To the Editor: The genus Alphavirus (family Togaviridae) comprises 29 viral species (1), grouped in at least 7 antigenic complexes by their serologic cross-reactivity (2). They are maintained in nature through enzootic cycles involving arthropods as vectors with subsequent amplification in small mammals or birds, and epizootic cycles between mosquitoes and large mammals such as horses or humans.
Few reports have been made of the circulation of alphaviruses in Uruguay. A serologic study conducted in 1970 found antibodies to western (WEEV) and eastern equine encephalitis viruses by using complement fixation and hemagglutination inhibition tests in serum specimens from children (3). In 1972–1973, epizootics in horses caused by WEEV were reported in Argentina and Uruguay, and WEEV was isolated from a sick horse (4).
We report a fatal case of viral encephalitis in April 2009 in Montevideo, Uruguay, in a previously healthy 14-year-old boy. Four days before he sought treatment, he had fever, asthenia, and headaches. At hospital admission (April 12, 2009), he was febrile and without neurologic signs; amoxicillin treatment was initiated. Results of a computed tomography scan of the brain were normal.
On day 1, headache, vomiting, neck stiffness, and partial left seizures on the left side developed. Also exhibited were consciousness depression (Glasgow Coma Scale 12 points), hyperreflexia, and bilateral Babinski sign. A cerebrospinal fluid (CSF) sample was negative for bacteria in cultures. An electroencephalogram showed diffuse brain suffering. The patient was brought to the intensive care unit with a clinical diagnosis of viral encephalitis. Over the next 24–36 hours, intracranial hypertension developed, and medical treatment was given (sedation, hyperventilation, mannitol, and barbiturates). Conscience depression progressed to a deeper level, and a computed tomography scan of the brain showed dilatation of the temporal ventricles and compression of the peritroncal and sylvian cisterns. During the next 48 hours, the coma level went deeper, reaching 6 on the Glasgow Scale. Another CSF specimen was taken, and PCR results were negative for herpesvirus and enterovirus. Glasgow Coma Scale level was 3 on April 15, and a decompressive craniectomy was done. Seventy-two hours after admission, the patient died.
The patient’s plasma and CSF were tested for antibodies to dengue and West Nile viruses (immunoglobulins M and G) through ELISA (Focus Technologies, Cypress, CA, USA) and for St. Louis encephalitis and dengue virus by M antibody-capture–ELISA (5). RNA was extracted from plasma and CSF, followed by a generic nested reverse transcription–PCR (RT-PCR) for flaviviruses (6). Serologic and molecular test results were negative for the above-mentioned pathogens. Then we performed a generic nested RT-PCR (7), which amplifies 448 bp at first round and 195 bp (second round) of the alphavirus NSP4 gene. Also, a heminested PCR was done (products 372 bp and 303 bp); RNA from Venezuelan equine encephalitis virus Tc-83 (provided by M. Contigiani, Universidad de Córdoba) was used as positive control. The target region is informative enough to allow the precise identification of the most relevant alphaviruses by sequencing and phylogenetic analysis. Alphavirus genome amplification was achieved for the CSF specimen collected at admission to the hospital. Plasma and a second CSF specimen were PCR negative. To confirm these findings, another nested RT-PCR reaction targeting the NSP1 gene was done as described previously (8). A 208-bp amplicon, which corresponded to the expected size for WEEV, was obtained from plasma and the first CSF specimen.
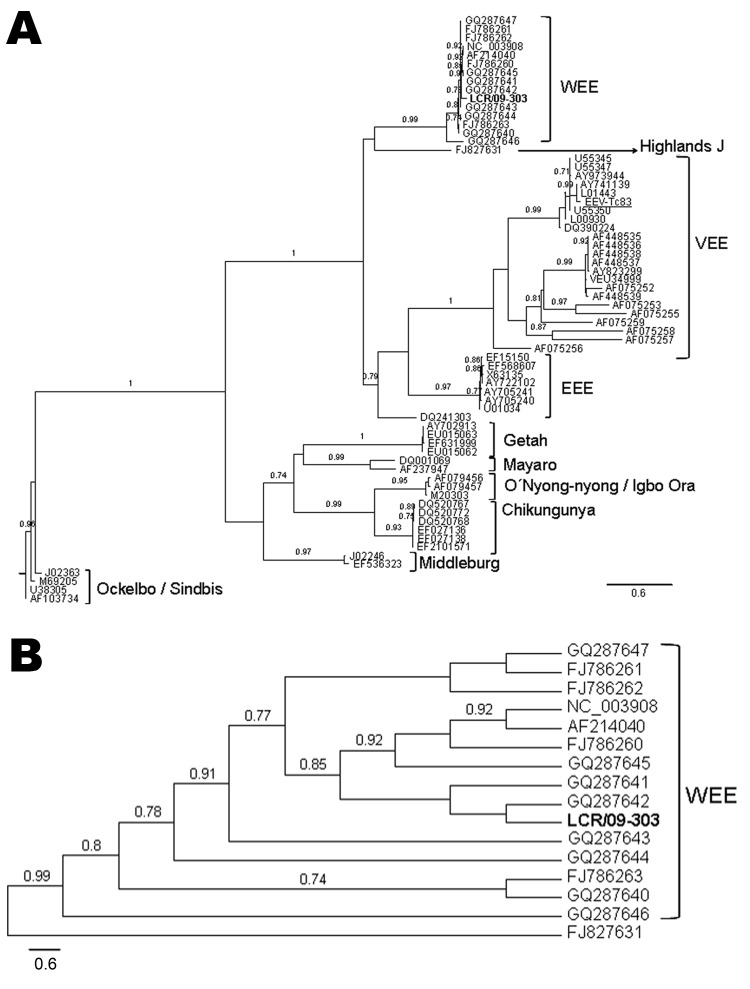
Sequence analyses were conducted on the NSP4 fragments. Maximum likelihood (9) and Bayesian (10) phylogenetic analyses gave similar results. The Figure, panel A, shows that sample LCR/09-303 is part of a well-supported clade (aLRT = 0.99), which groups WEEVs. The sequence LCR/09-303 is a sister taxon to sequences GQ287641 and GQ287642, with poor support (Figure, panel B) (Table A1). These are reference WEEV USA strains (Imperial and Kern) obtained from Culex tarsalis mosquitoes. Our sample and the mentioned sequences are part of a well-supported clade (aLRT = 0.85), together with GQ287645, AF214040, and FJ786260. These are also USA strains; 2 were isolated from infected horses and 1 from Cx. tarsalis mosquitoes. Notably, our sequence is distantly related to GQ287646, which was isolated from Culex spp. mosquitoes in Chaco, Argentina. The nucleotide sequence of the positive control VEEV-Tc83 is correctly placed in the VEEV clade.
Figure.
A) Phylogenetic tree obtained by maximum likelihood analysis of sequences corresponding to the alphavirus NSP4 gene. Alignment used in the analysis had 448 bp and was conducted by using BioEdit software version 7.0.9.0 (www.mbio.ncsu.edu/BioEdit/BioEdit.html). Estimation of the suitable model of nucleotide substitution was carried out by using Modelgenerator (http://bioinf.may.ie/software/modelgenerator). Phylogenetic analysis was run on the PhyML web server (www.atgc-montpellier.fr/phyml), with the following settings: nucleotide substitution model: general time reversible + proportion invariant + Γ; proportion of invariable sites: 0.39; gamma distribution parameter α: 0.67; node support: approximate likelihood-ratio test (only values over 0.70 are shown). Sequences included in the analysis were the following (GenBank accession numbers for individual isolates provided when applicable): human encephalitis case-patient: LCR/09-303 (boldface); reverse transcription–PCR positive control Venezuelan equine encephalitis virus [VEEV] Tc83 (282 nt), FJ786261; western equine encephalitis virus (WEEV): AF214040, GQ287647, GQ287646, GQ287645, GQ287644, GQ287643, GQ287642, GQ287641, GQ287640, FJ786263, FJ786262, FJ786260, NC003908; Highlands J virus, FJ827831; Venezuelan equine encephalitis virus (VEEV), L01443, DQ390224, AF075255, AY823299, AF448539, AF448538, AF448537, AF448536, AF448535, AF075252, U34999, AF075259, AF075256, AF075253, AF075257, AF075258, AY973944. L00930, AY741139, U55350, U55347, U55345; eastern equine encephalitis virus (EEEV), AY722102, U01034, EF568607, EF15150, AY705241, AY705240, X63135, DQ241303; Getah virus, EU015063, EU015062, EF631999, AY702913; Mayaro virus, AF237947, DQ001069; M20303; O’nyong-nyong virus, AF079456; Igbo Ora virus, AF079457; chikungunya virus, EF210157, EF027138, EF027136, DQ520772, DQ520768, DQ520767; Middleburg virus, EF536323, J02246; Ockelbo virus, M69205: Sindbis virus, AF103734, U38305, J02363, M69205. B) Detail of the WEEV clade, showing the relationships between the sample LCR/09-303 and the rest of the WEEV isolates included in the analysis. Scale bars indicate expected nucleotide changes per site.
Clinical and laboratory findings showed that the illness described here was compatible with viral encephalitis. Using a generic RT-PCR assay on an early CSF sample, we amplified a partial sequence (NSP4 gene) of an alphavirus. Phylogenetic analyses showed that the patient’s sequence grouped with sequences from WEEV, with high statistical support. A second RT-PCR assay on the NSP1 gene enabled us to obtain an amplification of 208 bp, which is consistent with the expected size for WEEV. Therefore, we concluded that the fatal disease was likely caused by WEEV. Since the 1970s, to our knowledge, the presence of WEEV (or other alphaviruses) in Uruguay has not been documented. Moreover, no recent reports have been made of genome detection of WEEV in encephalitis cases in the region.
Although the case described here may be rare, the etiology of many viral encephalitides in Uruguay remains unknown. Serologic studies in horses and studies to detect arboviruses in mosquito populations are being conducted to investigate the status of arbovirus infections in Uruguay.
Acknowledgments
We thank José C. Russi for his invaluable scientific advice during all the stages of this work and Gabriela Algorta for her help and interest in our research.
Table A1. Strain designation, place and date of isolation, and encephalitis virus species.
| Strain designation |
Place of isolation |
Year |
Virus |
| TBT 235 | Texas, USA | 1971 | Western equine encephalitis |
| 71V-1658 | Alberta, Canada | 1999 | Western equine encephalitis |
| 85-452NM | New Mexico, USA | 1985 | Western equine encephalitis |
| AG80-646 | Chaco, Argentina | 1980 | Western equine encephalitis |
| 71V1658 | Oregon, USA | 1971 | Western equine encephalitis |
| BFS-2005 | California, USA | 1971 | Western equine encephalitis |
| Montana-64 | Montana, USA | 1967 | Western equine encephalitis |
| Kern | California, USA | 1974 | Western equine encephalitis |
| Imperial | California, USA | 2005 | Western equine encephalitis |
| McMillan | Ontario, Canada | 1941 | Western equine encephalitis |
| ON41-McMillan | Ontario, Canada | 1941 | Western equine encephalitis virus |
| TBT 235 (2) | Texas, USA | 1971 | Western equine encephalitis virus |
| CO92-1356 | USA | 1992 | Western equine encephalitis |
| 71V-1658 | Alberta, Canada | 1999 | Western equine encephalitis |
| 585-01 | Georgia, USA | 2001 | Highands J virus |
| TC-83 | Venezuela | 1986 | Venezuelan equine encephalitis |
| 8131 | Peru | 2008? | Venezuelan equine encephalitis |
| 71D-1252 | Peru | 1971 | Venezuelan equine encephalitis |
| MX01-22 | Mexico | 2004 | Venezuelan equine encephalitis |
| 80U76 | Unknown | 2001? | Venezuelan equine encephalitis |
| OAX142 | Mexico | 1996 | Venezuelan equine encephalitis |
| CPA201 | Mexico | 1993 | Venezuelan equine encephalitis |
| OAX131 | Mexico | 1996 | Venezuelan equine encephalitis |
| CPA152 | Mexico | 1996 | Venezuelan equine encephalitis |
| Mena II | Panama | 1962 | Venezuelan equine encephalitis |
| 68U201 | Unknown | 1968 | Venezuelan equine encephalitis |
| Cabassou CaAr 508 | Guyana | 1968 | Venezuelan equine encephalitis |
| Pixuna BeAr 35645 | Brazil | 1961 | Venezuelan equine encephalitis |
| Mucambo BeAn 8 | Brazil | 1964 | Venezuelan equine encephalitis |
| 78V-3531 | Brazil | 1968 | Venezuelan equine encephalitis |
| AG80-663 | Argentina | 1980 | Venezuelan equine encephalitis |
| 254934 | Venezuela | 2000 | Venezuelan equine encephalitis |
| 3880 | Panamá | 1988? | Venezuelan equine encephalitis |
| V3526 | USA | 2003? | Venezuelan equine encephalitis |
| 3908 | Venezuela | 1995 | Venezuelan equine encephalitis |
| 6119 | Venezuela | 1995 | Venezuelan equine encephalitis |
| PMCHo5 | Venezuela | 1995 | Venezuelan equine encephalitis |
| PE6 | USA | 2004? | Eastern equine encephalitis |
| 82V2137 | USA | 1982 | Eastern equine encephalitis |
| NJ/60 | USA | Unknown | Eastern equine encephalitis |
| FL93-939 | USA | 1993 | Eastern equine encephalitis |
| Florida91-4697 | USA | 1991 | Eastern equine encephalitis |
| Georgia 97 | USA | 1997 | Eastern equine encephalitis |
| ssp. North American variant | USA | 1991? | Eastern equine encephalitis |
| PE-3.0815 | Peru | 1996 | Eastern equine encephalitis |
| YN0540 | China | 2005 | Getah |
| HB0234 | China | 2002 | Getah |
| LEIV 17741 MPR | Mongolia | Unknown | Getah |
| Getah virus, swine | Korea | Unknown | Getah |
| Mayaro virus | Brazil | Unknown | Mayaro |
| MAYLC | French Guiana | Unknown | Mayaro |
| Gulu strain | Africa | Unknown | O´nyong-nyong |
| SG650 | Uganda | 1996–1997 | O´nyong-nyong |
| IBH10964 | Nigeria | 1966 | Igbo Ora |
| DRDE-06 | India | 2006 | Chikungunya |
| IND-06-TN1 | India | 2006 | Chikungunya |
| IND-06-MH2 | India | 2006 | Chikungunya |
| IND06KA3 | India | 2006 | Chikungunya |
| IND06KA9 | India | 2006 | Chikungunya |
| IND06KA12 | India | 2006 | Chikungunya |
| MIDV857 | Zimbabwe | 1993 | Middelburg |
| Middleburg strain | Unknown | Unknown | Middelburg |
| Edsbyn 82-5 | Sweden | Unknown | Ockelbo |
| YN87448 | China | 1998? | Sindbis |
| S.A.AR86 | South Africa | Unknown | Sindbis |
| HR wt strain | Unknown | Unknown | Sindbis |
Footnotes
Suggested citation for this article: Delfraro A, Burgueño A, Morel N, González G, García A, Morelli J, et al. Fatal human case of Western equine encephalitis, Uruguay [letter]. Emerg Infect Dis [serial on the Internet]. 2011 May [date cited]. http://dx.doi.org/10.3201/eid1705.101068
References
- 1.Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, eds. Virus taxonomy. VIIIth report of the International Committee on Taxonomy of Viruses. Philadelphia: Elsevier Academic Press; 2005. [Google Scholar]
- 2.Griffin DE. Alphaviruses. In: Knipe DM, Howley PM, editors. Fields virology, 5th ed. vol. 1. Philadelphia: Lippincott Williams and Wilkins; 2007. [Google Scholar]
- 3.Somma Moreira RE, Campione-Piccardo J, Russi JC, Hortal de Giordano M, Bauzá CA, Peluffo G, et al. Arbovirus en el Uruguay. Arch Pediatr Urug. 1970;41:359–63. [Google Scholar]
- 4.Acha P, Szyfres B. Clamidiosis, rickettsiosis y virosis. In: Zoonosis y enfermedades trasmisibles comunes al hombre y a los animales, 3rd ed., vol. 2. Washington: World Health Organization; 2003. p. 425. [Google Scholar]
- 5.Martin DA, Muth DA, Brown T, Johnson AJ, Karabatsos N, Roehrig JT. Standardization of immunoglobulin M capture enzyme-linked immunosorbent assays for routine diagnosis of arboviral infections. J Clin Microbiol. 2000;38:1823–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sánchez-Seco MP, Rosario D, Domingo C, Hernández L, Valdés K, Guzmán MG, et al. Generic RT-nested-PCR for detection of flaviviruses using degenerated primers and internal control followed by sequencing for specific identification. J Virol Methods. 2005;126:101–9. 10.1016/j.jviromet.2005.01.025 [DOI] [PubMed] [Google Scholar]
- 7.Sánchez-Seco MP, Rosario D, Quiroz E, Guzmán G, Tenorio A. A generic nested-RT-PCR followed by sequencing for detection and identification of members of the alphavirus genus. J Virol Methods. 2001;95:153–61. 10.1016/S0166-0934(01)00306-8 [DOI] [PubMed] [Google Scholar]
- 8.Bronzoni RVM, Baleotti FG, Ribeiro Nogueira RM, Nunes M, Figueiredo LTM. Duplex reverse transcription-PCR followed by nested PCR assays for detection and identification of Brazilian alphaviruses and flaviviruses. J Clin Microbiol. 2005;43:696–702. 10.1128/JCM.43.2.696-702.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. 10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- 10.Ronquist F, Huelsenbeck JP, van der Mark P. MrBayes: Bayesian inference of phylogeny, v. 3.1. 2005. [cited 2010 Jul 1]. http://mrbayes.csit.fsu.edu