped with a stir bar was charged with 3 (350 mg, 14.1 μmol) and DMF (5.00 g). Stock solutions of HOBT, HBTU and CONH-lysine-DOTA-Boc were prepared in dry DMF. After the polymer was allowed to dissolve completely, HBTU (56.1 mg, 0.148 mmol), HOBt (19.9 mg, 0.148 mmol) and DIPEA (6.19 mg, 0.148 mmol) were added at intervals of 30 min. DOTA-lysine (51.7 mg, 73.8 μmol) was added and the solution was stirred for 24 h at RT. The reaction mixture was then transferred to dialysis tubing (MWCO ca.6000–8000 Da) presoaked in nanopure water. Extensive dialysis against nanopure water followed by lyophilization afforded a white powder of 3 (235 mg, 89.0%, 3 DOTA-lysine per chain, 60% coupling efficiency). Mn=26.7 kDa. IR: 3400-2570, 1715, 1580, 1477, 1250, 1160, 978, 830, 750 cm−1. 1H NMR (DMF-d7) δ: 0.86 (t, J = 6.6 Hz, -CH3CH2- of ODA, DA and chain transfer agent, CH3- tert-butyl groups of DOTA), 1.15–1.4 (br, -CH2-alkyl chain of PODA, PDA, CTA and HOOCC(CH3)2-), 1.95–2.1 (br, -CH2CH- backbone of PODA, PDA and CTA) 2.05–2.20 (br, -CH2CH- backbone of PODA, PDA and CTA), 2.3–2.4 (br, DOTA), 2.95–3.10 (br, DOTA), 4.00 (br, -OCH2CH2-, PODA, PDA) ppm. 13C NMR (DMF-d7) δ: 14.5, 23.5, 27.0, 29.5, 30.5, 31.0, 33.0, 35.1–37.5, 41.5–43.0, 53.0, 56.0, 65.0, 82.0, 174.1–175.1, 177.6 ppm. DSC: (Tg) = 131.8 °C. (Tm) = 1.0 °C, (Tc) = −3.0 °C. TGA: Tonset = 112 °C, Tdecomposition: (112–190°C) 9 % mass loss; (190–247 °C) 7% mass loss; (264–278 °C) 7% mass loss; (278–395 °C) 38% mass loss; (395–500 °C) 19% mass loss; 20% mass remaining.
Synthesis of poly(acrylic acid)131-g-(PEG3 kDa-Boc)3-g-(mPEG2 kDa)3-g-(CONH-lysine-DOTA-Boc)3-b-poly(octadecyl acrylate16-co-decyl acrylate43) (PAA131-g-(PEG3 kDa-Boc)3-g-(mPEG2 kDa)3-g-(CONH-lysine-DOTA-Boc)3-b-P(ODA16-co-DA43)) (5)
A reaction mixture of 4 (97.0 mg, 3.63 μmol), HBTU (19.4 mg, 51.1 μmol), HOBt (12.1 mg, 51.1 μmol), DIPEA (6.58 mg, 51.1 μmol), 2 kDa mPEG-NH2 (25.8 mg, 12.8 μmol), and 3 kDa Boc-NH-PEG-NH2 (82.3 mg, 12.8 μmol) in DMF was allowed to undergo reaction under the general procedure outlined for amidation above to yield an amorphous white solid of 5, PAA131-g-(PEG3 kDa-Boc)3-g-(mPEG2 kDa)3-g-(CONH-lysine-DOTA-Boc)3-b-P(ODA16-co-DA43) (133 mg, 88%), having ca. 3 of each type of PEG per chain, 75 % coupling efficiency. (Mn)NMR = 41.6 kDa. IR: 3600-2580, 1715, 1543, 1455, 1250, 1160, 1114, 966, 807, 760 cm−1. 1H NMR (DMF-d7) δ: 0.86 (t, J = 6.6 Hz, -CH3CH2- of ODA, DA and chain transfer agent, CH3- tert-butyl groups of DOTA), 1.15–1.4 (br, CH2-alkyl chain of PODA, PDA, CTA and HOOCC(CH3)2-), 1.95–2.10 (br, -CH2CH- backbone of PODA, PDA and CTA) 2.05–2.20 (br, 3 H, -CH2CH- backbone of PODA, PDA and CTA), 2.30–2.4 (br, DOTA), 2.95–3.10 (br, DOTA), 3.20 (s, -OCH3, PEG), 3.50 (-CH2-,br, PEG), 4.00 (br, -OCH2CH2-, PODA, PDA) ppm. 13C NMR (DMF-d7) δ: 14.5, 23.5, 27.0, 29.5, 30.5, 31.0, 33.0, 35.1–37.5, 41.5–43.0, 52.0, 56.0, 58.6, 65.0, 70.8, 82.0, 174.6, 176.8 ppm. DSC: (Tm) = −43.8 °C, (Tm) (ODA-co-DA) = 8.2 °C, (Tm) (PEO) = 33.6 °C, (Tc) = −44.3 and 3.8 °C. TGA: Tonset = 150 °C, Tdecomposition: (150–438 °C) 68% mass loss; (438–500 °C) 6% mass loss; 26% mass remaining.
Synthesis of poly(acrylic acid)131-g-(PEG3 kDa-NH2)3-g-(mPEG2 kDa)3-g-(CONH-lysine-DOTA-COOH)3-b-poly(octadecyl acrylate16-co-decyl acrylate43) (PAA131-g-(PEG3 kDa-NH2)3-g-(mPEG2 kDa)3-g-(CONH-lysine-DOTA-COOH)3-b-P(ODA16-co-DA43)) (6)
In a round bottom flask was placed 5 (90.0 mg, 2.16 μmol) and TFA (6.00 mL) was added dropwise. The reaction mixture was allowed to stir for 24 h at RT. The TFA was then removed under a flow of N2, the polymer was dissolved in THF, the solution was then dialyzed against nanopure water, and the polymer was isolated by lyophilization to obtain 6 as a fluffy white powder (55 mg, 65%). (Mn) NMR = 41.0 kDa. IR: 3600–2600, 1715, 1590, 1430, 1250, 1135, 945, 820, 760 cm−1. 1H NMR (DMF-d7) δ: 0.86 (t, J = 6.6 Hz, -CH3CH2- of ODA, DA and chain transfer agent), 1.15–1.4 (br, CH2- alkyl chain of PODA, PDA, CTA and HOOCC(CH3)2-), 1.95–2.1 (br, -CH2CH- backbone of PODA, PDA and CTA), 2.05–2.20 (br, -CH2CH- backbone of PODA, PDA and CTA), 2.3–2.4 (br, DOTA), 2.95–3.10 (br, DOTA), 3.20 (s, -OCH3, PEG), 3.50 (-CH2-,br, PEG), 4.00 (br, 4 H, -OCH2CH2-, PODA, PDA) ppm. 13C NMR (DMF-d7) δ: 14.5, 23.5, 27.0, 29.5, 30.5, 31.0, 33.0, 35.1–37.5, 41.5–43.0, 52.0, 58.0, 58.6, 65.0, 70.5, 174.1–175.1, 176.8 ppm. DSC: (Tm) = −43.5 °C, (Tm)(ODA-co-DA) = 10.8 °C, (Tm)(PEO) = 29.8 °C, (Tc) = −35.2 and 3.9 °C. TGA: Tonset = 150 °C, Tdecomposition: (150–438 °C) 68% mass loss; (438–500 °C) 6% mass loss; 26% mass remaining.
Synthesis of poly(acrylic acid)131-g-(PEG3 kDa- NHCO-C2H4-SCOCH3)3-g-(mPEG2 kDa)3-g-(CONH-lysine-DOTA-COOH)3-b-poly(octadecyl acrylate16-co-decyl acrylate46) (PAA131-g-(PEG3 kDa-NHCO-C2H4-SCOCH3)3–g-(mPEG2 kDa)3-(CONH-lysine-DOTA-COOH)3-b-P(ODA16-co-DA43)) (7)
In a round bottom flask, 6 (30 mg, 0.68 μmol) was dissolved in dry DMF (4.00 mL), SATP (4.74 mg, 20.5 μmol), DIPEA (20 μL, 0.1 μmol) was added and the reaction mixture was stirred at RT. The reaction mixture was then transferred into presoaked dialysis tubing (MWCO ca. 6000–8000 Da) to remove small molecule impurities. After extensive dialysis and lyophilization 7 was obtained (25 mg, 82%). (Mn) NMR = 41.7 kDa. IR: 3600-2600, 1715, 1590, 1415, 1250, 1120, 920, 820, 730 cm−1. 1H NMR (DMF-d7) δ: 0.86 (t, J = 6.6 Hz, -CH3CH2 of ODA, DA and chain transfer agent), 1.15–1.40 (br, CH2- alkyl chain of PODA, PDA, CTA and HOOCC(CH3)2), 1.95–2.10 (br, -CH2CH- backbone of PODA, PDA and CTA), 2.05–2.20 (br, -CH2CH- backbone of PODA, PDA and CTA ), 2.30–2.40 (br, DOTA), 2.71–2.75 (br, -CH2S-), 2.95–3.10 (br, DOTA), 3.50 (-OCH2-CH2O-,br, PEG) 4.00 ( br, -OCH2CH2-, PODA, PDA) ppm. 13C NMR (DMF-d7) δ: 14.5, 23.5, 27.0, 29.5, 30.5–31.5, 33.0, 35.1–37.5, 41.5–43.0, 55.8, 57.5, 65.0, 70.5, 174.1–175.1, 176.5 ppm. DSC: (Tm) = −44.3 °C, (Tm)(ODA-co-DA) = 11.3 °C, (Tm)(PEO) = 27.8 °C, (Tc) = −36.4 and 6.1 °C. TGA: Tonset = 170 °C, Tdecomposition: (170–283 °C) 4% mass loss; (283–460 °C) 87.3% mass loss; 8.7% mass remaining.
Procedure for the preparation of micelles (8)
To 7 in DMF (8.0 mg, 1.0 mgmL−1) was added an equal volume of nanopure water dropwise via a syringe pump over the course of 3 h. The mixture was allowed to stir for 16 h at RT and was then transferred to presoaked dialysis tubing (MWCO ca. 6000–8000 Da) and dialyzed against nanopure water for ca. 4 d to remove the organic solvent and give a final polymer concentration of 0.32 mgmL−1. (Dh)n (DLS) = 43 ± 8 nm; (Dh)vol (DLS) = 63 ± 6 nm; (Dh)int (DLS) = 124 ± 20 nm; Dav (TEM) = 36 ± 4 nm.
Procedure for the preparation of SCKs (9)
To a solution of micelles (8) (15 mL, 0.32 mgmL−1) was added 2, 2′-(ethylenedioxy)bis(ethylamine) (0.63 mg, 4.3 μmol, to achieve nominally crosslinking through 20% of the COOH groups) from a stock solution via a syringe pump over 30 min. The reaction mixture was allowed to stir at RT for 30 min and EDCI (1.17 mg, 3.93 μmol) was added as a solution in water via syringe pump over 15 min. The reaction mixture was allowed to stir at RT for 24 h and was then transferred to presoaked dialysis tubing (MWCO ca. 6000–8000 Da) and dialyzed for ca. 4 d to afford the SCK solution with a final concentration of 0.29 mgmL−1. (Dh)n (DLS) = 48 ± 7 nm; (Dh)vol (DLS) = 79 ± 10 nm; (Dh)int (DLS) = 123 ± 9 nm; Dav (TEM) = 41 ± 4 nm.
Conjugation of Alexa Fluor 633 hydrazide to SCKs (10)
To a SCK solution (8.0 mL, 0.29 mgmL−1) at 0 °C was added a stock aqueous solution of EDCI (2.41 mg, 8.10 μmol) over a period of 3 min, and the mixture was allowed to stir for 20 min. Sulfo-NHS (1.78 mg, 8.10 μmol) was added from a stock solution prepared in nanopure water and the reaction was stirred for 2 h. The pH of the solution was adjusted to 7.2 using PBS buffer (pH 8.86, 0.2 M) and Alexa Fluor 633 hydrazide (72.0 μg, 0.064 μmol) was added to the reaction mixture from a stock solution in DMSO. The reaction mixture was allowed to stir for 24 h in the dark and was then transferred to presoaked dialysis tubing (MWCO ca. 6000–8000 Da) and dialyzed against NP water for 5 d in an Al-foil-wrapped beaker. The concentration of dye was found to be 12.0 μM from UV-vis spectroscopy. (Dh)n (DLS) = 60 ± 16 nm; (Dh)vol (DLS) = 79 ± 30 nm; (Dh)int (DLS) = 125 ± 53 nm; Dav (TEM) = 40 ± 4 nm.
Deprotection of masked thiols of Alexa Fluor 633 hydrazide labeled SCKs (11)
Hydroxyl amine hydrochloride (75.0 μL, 0.5 M) in aqueous buffer (100 mM, 150 mM NaCl) was added to a stirred solution of SCK 10 (5.0 mL) suspended in buffer (100 mM, 150 mM NaCl) and the reaction mixture was allowed to stir for 3 h at RT. The solution was assayed by Ellman’s assay and the thiol content was determined to be 33 ± 4 μM. The solution was immediately used for conjugation with maleimide-functionalized PNAs and CPPs.
Synthesis of maleimide-PNA conjugates (12)
PNAs were synthesized continuously on universal support XAL-PEG-PS resin at a 2 μmol scale with standard solid phase Fmoc chemistry on an Expedite 8909 Synthesizer. After completion of automated synthesis, PNAs were cleaved from the solid support and the bases were deprotected using trifluoroacetic acid: m-cresol (4:1, v/v) for 3 h and then precipitated with diethyl ether. The crude products were purified by HPLC and characterized by MALDI-TOF. PNAs were then dissolved in NMP: dimethylsulfoxide (DMSO) (7:3, v/v) with 15 min vigorous stirring. Maleimide dissolved in NMP was added to the PNA solution followed by addition of DIEA (16 μL) and the reaction mixture was maintained at room temperature for 2.5 h with gentle shaking. Excess cold diethyl ether (anhydrous) was added to precipitate the PNA-maleimide conjugates. The PNA-maleimide conjugates were dried under nitrogen and characterized by MALDI-TOF mass spectrometry.
General procedure for conjugation of SCKs with iNOS PNAs with and without TAT (13)
To a SCK solution (750 μL, 0.29 mgmL−1) suspended in buffer (100 mM PBS, 150 mM NaCl, pH 7.2), citric acid buffer (pH 5.12) was used to adjust the pH to 6.5. A concentrated solution of iNOS PNA (Maleimide-TTTCCTTTTCCTCTTTCA or Maleimide-TGTCCTCCTTTTCTTTCA) (5.0 nmoles in DMF) in (100 mM PBS, 150 mM NaCl, pH 7.4) was added to the SCK solution to prepare SCKs with mismatch or match PNAs. Additionally, TAT (Maleimide-GGYGRKKRRQRRR) (6.7 nmoles from a stock solution in DMF) was added to the reaction mixture of SCKs with either match or mismatch PNA sequences to prepare PNA-SCK-TAT conjugates. The reaction mixture was stirred at room temperature for 6 h and remaining thiols were capped by addition of 4-maleimido-butyric acid (1 mg suspended in 100 mM PBS, 150 mM NaCl, pH 7.4). The reaction was stirred for 16 h before being purified by Sephadex®G-50 gel column. Dav (TEM) = 28 ± 5 nm.
Preparation of biotinylated iNOS mRNA (14)
Image CloneID 4978648 containing the iNOS mRNA sequence (Gene Bank BC062378, ATCC cloning NO. 10470196) was obtained from ATCC. The plasmid DNA was purified and linearized at the unique XbaI site before transcription. Transcription reactions were carried out using the Promega RiboMAX Large Scale RNA production system utilizing the SP6 promoter (Promega). Transcription proceeded with SP6 RNA polymerase, NTPs and Bio-UTP (Roche) for 4 h at 37 °C, and followed by DNase I treatment, phenol:chloroform: isoamylalcohol (25:24:1) extraction and ethanol precipitation. The final purified iNOS RNA was quite homogeneous and corresponded in size to the expected 4 Kb transcript.
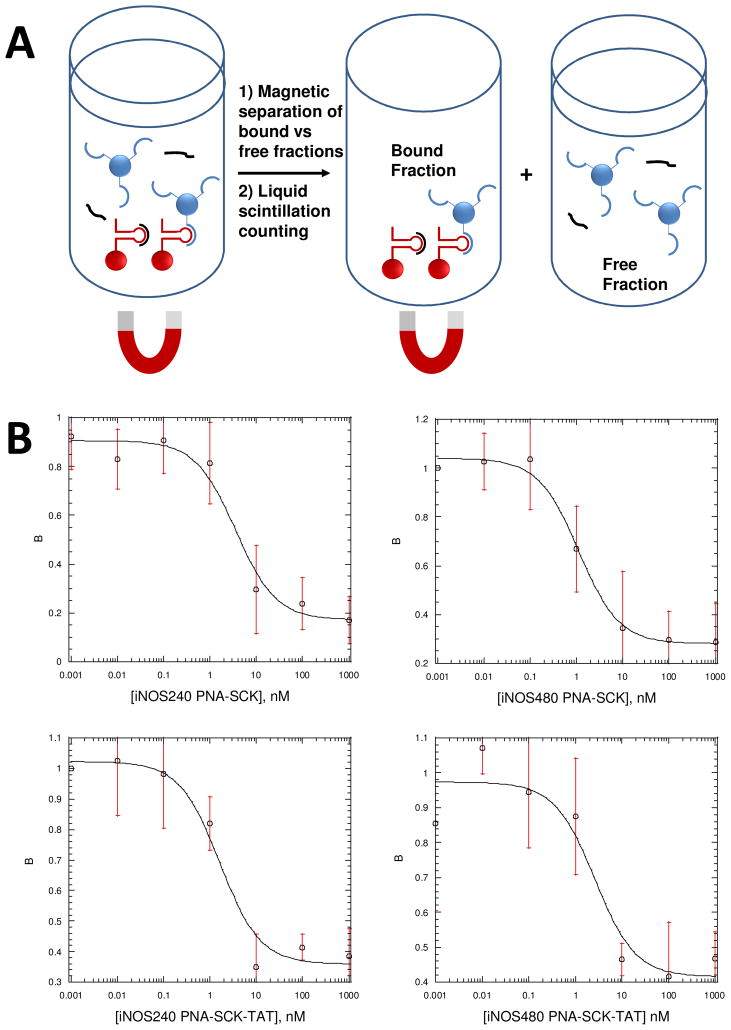
Binding affinity of ODNs to iNOS mRNA by a Dynabead-based binding assay
The radiolabeled ODN (100 pM) was incubated with biotinylated mRNA (0.01, 0.1, 1, 10, 100 nM) and 1 μL of RNase inhibitor for 4h at 37°C in a total volume of 100 μL. Streptavidin coated Dynabeads were added and mixed for 30 min. The beads were then separated by a magnet, washed twice and resuspended in 100 μL of hybridization buffer (5m M Tris-HCl, pH 7.0, 1 mM EDTA, 0.1 M NaCl). Solutions were then counted by liquid scintillation to give bound fractions directly.
The dissociation constants were determined by non-linear fitting of the fraction bound versus RNA concentration data to the analytical expression shown below for a two state binding equilibrium using the Kaleidagraph program.
| (1) |
where FB is the experimentally determined fraction of bound ODN, NSB is the fraction of ODN that is nonspecifically bound, SB is the fraction of ODN that can be specifically bound and was set equal to (1-NSB), [L] is total ODN concentration, Kd is dissociation constant of the ODN and [RNA] is total RNA concentration.
Binding affinity of iNOS PNA-SCK-TAT for iNOS mRNA by a competition assay
Biotinylated mRNA (10 pM) was incubated with 1000 pM radiolabeled ODN and 1 μL of RNase inhibitor for 4 h at 37 °C, to which unlabeled competitor PNAs (0, 0.001, 0.01, 0.1, 1, 10 nM) or iNOS PNA-SCK-TAT (0, 0.01, 0.1, 1, 10, 100 and 1000 nM, concentration calculated based on PNA concentration) were added. The streptavidin coated Dynabeads were added and mixed for another 30 min at 37 °C. Following incubation, the reaction mixture was separated by a magnetic field, washed twice and resuspended in hybridization buffer (5 mM Tris-HCl, pH 7.0, 1 mM EDTA, 0.1 M NaCl). The solution containing bound ODNs was counted by liquid scintillation. Additionally, SCK-TAT alone served as a control group.
The fraction of antisense ODN bound to the RNA (B) was then plotted against the competitor PNA concentration ([PNA]), and an IC50 value is determined (the concentration of competitor PNA that reduces binding of the ODN by 50%) by fitting the data according to a standard equation. Once the IC50 values are obtained, the Kds for the PNAs can be determined using a simple relationship involving the IC50 and the Kd for the antisense ODN.
| (2) |
| (3) |
RESULTS AND DISCUSSION
Design of the PNA nanoparticles
The PNA nanoparticles were designed to meet a number of criteria for future applications as imaging and knock down agents for iNOS expression. The first was that the nanoparticle be able to enter cells efficiently so as to be able to engage the iNOS mRNA in the cytoplasm. The second was that the attached PNAs would be able to freely interact with the mRNA. The third was that the nanoparticle would be capable of being detected by both fluorescent and PET imaging modalities. The fourth was that the nanoparticles avoid detection by the mononuclear phagocytic system (MPS), also referred to as the reticuloendothelial system (RES), which would otherwise result in their rapid clearance from the blood. To meet these criteria, a PEGylated shell crosslinked nanoparticle was chosen in which the PNAs and the TAT peptide would be attached to the ends of the PEG chains, while the imaging agents would be conjugated to the polyacrylate shell. PEGylation has been previously shown to impart stealth character to SCKs and prolong their retention in the blood 44, 46, presumably by shielding the polyacrylate core from components of the MPS. The TAT peptide was chosen to facilitate endocytosis of the SCKs 63, 64, and to enable TAT to interact with the cell surface, it was attached to the ends of the PEG chains, using a thiol coupling strategy 49. Likewise, the PNAs were designed to also be attached to the ends of the PEG chains to enable them to interact with the mRNA in the cytoplasm. The imaging agents were designed to be attached to the polyacrylate shell to minimize their exposure to the MPS. The polyacrylate shell was also expected to serve additional functions to serve as a robust network for maintaining the nanoparticle structure and to, potentially, facilitate rupture of the endosome upon lowering of the pH during endosomal acidification 65.
Synthesis of the SCK nanoparticles
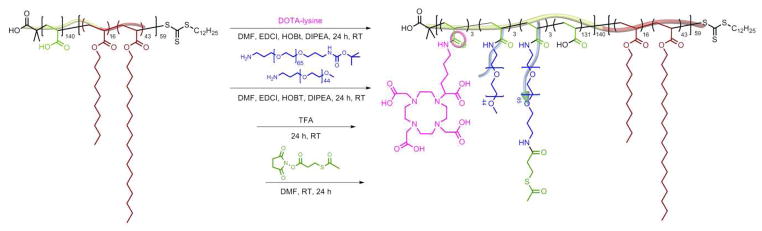
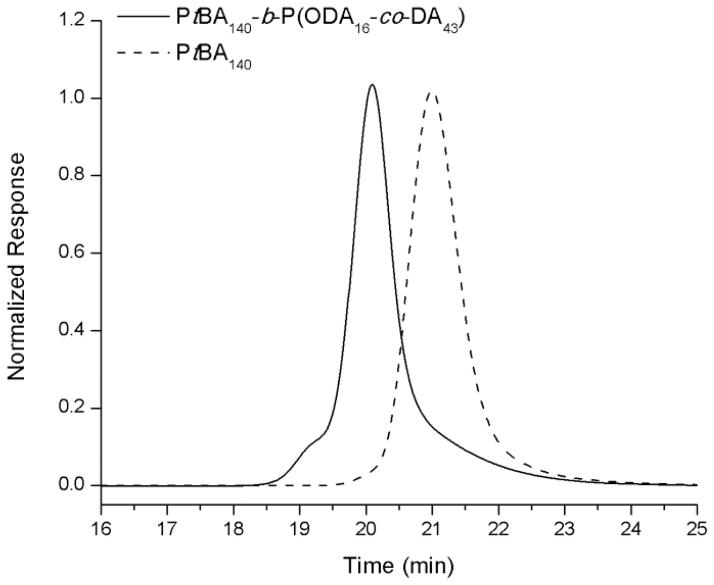
The block copolymer precursor used in this study was prepared by a two-step reversible addition-fragmentation chain transfer (RAFT) polymerization process, followed by deprotection (Scheme 1), as described in previous work by our group 58. S-1-dodecyl-S′-(α, α′-dimethyl-α″-acetic acid) trithiocarbonate (DDMAT) was used as a chain transfer agent to synthesize an initial PtBA140 1 homopolymer, which served as a macrochain transfer agent. This macrochain transfer agent was then employed during the copolymerization of octadecyl acrylate and decyl acrylate to afford PtBA140-b-P(ODA16-co-DA43), 2, having a narrow molecular weight distribution (PDI < 1.10) (Figure 1). The tert-butyl protecting groups were removed via acidolysis with TFA in CH2Cl2 to yield the amphiphilic block copolymer of PAA140-b-P(ODA16-co-DA43), 3. The PAA140-b-P(ODA16-co-DA43) polymer was further functionalized with chelators and PEO grafts to provide for terminal coupling sites for attachment of the biologically-active ligands (PNA and TAT) and to allow for direct, future translation to in vivo studies. The block copolymer was first functionalized with Boc-protected DOTA-lysine via amidation of acrylic acid repeat units of the polymer to afford 4 with ca. 3 DOTA-lysines per polymer backbone. DOTA serves as a chelator for 64Cu which will allow the resulting nanoparticles to be used for PET imaging in vivo 44–46, 66. Furthermore, different lengths of PEO grafts (2 kDa and 3 kDa) were appended to the polymer backbone to obtain 5, using similar amidation chemistry to yield ca. 3 of each type of PEO per polymer chain. The 2 kDa PEO grafts provide stealth properties for the nanoparticles, while the 3 kDa PEO chains, after chain terminus deprotection and modification, additionally, provide reactive sites. The Boc-protecting groups from DOTA and 3 kDa PEOs were removed using TFA in a global deprotection step revealing amines at their termini. In the last step, methyl ester protected sulfohydryl groups were introduced at the NH2- termini of the 3 kDa PEOs by reaction with SATP to yield functional block graft copolymers, 7 (Table 1). After each chemical transformation of the amphiphilic block copolymers, purification was performed via extensive dialysis against nanopure water and the polymers were isolated by lyophilization.
Scheme 1.
Synthesis of amphiphilic functional block graft copolymer, 7.
Figure 1.
GPC traces for PtBA 1 and PtBA-b-P(ODA-co-DA) 2.
Table 1.
Summary of polymers prepared.
| Polymer | DOTA | PEG | SCOCH3 |
Mn [Da] 1H NMR |
Mn [Da] GPC |
Mw [Da] GPC |
PDI GPC |
|---|---|---|---|---|---|---|---|
| 1 PtBA140 | - | - | - | 18,300 | 12,000 | 13,200 | 1.1 |
| 2 PtBA140-b-P(ODA16-co-DA43) | - | - | - | 32,600 | 20,800 | 25,500 | 1.2 |
| 3 PAA140-b-P(ODA16-co-DA43) | - | - | - | 24,700 | - | - | - |
| 4 PAA137-g-(CONH-lysine-DOTA-Boc)3-b-P(ODA16-co-DA43) | 3 | - | - | 26,700 | - | - | - |
| 5 PAA131-g-(PEG3 kDa-Boc)3-g-(mPEG2 kDa)3-g-(CONH-lysine-DOTA-Boc)3-b-P(ODA16-co-DA43) | 3 | 3 | - | 41,600 | - | - | - |
| 6 PAA131-g-(PEG3 kDa-NH2)3-g-(mPEG2 kDa)3-g-(CONH-lysine-DOTA-COOH)3-b-P(ODA16-co-DA43) | 3 | 3 | - | 41,000 | - | - | - |
| 7 PAA131-g-(PEG3 kDa-NHCO-C2H4-SCOCH3)3–g-(mPEG2 kDa)3-(CONH-lysine-DOTA-COOH)3-b-P(ODA16-co-DA43) | 3 | 3 | 3 | 41,700 | - | - | - |
Preparation of functionalized and labeled nanoparticles
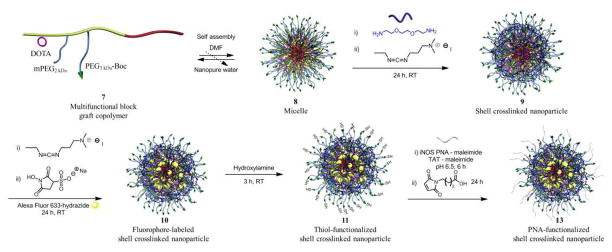
SCKs were prepared by the solution-state assembly of the amphiphilic block graft copolymers in water followed by crosslinking reactions selectively throughout the hydrophilic shell layer (Scheme 2). After dissolving 7 in DMF at a concentration of 1 mg·mL−1, self assembly was induced by gradual addition of an equal amount of water over a period of 3 h. The aqueous micelle solution was allowed to stir overnight and was then transferred to dialysis tubing (MWCO 6–8 kDa) and dialyzed against nanopure water for 4 d to remove organic solvent. The SCKs were obtained by crosslinking approximately 20% of the acrylic acid segments by amidation with the amine groups of the crosslinker (2, 2′-ethylenedioxy)bis(ethylamine), in the presence of EDCI, followed by extensive dialysis to remove unreacted small molecule starting materials and reaction by products.
Scheme 2.
Self assembly of the multi-functional block graft copolymer 7 into micelle 8, crosslinking into SCK 9, conjugation of Alexa Fluor 633 hydrazide 10, and functionalization with PNAs and TAT to give the final bio-synthetic hybrid nanostructure 13.
Alexa Fluor 633 hydrazide was attached to the SCKs, to serve as a fluorescent probe, by performing post-assembly conjugation of the hydrazide group of the dye to EDCI-activated acrylic acids in the shell of the SCKs at 0 °C. The reaction was carried out for 24 h in the dark and the fluorescently-labeled SCKs were purified by extensive dialysis for 5 d against nanopure water to remove unconjugated dye molecules.
Bioconjugation of TAT and PNAs to the SCK nanoparticles
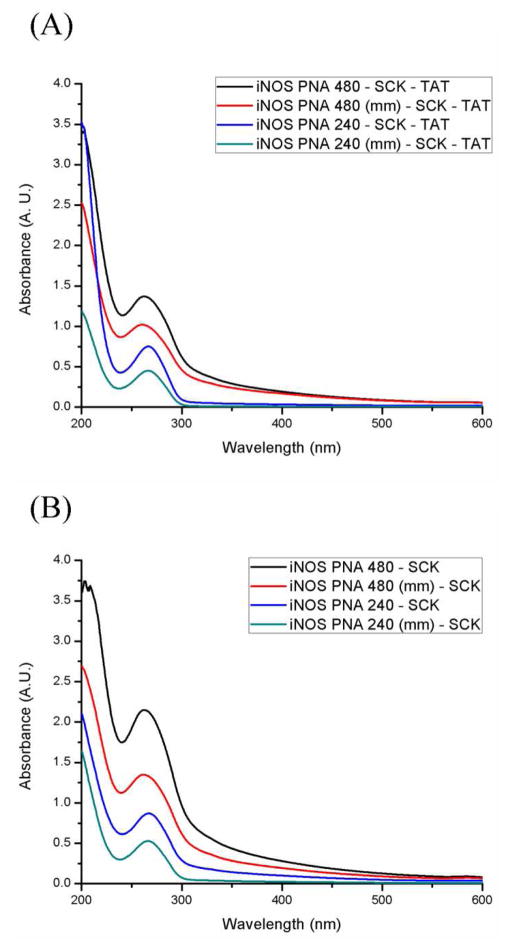
By introducing a masked thiol on to the end of the 3 kDa PEO terminus, a surface functional nanoparticle was designed and prepared that could undergo conjugation with biomacromolecules, e.g., proteins 49, peptides and nucleic acid sequences, through efficient reactions, such as thiol-maleimide, thiol-haloacetyl or disulfide couplings. The acetyl protecting groups of the masked thiols were removed by reaction with the addition of a 0.1 M hydroxylamine solution for 4 h (Scheme 2). The thiol content of the nanoparticles was then evaluated using Ellman’s assay and was determined to be 33 ± 4 μM. Ellman’s assay of SCKs before reacting with hydroxylamine showed undetectable concentration of –SH. The resulting reactive –SH sites were expected to be presented on the surface of the SCK nanostructures, allowing for surface availability of biologically-active ligands, and were therefore, utilized for conjugation of the PNAs as mRNA recognition elements and TAT CPPs for cell entry. These SCKs were subjected to reaction, coincidentally, with PNAs and TAT that were functionalized with maleimide, at pH 6.5 for 6 h at 0 °C. Two anti-iNOS PNAs, PNA240 and PNA480, and mismatch controls were selected for conjugation to the SCKs (Table 2), from a set of PNAs previously shown to have similar affinity for iNOS mRNA, but differing abilities to inhibit iNOS expression 67. In each case, the unreacted thiols were capped using 4-maleimido butyric acid. The nanoparticle conjugates were purified with Sephadex G-50 columns and the amounts of PNAs incorporated into the SCK samples were calculated by UV-vis spectroscopy (Figures 2A and B) using the inherent wavelength of 260 nm for PNAs.
Table 2.
ODN and PNA sequences used.
| Sequence ID | Sequence (5′-3′ or amino-carboxy) |
|---|---|
| 240 | 5′-TGT CCT TTT CCT CTT TCA-3′ |
| 240 (mm) | 5′-TGT CCT CCT TTT CTT TCA-3′ |
| 480 | 5′-TGA AAT CCG ATG TGG CCT-3′ |
| 480(mm) | 5′-TAG AAT CCA GTG GTG CCT-3′ |
Figure 2.
(A) UV-vis spectra of iNOS PNA-SCK conjugates, (B). UV-vis spectroscopy of iNOS PNA-SCK-TAT conjugates.
Characterization of nanoparticles
The size and shape of the SCKs were determined using DLS and TEM. DLS analysis showed that the hydrodynamic diameters of the SCKs were slightly larger than were the diameter values determined by TEM, due to the hydration of the hydrophilic shell in aqueous conditions. The micelles and SCKs were 48 ± 7 nm by DLS and 41 ± 4 nm by TEM (Figure 3). The average core diameter of the nanoparticles after conjugation with PNAs and TAT were determined using TEM (Supplementary Figure 1) and were found to be ca. 28 ± 5 nm.
Figure 3.
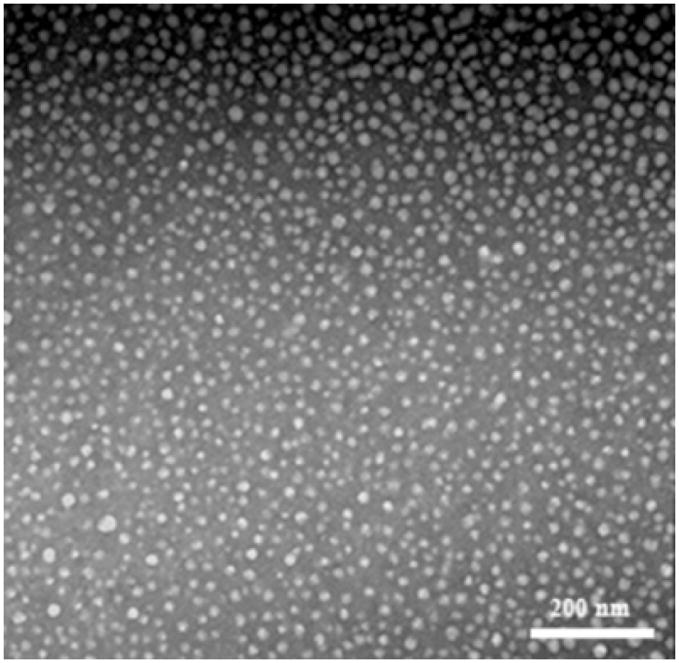
TEM image of SCKs 9.
Binding affinity of the PNA and PNA-SCK conjugates for iNOS mRNA
The binding affinities of the PNA and PNA-SCK conjugates were determined by a competition assay against radiolabeled ODNs targeting the same site on the mRNA. To carry out this assay, we first determined the binding affinity of the ODNs targeting the same site as the PNA using a Dynabead assay that we had previously developed for this purpose 68. The target mRNA was produced by transcription from an iNOS cDNA clone in vitro with Bio-dUTP to incorporate biotin groups into the RNA to enable it to be retrieved with streptavidin coated magnetic beads. The 5′-radiolabeled antisense ODNs were then incubated with the iNOS RNA and allowed to equilibrate for 4 h, after which the Dynabeads were added to bind to the mRNA, which could then be immobilized with a magnetic field. The solution containing the free ODNs was removed from the reaction tube and both the free and bound fractions were quantified by liquid scintillation counting. The ratio of bound to free ODN vs concentration was then fit to a standard equation (Equation 1), from which the dissociation constant (Kd) could be determined. As can be seen in Table 3, ODNs designed on the basis of binding sites identified by the RT-ROL mapping assay (iNOS240, iNOS480) 67 showed very high affinity with dissociation constants (Kd’s) of 7.3 ± 1.2 and 5.2 ± 2.5 nM (Table 3).
Table 3.
Binding affinities of ODNs, PNAs, PNA-SCK and iNOS PNA-SCK-TAT conjugates.
| Sequence ID | ODN (nM) | PNA (nM) | PNA-SCK (nM) | PNA-SCK-TAT (nM) |
|---|---|---|---|---|
| 240 | 7.3 ± 1.2 | 0.10 ± 0.052 | 3.2 ± 2.0 | 1.4 ± 0.5 |
| 240mm | - | >10000 | >1000 | >1000 |
|
| ||||
| 480 | 5.2 ± 2.5 | 0.086 ± 0.026 | 1.1 ± 0.3 | 1.7 ± 1.0 |
| 480mm | - | >10000 | >1000 | >1000 |
| No PNA | - | - | >1000 | >1000 |
Once the binding affinity of the ODNs was established, the binding affinity of PNAs and PNA-SCK conjugates to iNOS mRNA could be determined by a competition assay in which the concentration of ODN was held constant and the concentration of the PNA or PNA-SCK was varied (Figure 4). The Kd could then be calculated from the IC50. As can be seen in Table 3, the two PNAs, PNA-240 and PNA-480 showed similar Kd’s of 101 ± 52 and 86 ± 26 pM, while the mismatched PNAs had Kd’s of > 10 nM. When conjugated to the SCKs, the binding affinity of the PNAs appeared to decrease about 30 to 10-fold, respectively, to 3.2 ± 2.0 and 1.1 ±0.3 nM, while the mismatched PNAs showed no significant affinity. The decrease in affinity of the PNAs upon conjugation could be due to a number of factors, such as electrostatic repulsion between the SCK and the RNA, or steric interference by a bound RNA molecule. The binding affinity of the iNOS PNA-SCK-TAT conjugates, which could have been expected to be higher due to the positive charge of the TAT peptide, was not significantly different from the conjugates lacking the TAT peptide.
Figure 4.
Dynabead competition assay to determine iNOS mRNA binding affinity of SCK-PNA conjugates. A) Biotinylated iNOS mRNA (10 pM) bound to streptavidin coated Dynabeads were incubated with 1 nM 5′-radiolabeled 240 or 480 antisense ODN and increasing concentrations of the corresponding SCK-PNA conjugate. After incubation for 4 h, the bound versus free fractions were separated by a magnet and the radioactivity in each fraction was quantified by liquid scintillation counting. B) Plots of the average fraction of mRNA bound by radioactively labeled ODN vs. the SCK-PNA conjugates from three experiments fit to the analytical expression 2) described in the experimental section from which the IC50 was extracted. The data in Table 3 was obtained from averaging fits to each independent experiment (see supporting information).
CONCLUSIONS
SCKs covalently conjugated with iNOS PNAs and CPPs have been prepared and their binding affinities with complementary mRNAs post-conjugation were evaluated. Specific effort was made to have these biologically-active units presented from the outer most surface of the SCKs, by employing well-defined multi-step polymerization and polymer modification reactions that involved combinations of stealthy PEG chain grafts. The binding affinity measurements demonstrated that the PNAs retain molecular recognition selectivity even after undergoing coupling onto the nanoscopic SCK objects. The shapes and sizes of SCKs remained almost the same in the resulting nanoconjugates. By partitioning various optical and radiolabeling sites and reactive functionalities for conjugation of biological ligands within a size-, shape- and flexibility-controlled nanoscopic framework, these bio-synthetic hybrid materials provide preliminary steps towards the development of hierarchical nanostructures that hold great promise as antisense imaging agents and therapeutics. The availability of particles with tunable compositions and properties will allow us to fine-tune the PNA-RNA molecular recognition events, while balancing the electrostatic binding and repulsion 55, 56. With the high levels of sophistication that are possible via simple iterations of controlled polymer chemistries, there are several functions whose biological performances are yet to be evaluated. Extended in vitro and in vivo studies are underway.
Supplementary Material
Acknowledgments
This material is based upon work supported by the National Heart Lung and Blood Institute of the National Institutes of Health as a Program of Excellence in Nanotechnology (HHSN268201000046C) and by the Office of Naval Research (N00014-10-1-0527). The Welch Foundation is gratefully acknowledged for support through the W. T. Doherty-Welch Chair in Chemistry, Grant No. A-0001.
Footnotes
Additional TEM images and binding affinity data. This information is available free of charge via the internet at http://pubs.acs.org/.
References
- 1.Rubenfeld GD, Caldwell E, Peabody E, Weaver J, Martin DP, Neff M, Stern EJ, Hudson LD. Incidence and Outcomes of Acute Lung Injury. New England Journal of Medicine. 2005;353:1685–1693. doi: 10.1056/NEJMoa050333. [DOI] [PubMed] [Google Scholar]
- 2.Goodman RB, Pugin J, Lee JS, Matthay MA. Cytokine-mediated inflammation in acute lung injury. Cytokine & growth factor reviews. 2003;14:523–535. doi: 10.1016/s1359-6101(03)00059-5. [DOI] [PubMed] [Google Scholar]
- 3.Hemmrich K, Kröncke KD, Suschek CV, Kolb-Bachofen V. What sense lies in antisense inhibition of inducible nitric oxide synthase expression? Nitric Oxide. 2005;12:183–199. doi: 10.1016/j.niox.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Guo FH, Deraeve HR, Rice TW, Stuehr DJ, Thunnissen FBJM, Erzurum SC. Continuous Nitric-Oxide Synthesis by Inducible Nitric-Oxide Synthase in Normal Human Airway Epithelium in-Vivo. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:7809–7813. doi: 10.1073/pnas.92.17.7809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sittipunt C, Steinberg KP, Ruzinski JT, Myles C, Zhu S, Goodman RB, Hudson LD, Matalon S, Martin TR. Nitric oxide and nitrotyrosine in the lungs of patients with acute respiratory distress syndrome. Am J Respir Crit Care Med. 2001;163:503–10. doi: 10.1164/ajrccm.163.2.2004187. [DOI] [PubMed] [Google Scholar]
- 6.Hosogi S, Iwasaki Y, Yamada T, Komatani-Tamiya N, Hiramatsu A, Kohno Y, Ueda M, Arimoto T, Marunaka Y. Effect of inducible nitric oxide synthase on apoptosis in Candida-induced acute lung injury. Biomedical Research-Tokyo. 2008;29:257–266. doi: 10.2220/biomedres.29.257. [DOI] [PubMed] [Google Scholar]
- 7.Mehta S. The effects of nitric oxide in acute lung injury. Vascular Pharmacology. 2005;43:390–403. doi: 10.1016/j.vph.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 8.Zhang J, McCarthy TJ, Moore WM, Currie MG, Welch MJ. Synthesis and evaluation of two positron-labeled nitric oxide synthase inhibitors, S-[11C]methylisothiourea and S-(2-[18F]fluoroethyl)isothiourea, as potential positron emission tomography tracers. Journal of medicinal chemistry. 1996;39:5110–8. doi: 10.1021/jm960481q. [DOI] [PubMed] [Google Scholar]
- 9.Panda K, Chawla-Sarkar M, Santos C, Koeck T, Erzurum SC, Parkinson JF, Stuehr DJ. Visualizing inducible nitric-oxide synthase in living cells with a heme-binding fluorescent inhibitor. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:10117–10122. doi: 10.1073/pnas.0408972102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Towner RA, Smith N, Doblas S, Garteiser P, Watanabe Y, He T, Saunders D, Herlea O, Silasi-Mansat R, Lupu F. In vivo detection of inducible nitric oxide synthase in rodent gliomas. Free Radical Biology and Medicine. 2010;48:691–703. doi: 10.1016/j.freeradbiomed.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 11.Hesslinger C, Strub A, Boer R, Ulrich WR, Lehner MD, Braun C. Inhibition of inducible nitric oxide synthase in respiratory diseases. Biochemical Society transactions. 2009;37:886–91. doi: 10.1042/BST0370886. [DOI] [PubMed] [Google Scholar]
- 12.Lim MH, Xu D, Lippard SJ. Visualization of nitric oxide in living cells by a copper-based fluorescent probe. Nat Chem Biol. 2006;2:375–380. doi: 10.1038/nchembio794. [DOI] [PubMed] [Google Scholar]
- 13.Ouyang J, Hong H, Zhao Y, Shen H, Shen C, Zhang C, Zhang J. Bioimaging nitric oxide in activated macrophages in vitro and hepatic inflammation in vivo based on a copper–naphthoimidazol coordination compound. Nitric Oxide. 2008;19:42–49. doi: 10.1016/j.niox.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 14.Zhou D, Lee H, Rothfuss JM, Chen DL, Ponde DE, Welch MJ, Mach RH. Design and synthesis of 2-amino-4-methylpyridine analogues as inhibitors for inducible nitric oxide synthase and in vivo evaluation of [18F]6-(2-fluoropropyl)-4-methyl-pyridin-2-amine as a potential PET tracer for inducible nitric oxide synthase. Journal of medicinal chemistry. 2009;52:2443–53. doi: 10.1021/jm801556h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koppelhus U, Nielsen PE. Cellular delivery of peptide nucleic acid (PNA) Advanced Drug Delivery Reviews. 2003;55:267–280. doi: 10.1016/s0169-409x(02)00182-5. [DOI] [PubMed] [Google Scholar]
- 16.Nielsen PE, Egholm M, Berg RH, Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 17.Nielsen PE. PNA Technology. Mol Biotechnol. 2004;26:233–48. doi: 10.1385/MB:26:3:233. [DOI] [PubMed] [Google Scholar]
- 18.Lundin KE, Good L, Stromberg R, Graslund A, Smith CI. Biological activity and biotechnological aspects of peptide nucleic acid. Adv Genet. 2006;56:1–51. doi: 10.1016/S0065-2660(06)56001-8. [DOI] [PubMed] [Google Scholar]
- 19.Nielsen PE. Gene targeting and expression modulation by peptide nucleic acids (PNA) Current pharmaceutical design. 2010;16:3118–23. doi: 10.2174/138161210793292546. [DOI] [PubMed] [Google Scholar]
- 20.Shi N, Boado RJ, Pardridge WM. Antisense imaging of gene expression in the brain in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:14709–14. doi: 10.1073/pnas.250332397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rao PS, Tian X, Qin W, Aruva MR, Sauter ER, Thakur ML, Wickstrom E. 99mTc-peptide-peptide nucleic acid probes for imaging oncogene mRNAs in tumours. Nuclear medicine communications. 2003;24:857–63. doi: 10.1097/01.mnm.0000084583.29433.df. [DOI] [PubMed] [Google Scholar]
- 22.Sun X, Fang H, Li X, Rossin R, Welch MJ, Taylor JS. MicroPET imaging of MCF-7 tumors in mice via unr mRNA-targeted peptide nucleic acids. Bioconjugate chemistry. 2005;16:294–305. doi: 10.1021/bc049783u. [DOI] [PubMed] [Google Scholar]
- 23.Santangelo P, Nitin N, Bao G. Nanostructured probes for RNA detection in living cells. Ann Biomed Eng. 2006;34:39–50. doi: 10.1007/s10439-005-9003-6. [DOI] [PubMed] [Google Scholar]
- 24.Jia F, Figueroa SD, Gallazzi F, Balaji BS, Hannink M, Lever SZ, Hoffman TJ, Lewis MR. Molecular imaging of bcl-2 expression in small lymphocytic lymphoma using 111In-labeled PNA-peptide conjugates. Journal of nuclear medicine : official publication, Society of Nuclear Medicine. 2008;49:430–8. doi: 10.2967/jnumed.107.045138. [DOI] [PubMed] [Google Scholar]
- 25.Lendvai G, Estrada S, Bergstrom M. Radiolabelled oligonucleotides for imaging of gene expression with PET. Current medicinal chemistry. 2009;16:4445–61. doi: 10.2174/092986709789712844. [DOI] [PubMed] [Google Scholar]
- 26.Iyer AK, He J. Radiolabeled oligonucleotides for antisense imaging. Current organic synthesis. 2011;8:604–614. doi: 10.2174/157017911796117241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhilina ZV, Ziemba AJ, Ebbinghaus SW. Peptide nucleic acid conjugates: synthesis, properties and applications. Curr Top Med Chem. 2005;5:1119–31. doi: 10.2174/156802605774370892. [DOI] [PubMed] [Google Scholar]
- 28.Abes R, Arzumanov AA, Moulton HM, Abes S, Ivanova GD, Iversen PL, Gait MJ, Lebleu B. Cell-penetrating-peptide-based delivery of oligonucleotides: an overview. Biochem Soc Trans. 2007;35:775–9. doi: 10.1042/BST0350775. [DOI] [PubMed] [Google Scholar]
- 29.Thurmond KB, Kowalewski T, Wooley KL. Shell cross-linked knedels: A synthetic study of the factors affecting the dimensions and properties of amphiphilic core-shell nanospheres. Journal of the American Chemical Society. 1997;119:6656–6665. [Google Scholar]
- 30.Thurmond KB, Huang HY, Clark CG, Kowalewski T, Wooley KL. Shell cross-linked polymer micelles: stabilized assemblies with great versatility and potential. Colloids and Surfaces B-Biointerfaces. 1999;16:45–54. [Google Scholar]
- 31.Remsen EE, Thurmond KB, Wooley KL. Solution and surface charge properties of shell cross-linked knedel nanoparticles. Macromolecules. 1999;32:3685–3689. [Google Scholar]
- 32.Thurmond KB, Kowalewski T, Wooley KL. Water-soluble knedel-like structures: The preparation of shell-cross-linked small particles. Journal of the American Chemical Society. 1996;118:7239–7240. [Google Scholar]
- 33.Samarajeewa S, Shrestha R, Li Y, Wooley KL. Degradability of Poly(Lactic Acid)-Containing Nanoparticles: Enzymatic Access through a Cross-Linked Shell Barrier. J Am Chem Soc. 2012;134:1235–42. doi: 10.1021/ja2095602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pochan DJ, Chen ZY, Cui HG, Hales K, Qi K, Wooley KL. Toroidal triblock copolymer assemblies. Science. 2004;306:94–97. doi: 10.1126/science.1102866. [DOI] [PubMed] [Google Scholar]
- 35.Pochan DJ, Cui HG, Chen ZY, Wooley KL. Origins of toroidal micelle formation through charged triblock copolymer self-assembly. Soft Matter. 2009;5:1269–1278. [Google Scholar]
- 36.Pochan DJ, Zhong S, Cui HG, Chen ZY, Wooley KL. Helix self-assembly through the coiling of cylindrical micelles. Soft Matter. 2008;4:90–93. doi: 10.1039/b715459c. [DOI] [PubMed] [Google Scholar]
- 37.Pochan DJ, Zhu JH, Zhang K, Wooley KL, Miesch C, Emrick T. Multicompartment and multigeometry nanoparticle assembly. Soft Matter. 2011;7:2500–2506. [Google Scholar]
- 38.Cui HG, Chen ZY, Zhong S, Wooley KL, Pochan DJ. Block copolymer assembly via kinetic control. Science. 2007;317:647–650. doi: 10.1126/science.1141768. [DOI] [PubMed] [Google Scholar]
- 39.Sorrells JL, Shrestha R, Neumann WL, Wooley KL. Porphyrin-crosslinked block copolymer assemblies as photophysically-active nanoscopic devices. Journal of Materials Chemistry. 2011;21:8983–8986. [Google Scholar]
- 40.Becker ML, Bailey LO, Wooley KL. Peptide-derivatized shell-cross-linked nanoparticles. 2 Biocompatibility evaluation. Bioconjugate Chemistry. 2004;15:710–717. doi: 10.1021/bc049945m. [DOI] [PubMed] [Google Scholar]
- 41.Becker ML, Remsen EE, Pan D, Wooley KL. Peptide-derivatized shell-cross-linked nanoparticles. 1 Synthesis and characterization. Bioconjugate Chemistry. 2004;15:699–709. doi: 10.1021/bc049946e. [DOI] [PubMed] [Google Scholar]
- 42.Joralemon MJ, Murthy KS, Remsen EE, Becker ML, Wooley KL. Synthesis, characterization, and bioavailability of mannosylated shell cross-linked nanoparticles. Biomacromolecules. 2004;5:903–913. doi: 10.1021/bm0344710. [DOI] [PubMed] [Google Scholar]
- 43.Joralemon MJ, Smith NL, Holowka D, Baird B, Wooley KL. Antigen-decorated shell cross-linked nanoparticles: Synthesis, characterization, and antibody interactions. Bioconjugate Chemistry. 2005;16:1246–1256. doi: 10.1021/bc0501505. [DOI] [PubMed] [Google Scholar]
- 44.Sun XK, Rossin R, Turner JL, Becker ML, Joralemon MJ, Welch MJ, Wooley KL. An assessment of the effects of shell cross-linked nanoparticle size, core composition, and surface PEGylation on in vivo biodistribution. Biomacromolecules. 2005;6:2541–2554. doi: 10.1021/bm050260e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Welch MJ, Sun G, Xu J, Hagooly A, Rossin R, Li Z, Moore DA, Hawker CJ, Wooley KL. Strategies for optimized radiolabeling of nanoparticles for in vivo PET Imaging. Advanced Materials. 2007;19:3157. [Google Scholar]
- 46.Sun G, Hagooly A, Xu J, Nystrom AM, Li ZC, Rossin R, Moore DA, Welch MJ, Wooley KL. Facile, efficient approach to accomplish tunable chemistries and variable biodistributions for shell cross-linked nanoparticles. Biomacromolecules. 2008;9:1997–2006. doi: 10.1021/bm800246x. [DOI] [PubMed] [Google Scholar]
- 47.Lin LY, Lee NS, Zhu J, Nystrom AM, Pochan DJ, Dorshow RB, Wooley KL. Tuning core vs. shell dimensions to adjust the performance of nanoscopic containers for the loading and release of doxorubicin. J Control Release. 2011;152:37–48. doi: 10.1016/j.jconrel.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nystrom AM, Xu ZQ, Xu JQ, Taylor S, Nittis T, Stewart SA, Leonard J, Wooley KL. SCKs as nanoparticle carriers of doxorubicin: investigation of core composition on the loading, release and cytotoxicity profiles. Chemical Communications. 2008:3579–3581. doi: 10.1039/b805428b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nystrom AM, Wooley KL. Thiol-functionalized shell crosslinked knedel-like (SCK) nanoparticles: A versatile entry for their conjugation with biomacromolecules. Tetrahedron. 2008;64:8543–8552. doi: 10.1016/j.tet.2008.04.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang S, Li Z, Samarajeewa S, Sun G, Yang C, Wooley KL. Orthogonally Dual-Clickable Janus Nanoparticles via a Cyclic Templating Strategy. J Am Chem Soc. 2011;133:11046–9. doi: 10.1021/ja203133h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pan D, Turner JL, Wooley KL. Folic acid-conjugated nanostructured materials designed for cancer cell targeting. Chemical Communications. 2003:2400–2401. doi: 10.1039/b307878g. [DOI] [PubMed] [Google Scholar]
- 52.Zhang K, Fang HF, Chen ZY, Taylor JSA, Wooley KL. Shape effects of nanoparticles conjugated with cell-penetrating peptides (HIV Tat PTD) on CHO cell uptake. Bioconjugate Chemistry. 2008;19:1880–1887. doi: 10.1021/bc800160b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Turner JL, Becker ML, Li XX, Taylor JSA, Wooley KL. PNA-directed solution- and surface-assembly of shell crosslinked (SCK) nanoparticle conjugates. Soft Matter. 2005;1:69–78. doi: 10.1039/b417653g. [DOI] [PubMed] [Google Scholar]
- 54.Fang H, Zhang K, Shen G, Wooley KL, Taylor JS. Cationic shell-cross-linked knedel-like (cSCK) nanoparticles for highly efficient PNA delivery. Mol Pharm. 2009;6:615–26. doi: 10.1021/mp800199w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang K, Fang HF, Wang ZH, Li Z, Taylor JSA, Wooley KL. Structure-activity relationships of cationic shell-crosslinked knedel-like nanoparticles: Shell composition and transfection efficiency/cytotoxicity. Biomaterials. 2010;31:1805–1813. doi: 10.1016/j.biomaterials.2009.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang K, Fang H, Wang Z, Li Z, Taylor JS, Wooley KL. Structure-activity relationships of cationic shell-crosslinked knedel-like nanoparticles: shell composition and transfection efficiency/cytotoxicity. Biomaterials. 2010;31:1805–13. doi: 10.1016/j.biomaterials.2009.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang K, Fang H, Wang Z, Taylor JS, Wooley KL. Cationic shell-crosslinked knedel-like nanoparticles for highly efficient gene and oligonucleotide transfection of mammalian cells. Biomaterials. 2009;30:968–77. doi: 10.1016/j.biomaterials.2008.10.057. [DOI] [PubMed] [Google Scholar]
- 58.Nystrom AM, Wooley KL. Construction of thermoresponsive SCKs through tuning the crystalline melting point of the core domain. Soft Matter. 2008;4:849–858. doi: 10.1039/b719314a. [DOI] [PubMed] [Google Scholar]
- 59.O’Connor RD, Zhang Q, Wooley KL, Schaefer J. Crystallization of poly(epsilon-caprolactone) under nanoparticle confinement. Helvetica Chimica Acta. 2002;85:3219–3224. [Google Scholar]
- 60.Zhang Q, Clark CG, Wang M, Remsen EE, Wooley KL. Thermally-induced (re)shaping of core-shell nanocrystalline particles. Nano Letters. 2002;2:1051–1054. [Google Scholar]
- 61.Zhang Q, Wang M, Wooley KL. Nanoscopic confinement of semi-crystalline polymers. Current Organic Chemistry. 2005;9:1053–1066. [Google Scholar]
- 62.Lai JT, Filla D, Shea R. Functional polymers from novel carboxyl-terminated trithiocarbonates as highly efficient RAFT agents. Macromolecules. 2002;35:6754–6756. [Google Scholar]
- 63.Liu J, Zhang Q, Remsen EE, Wooley KL. Nanostructured materials designed for cell binding and transduction. Biomacromolecules. 2001;2:362–8. doi: 10.1021/bm015515c. [DOI] [PubMed] [Google Scholar]
- 64.Zhang K, Fang H, Chen Z, Taylor JSA, Wooley KL. Shape Effects of Nanoparticles Conjugated with Cell-Penetrating Peptides (HIV Tat PTD) on CHO Cell Uptake. Bioconjugate Chem. 2008;19:1880–1887. doi: 10.1021/bc800160b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pack DW, Hoffman AS, Pun S, Stayton PS. Design and development of polymers for gene delivery. Nature Reviews Drug Discovery. 2005;4:581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 66.Rossin R, Pan DPJ, Qi K, Turner JL, Sun XK, Wooley KL, Welch MJ. Cu-64-labeled folate-conjugated shell cross-linked nanoparticles for tumor imaging and radiotherapy: Synthesis, radiolabeling, and biologic evaluation. Journal of Nuclear Medicine. 2005;46:1210–1218. [PubMed] [Google Scholar]
- 67.Fang H, Shen Y, Taylor JS. Native mRNA antisense-accessible sites library for the selection of antisense oligonucleotides, PNAs, and siRNAs. RNA. 2010;16:1429–35. doi: 10.1261/rna.1940610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fang H, Yue X, Li X, Taylor JS. Identification and characterization of high affinity antisense PNAs for the human unr (upstream of N-ras) mRNA which is uniquely overexpressed in MCF-7 breast cancer cells. Nucleic Acids Res. 2005;33:6700–11. doi: 10.1093/nar/gki968. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.