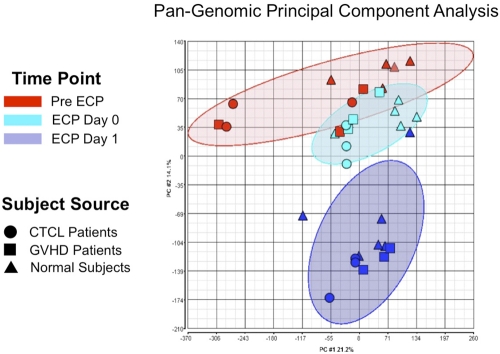
Figure 6.
ECP reproducibly produced distinctive pan-genomic transcriptome activation in processed monocytes. Principal component analysis (PCA) of global gene expression data are graphed, using the 2 principal variance components. Each symbol represents the entire transcriptome for a single sample (N = 34: 11 pre ECP (red), 11 ECP Day 0 (green), and 11 from ECP-processed monocytes Day 1 (blue). The position of each point is calculated as compression of the full set of 40 000 data points, generated using the pan-genomic battery of probes: cells obtained from normal subjects (triangles), CTCL patients (circles), and GVHD patients (squares). The analysis reveals that the samples segregate based on treatment irrespective of whether the processed monocytes derived from patients or normal subjects. The data grouping is highlighted by the ellipses in lighter red for the pre ECP, green for the immediately post-ECP, and blue for the post-18–hour ECP monocytes. This data reveal that ECP's complex impact on the monocyte transcriptome is high predictable and distinctive after 18-hour incubation.