Abstract
In conjunction with efforts to assess pathogen exposure in feral pigs from the southeastern United States, we amplified Bartonella henselae, B. koehlerae, and B. vinsonii subsp. berkhoffii from blood samples. Feral pigs may represent a zoonotic risk for hunters or butchers and pose a potential threat to domesticated livestock.
Keywords: swine, pigs, Bartonella vinsonii subsp. berkhoffii, Bartonella henselae, Bartonella koehlerae, bacteremia, bacteria, zoonoses, United States, dispatch
Bartonella spp. are intravascular, gram-negative bacteria that infect a diverse array of wild and domestic animals. These bacteria appear to induce a wide range of symptoms in humans and can cause similar disease manifestations in animals (1,2). An increasing number of Bartonella spp. are regarded as zoonotic pathogens, which creates a public health concern for human and veterinary medicine (3).
Feral pigs (Sus scrofa), nonnative, ancestral species derived from domesticated pigs in Europe, inhabit 39 states. As their geographic distribution expands and their numbers increase, these animals are causing substantial economic and ecologic damage, which has required implementation of specific damage management programs (4). Hunters and butchers coming in contact with blood from feral pigs may be at risk for infection with Bartonella spp (3). We report the molecular detection of 3 zoonotic Bartonella spp. in feral pigs harvested by hunters in Johnston County, North Carolina, USA.
The Study
During 2007–2009, a total of 135 EDTA-anticoagulated whole blood samples were obtained from 76 hunter-harvested juvenile and adult feral pigs (39 males). Blood samples were aspirated postmortem from the carotid artery, heart, or orbital venous sinus, resulting in >1 blood sample for 57 feral pigs. Specimens were stored frozen at –20°C until analysis.
DNA was extracted from EDTA anticoagulated whole blood with QIAGEN MagAttract DNA Blood Mini M48 Kits and QIAGEN BioRobot M48 Workstation (QIAGEN, Valencia, CA, USA) according to the manufacturer’s instructions. All 135 samples were initially screened for the Bartonella 16S–23S internal transcribed spacer (ITS) region by using oligonucleotides 438s (5′-GGT TTT CCG GTT TAT CCC GGA GGG C-3′) and 1100as (5′-GAA CCG ACG ACC CCC TGC TTG CAA AGC A-3′) as forward and reverse primers, respectively (5–7). Samples with positive ITS results were subsequently screened with citrate synthase, RNA polymerase B (rpoB), and a B. koehlerae–specific PCR (6).
For this study, a newly designed forward ITS primer (Bkoehl-1s (5′-CTT CTA AAA TAT CGC TTC TAA AAA TTG GCA TGC-3′) was used in conjunction with the 1100as reverse primer. Amplification was performed in a 25-µL final volume reaction containing 12.5 µL of Tak-Ex Premix (Fisher Scientific, Pittsburgh, PA, USA), 0.1 µL of 100 µmol/L of each forward and reverse primer (IDT DNA Technology, Coralville, IA, USA), 7.3 µL of molecular grade water, and 5 µL of DNA from each sample tested. Blood from a healthy dog was routinely used during DNA extraction and as a PCR negative (5 µL of extracted DNA) control. For positive controls, 5 µL of 0.001 pg/µL of B. henselae DNA (equivalent to 2.5 genome copies) was prepared by serial dilution in specific pathogen-free dog blood (7). No positive control was used for the B. koehlerae PCR. Conventional PCR was performed in an Eppendorf Mastercycler EPgradient (Eppendorf, Hamburg, Germany) under the following conditions: 1 denaturing cycle at 95°C for 2 min followed by 55 cycles at 94°C for 15 s, 68°C (Bartonella genus PCR) or 64°C (B. koehlerae PCR) for 15 s, and 72°C for 18 s. PCR was completed by an additional final cycle at 72°C for 30 sec. Products were analyzed by 2% agarose gel electrophoresis and detection by using ethidium bromide under UV light and sequenced either after purification of amplicons directly from the gel or from plasmid-clone minipreps by using QIAquick PCR purification kit or QIAGEN Miniprep Kit (QIAGEN), respectively, as described (6,7).
Sequence chromatograms and sequence analysis were examined by using ContigExpress software (Vector NTI Suite 10.1, Invitrogen Corp., Carlsbad, CA, USA) and BLAST version 2.0 (www.ncbi.nlm.nih.gov/Education/BLASTinfo/BLAST_algorithm.html) from GenBank. Bacteria species and strain identification was performed by using AlignX software (Vector NTI Suite 10.1, Invitrogen).
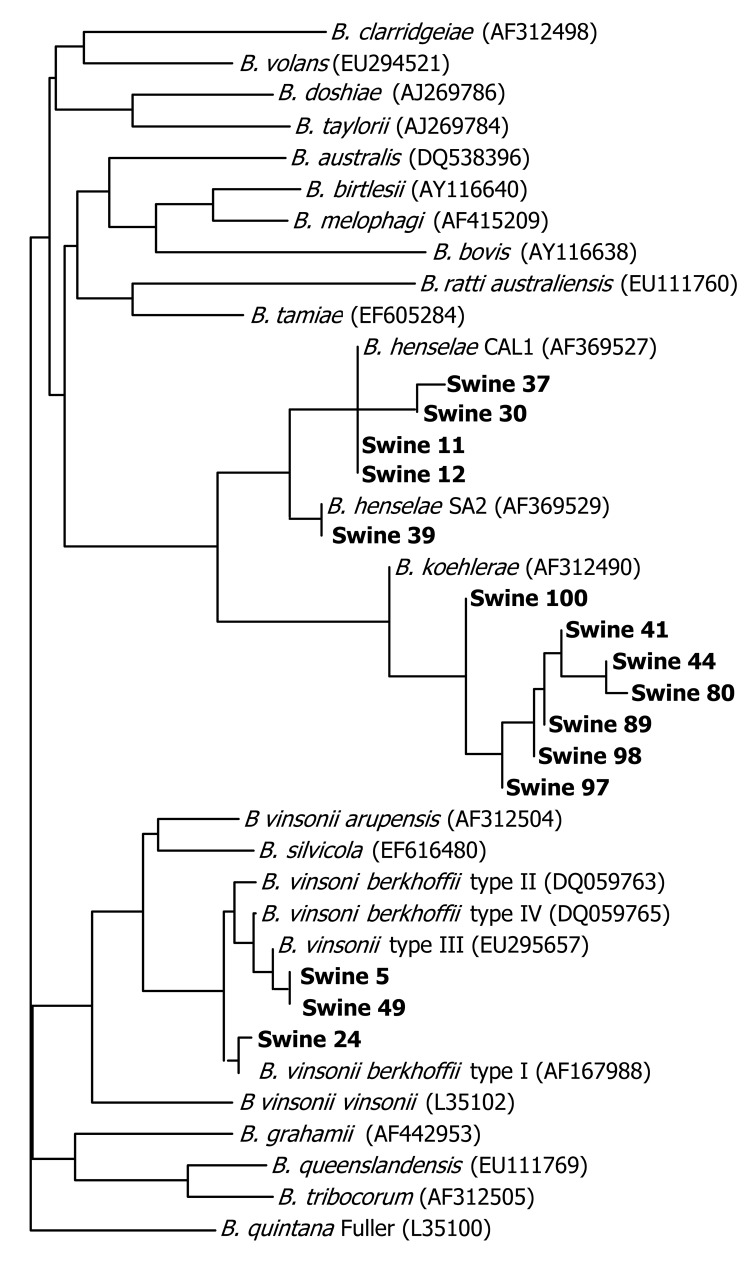
Of 76 feral pigs harvested from Johnston County, North Carolina, and tested by using the 438–1100 ITS PCR, amplicons consistent in size with a Bartonella spp. (400–600-bp amplicon size) were amplified and successfully sequenced from 15 (19.7%) animals. Two B. henselae strains, B. koehlerae and B. vinsonii subsp. berkhoffii genotypes I and III, were identified (Figure). Seven Bartonella PCR–positive samples aligned with B. koehlerae with sequence similarities of 99.2%, 99.4%, 99.8%, and 100% (4 animals) to GenBank sequence AF312490. Four sequences aligned with B. henselae strain Cal-1 (GenBank accession no. AF369527) with 98.7%, 99.1%, 99.4% (2 animals) sequence similarities. B. henselae strain SA2 (San Antonio 2, GenBank accession no. AF369529) was detected in an additional animal with sequence homology of 99.8%.
Figure.
Tree pair-wise alignment of Bartonella DNA sequences detected in feral pig blood samples. GenBank accession numbers are in parentheses. Boldface indicates sequences generated in this study compared with sequences previously submitted to GenBank.
Three feral pig sequences aligned with 2 genotypes of B. vinsonii subsp. berkhoffii (8): 2 animals with 100% homology to genotype III (GenBank accession no. DQ059764), and 1 animal with 99.6% homology with genotype I (GenBank accession no. AF167988). B. vinsonii subsp. berkhoffii genotype III (99.8% homologous to DQ059764) and B. koehlerae (99.1%, homologous to AF312490) sequences were amplified from the same sample. Two different primer sets amplified B. koehlerae DNA from 3 of 7 and 1 of 2 B. vinsonii subsp. berkhoffii genotype III–infected pigs, respectively. PCR specific for the rpoB gene resulted in amplification of B. vinsonii subsp. berkhoffii DNA from the only B. henselae SA2–infected pigs. In no instance was B. henselae (Cal1) amplified and sequenced by using 2 primer sets. Mesorhizobium sequences were obtained from most of the other rpoB PCR amplicons and from one 325s amplicon. Previously, we have reported nonspecific amplification of Mesorhizobium sequences by using other Bartonella spp. 16S–23S ITS primers (5).
Conclusions
We amplified and sequenced B. henselae, B. koehlerae, and B. vinsonii subsp. berkhoffii DNA using >1 primer sets from 19.7% of hunter-harvested feral pigs. The domestic cat is the primary reservoir for B. henselae and B. koehlerae, and fleas are the primary vector (1). Managers of the study site reported the presence of feral cats, but cat numbers and interactions with feral pigs were unknown. Although feral pigs in the southeastern United States are hosts for ticks that are potential Bartonella vectors (9,10), the pigs in this study were harvested during the winter so no ectoparasites were found.
Mesorhizobium, an environmental microbe, most likely introduced during sample collection under field conditions, also was amplified by using 3 primer sets. Although unlikely, ectoparasite feces or dirt containing Bartonella spp. could have been similarly introduced during venipuncture. For future studies in which molecular testing is anticipated, blood should be collected aseptically.
The 3 Bartonella spp. found in feral pigs, B. henselae, B. vinsonii subsp. berkhoffii, and B. koehlerae, are known zoonotic pathogens (3,11,12). Transmission of B. alsatica, which infects wild rabbits in Europe, has been reported in humans with endocarditis and lymphadenitis in association with butchering wild rabbits (13). Because hunters and butchers are exposed to large quantities of pig blood, potential exists for Bartonella spp. transmission through inadvertent cuts or scratches, which has occurred with other zoonotic pig pathogens, such as Brucella suis (14).
Another potential implication of these results involves the transmission of Bartonella spp. from feral to domesticated pigs (15). Ctenocephalides felis and C. canis fleas, known vectors of B. koehlerae and B. henselae, have been reported to infest young pigs (10,12). Measures to control ectoparasites are commonly used by large commercial pig operations, where transmission of Bartonella spp. is not likely to pose a production or zoonotic risk.
Acknowledgments
We thank Pedro Diniz, Mrudula Varanat, Barbara Hegarty, and the staff at Howell Woods Environmental Learning Center for assistance during this project.
Funding for A.W.B. was provided by the Merck-Merial Veterinary Scholars Program. This research was supported in part through a grant from the American College of Veterinary Internal Medicine Foundation and by the State of North Carolina. N.A.C. is the Novartis Fellow in Vector-borne Infectious Disease Research. Funding for field work was provided by the Fisheries and Wildlife Sciences Program at North Carolina State University, Howell Woods Environmental Learning Center, and Johnston Community College.
Biography
Dr Beard is a student at the North Carolina State University, College of Veterinary Medicine, Raleigh, North Carolina. His research interests include zoonotic diseases.
Footnotes
Suggested citation for this article: Beard AW, Maggi RG, Kennedy-Stoskopf S, Cherry NA, Sandfoss MR, DePerno CS, et al. Bartonella spp. in feral pigs, southeastern United States. Emerg Infect Dis [serial on the Internet]. 2011 May [date cited]. http://dx.doi.org/10.3201/eid1705.100141
References
- 1.Chomel BB, Boulouis HJ, Maruyama S, Breitschwerdt EB. Bartonella spp. in pets and effect on human health. Emerg Infect Dis. 2006;12:389–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chomel BB, Kasten RW, Williams C, Wey AC, Henn JB, Maggi R, et al. Bartonella endocarditis: a pathology shared by animal reservoirs and patients. Ann N Y Acad Sci. 2009;1166:120–6. 10.1111/j.1749-6632.2009.04523.x [DOI] [PubMed] [Google Scholar]
- 3.Boulouis HJ, Chang CC, Henn JB, Kasten RW, Chomel BB. Factors associated with the rapid emergence of zoonotic Bartonella infections. Vet Res. 2005;36:383–410. 10.1051/vetres:2005009 [DOI] [PubMed] [Google Scholar]
- 4.Campbell TA, Longa DB. Feral swine damage and damage management in forested ecosystems. For Ecol Manage. 2009;257(12):2319–26.
- 5.Maggi RG, Breitschwerdt EB. Potential limitations of the 16S–23S rRNA intergenic region for the molecular detection of Bartonella species. J Clin Microbiol. 2005;43:1171–6. 10.1128/JCM.43.3.1171-1176.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diniz PP, Maggi RG, Schwartz DS, Cadenas MB, Bradley JM, Hegarty B, et al. Canine bartonellosis: serological and molecular prevalence in Brazil and evidence of co-infection with Bartonella henselae and Bartonella vinsonii subsp. berkhoffii. Vet Res. 2007;38:697–710. 10.1051/vetres:2007023 [DOI] [PubMed] [Google Scholar]
- 7.Cherry NA, Maggi RG, Cannedy AL, Breitschwerdt EB. PCR detection of Bartonella bovis and Bartonella henselae in the blood of beef cattle. Vet Microbiol. 2009;135:308–12. 10.1016/j.vetmic.2008.09.063 [DOI] [PubMed] [Google Scholar]
- 8.Maggi RG, Chomel B, Hegarty BC, Henn J, Breitschwerdt EB. A Bartonella vinsonii berkhoffii typing scheme based upon 16S–23S ITS and Pap31 sequences from dog, coyote, gray fox, and human isolates. Mol Cell Probes. 2006;20:128–34. 10.1016/j.mcp.2005.11.002 [DOI] [PubMed] [Google Scholar]
- 9.Billeter SA, Levy MG, Chomel BB, Breitschwerdt EB. Vector transmission of Bartonella species with emphasis on the potential for tick transmission. Med Vet Entomol. 2008;22:1–15. 10.1111/j.1365-2915.2008.00713.x [DOI] [PubMed] [Google Scholar]
- 10.Cargill C, Davies PR. External parasites. In: Straw BE, Zimmerman JJ, D’Allaire S, Taylor DJ, editors. Diseases of swine, 9th ed. Ames (IA): Blackwell Publishing; 2006. p. 875–89. [Google Scholar]
- 11.Avidor B, Graidy M, Efrat G, Leibowitz C, Shapira G, Schattner A, et al. Bartonella koehlerae, a new cat-associated agent of culture-negative human endocarditis. J Clin Microbiol. 2004;42:3462–8. 10.1128/JCM.42.8.3462-3468.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Breitschwerdt EB, Kordick DL. Bartonella infection in animals: carriership, reservoir potential, pathogenicity, and zoonotic potential for human infection. Clin Microbiol Rev. 2000;13:428–38. 10.1128/CMR.13.3.428-438.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Angelakis E, Lepidi H, Canel A, Rispal P, Perraudeau F, Barre I, et al. Human case of Bartonella alsatica lymphadenitis. Emerg Infect Dis. 2008;14:1951–3. 10.3201/eid1412.080757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Centers for Disease Control and Prevention. Brucella suis infection associated with feral swine hunting—three states, 2007–2008. MMWR Morb Mortal Wkly Rep. 2009;58:618–21. [PubMed] [Google Scholar]
- 15.Wyckoff AC, Henke SE, Campbell TA, Hewitt DG, VerCauteren KC. Feral swine contact with domestic swine: a serologic survey and assessment of potential for disease transmission. J Wildl Dis. 2009;45:422–9. [DOI] [PubMed] [Google Scholar]