Abstract
Robust biomarkers are needed to identify donor kidneys with poor quality associated with inferior early and longer-term outcome. The occurrence of delayed graft function (DGF) is most often used as a clinical outcome marker to capture poor kidney quality. Gene expression profiles of 92 preimplantation biopsies were evaluated in relation to DGF and estimated glomerular filtration rate (eGFR) to identify preoperative gene transcript changes associated with short-term function. Patients were stratified into those who required dialysis during the first week (DGF group) versus those without (noDGF group) and subclassified according to 1-month eGFR of >45 mL/min (eGFRhi) versus eGFR of ≤45 mL/min (eGFRlo). The groups and subgroups were compared in relation to clinical donor and recipient variables and transcriptome-associated biological pathways. A validation set was used to confirm target genes. Donor and recipient characteristics were similar between the DGF versus noDGF groups. A total of 206 probe sets were significant between groups (P < 0.01), but the gene functional analyses failed to identify any significantly affected pathways. However, the subclassification of the DGF and noDGF groups identified 283 probe sets to be significant among groups and associated with biological pathways. Kidneys that developed postoperative DGF and sustained an impaired 1-month function (DGFlo group) showed a transcriptome profile of significant immune activation already preimplant. In addition, these kidneys maintained a poorer transplant function throughout the first-year posttransplant. In conclusion, DGF is a poor marker for organ quality and transplant outcome. In contrast, preimplant gene expression profiles identify “poor quality” grafts and may eventually improve organ allocation.
INTRODUCTION
Kidney transplantation is the treatment of choice for patients with end-stage renal disease. However, despite the significant survival benefit, poor long-term outcomes and insufficient number of donor organs remain the major problems in organ transplantation (1,2). This result has lead to the increasing pressure to use kidneys of potentially inferior quality with likely compromised short- and long-term function. Hence, reliable kidney quality and outcome markers are needed.
A wide number of publications have studied and suggested single as well as combinations of clinical- and histopathology-based variables and derived algorithms to assess organ tissue quality and to predict short- and long-term outcomes. However, none of these biomarkers have so far been proven to be useful in routine clinical decision-making for the individual donor organ and recipient (3,4). This result is partly due to the multitude of factors affecting donor organ quality, such as nephron supply, pre- and peritransplant insults and post-transplant immunologic and nonimmunologic variables (4). In addition, the lack of robust outcome measures further prevents the identification of predictive markers (5). Traditionally, the occurrence of delayed graft function (DGF) is used as a reference marker for poor organ quality and associated impaired short-and long-term outcome.
However, a major problem with DGF is the multitude of definitions itself, usually incorporating the need for hemodialysis during the first week after transplantation (6). The indication for dialysis, however, is subjective, and a number of factors such as postoperative hemodynamics independent of organ quality may affect the need for dialysis treatment. The lack of standardized definitions of DGF as well as the rather weak association between DGF and organ quality are reflected in the controversial results linking DGF with long-term allograft performance and contributes to the current paucity of biomarkers and therapies to affect early and long-term allograft outcome (3,7).
Recently, the value of molecular profiles in biopsies taken at time of transplantation to provide the missing information for a more complex understanding and evaluation of organ quality and long-term function have been reviewed (4). Earlier studies have investigated differences in gene expression between living and deceased donors (8–10), the effects of donor age on gene expression patterns (11) and genes associated with DGF (8–12). The identified transcriptome profiles have significantly enriched our understanding of the pathomechanisms associated with ischemia-reperfusion injury. However, none of these studies have so far produced a robust set of gene-associated transcripts applicable in the clinical routine to capture organ quality and outcome.
In the present study, a large patient cohort including a validation set are analyzed in regard to standard DGF diagnosis and glomerular filtration rate (GFR)-based outcome measures to discover new biomarkers in deceased donor biopsies associated with donor quality and short- and longer-term outcomes.
MATERIALS AND METHODS
Kidney Samples and Patient Enrollment
The study included 92 consecutive adult kidney transplant recipients of deceased donor kidneys (ages 17–70 years). The Institutional Review Board at Virginia Commonwealth University (VCU-IRB Protocol #HM11454) approved the study protocol. Written informed consent was obtained from all patients. No living donors, human immunodeficiency virus (HIV)-positive patients or retransplantation patients were included in the study. Allograft biopsies from kidneys preserved using both cold preservation and pump perfusion preservation were included. Kidney allograft tissue was obtained through an 18-gauge biopsy needle, and all samples were placed in RNAlater (Ambion) immediately after collection. Biopsies were collected at preimplantation time (post–cold ischemia time; n = 92). Estimated GFR (eGFR) was calculated using the abbreviated Modification of Diet in Renal Disease formula (13). DGF was defined as the need of dialysis during the first 7 d after kidney transplantation. The patients were stratified into patients undergoing post-transplant DGF versus patients without DGF (DGF versus noDGF) and then further subdivided into patients with an eGFR >45 mL/min versus patients with an eGFR =45 mL/min (eGFRhi versus eGFRlo) following the criteria described by Kainz et al. (14).
RNA Isolation, cDNA Synthesis and In Vitro Transcription for Labeled cRNA Probe
The sample preparation protocol follows the Affymetrix GeneChip® Expression Analysis Manual (Affymetrix, Santa Clara, CA, USA). Briefly, total RNA was reverse-transcribed using T7-polydT primer and converted into double-stranded cDNA (One-Cycle Target Labeling and Control Reagents; Affymetrix), with templates being used for an in vitro transcription reaction to yield biotin-labeled antisense cRNA. The labeled cRNA was chemically fragmented and made into the hybridization cocktail according to the Affymetrix GeneChip protocol, which was then hybridized to HG-U133A 2.0 GeneChips. The array image was generated by the high-resolution GeneChip® Scanner 3000 by Affymetrix.
Microarray Data Processing
Gene expression microarray data for 92 preimplantation (PI) samples were available. Probe level data were read into the R programming environment using the affy Bioconductor package, and the robust multiarray average method was used to obtain probe set expression summaries. Quality assessment of each GeneChip was performed by assessing the average background; scaling factor; percent present calls; the 3′:5′ ratio for GAPDH, β-actin and signal transducer and activator of transcription 1 (STAT1); and the overall 3′:5′ ratio with associated 95% confidence interval using the publicly available application (15,16). One microarray did not pass quality control, leaving a total of 91 arrays for the analysis. Probe level data were read into the R programming environment using the affy Bioconductor package, and the robust multiarray average method was used to obtain probe set expression summaries (17,18). All control and probe sets declared absent for all samples were removed, leaving 17,529 probe sets for statistical analysis of the PI biopsies.
Analysis of Differentially Expressed Genes among the Patient Groups
To identify genes differentially expressed between transplant patients with and without dialysis during the first week after kidney transplantation, for each probe set, a two-sample t test comparing the DGF versus noDGF groups was performed. Probe sets having a raw P value <0.01 were considered significant.
Subjects were subsequently classified into two groups on the basis of their GFR at 1 month, individuals having eGFR >45 (GFRhi) and individuals having eGFR =45 (GFRlo). A moderated t test was used to compare the two groups using the limma package, and a P value from the moderated t test <0.01 was used to identify significant probe sets. To identify probe sets exhibiting significant differential expression between the four groups, probe set level analysis of variance models were fit, considering probe set expression as the response and group (noDGF with 1 month eGFR >45; noDGF with 1 month eGFR =45; DGF with 1 month eGFR >45; and DGF with 1 month eGFR =45) as the fixed effect of interest. A group means parameterization of group was included as the fixed-effect term to facilitate extraction of linear contrasts of interest. Again, the limma package was used to fit the mixed-effects models using restricted maximum likelihood procedure, and an empirical Bayes method was applied to moderate probe set standard errors by borrowing information across the entire set of probe sets. To adjust for multiple comparisons, probe sets with a P value from the overall F test <0.01 were considered significant. Thereafter, among probe sets identified as differentially expressed via the F test, pair-wise comparisons of interest were performed. Specifically, we examined the following four pair-wise comparisons: (i) no DGF with 1 month eGFR =45 versus noDGF with 1 month eGFR >45, (ii) DGF with 1 month GFR =45 versus DGF with 1 month eGFR >45, (iii) DGF with 1 month eGFR =45 versus noDGF with 1 month GFR =45, and (iv) DGF with 1 month eGFR >45 versus noDGF with 1 month eGFR >45.
Biological and Functional Analysis
Gene ontology and gene interaction analyses were executed using ToppGene (http://toppgene.cchmc.org) (19). Gene lists containing Entrez GeneID numbers were used as input. Identification of significant biological processes and pathways was determined using the false discovery rate method (P < 0.01). The results obtained with ToppGene were confirmed using Ingenuity Pathway (IPA, www.ingenuity.com).
Validation of Five Genes in an Independent Set of Patients Using RT-qPCR
Initially, to validate our microarray findings, we carried out a reverse transcriptase–quantitative real-time polymerase chain reaction (RT-qPCR) for CCL5 (chemokine [C-C motif] ligand 5), ITGB2 (integrin β2 [complement component 3, receptor 3 and 4 subunit]) and EGF (epidermal growth factor) mRNAs from the same RNA samples that were subjected to microarray study. Moreover, an independent set of samples (validation set) was tested for CCL5, ITGB2, EGF, VCAN (versican) and CXCR4 (chemokine [C-X-C motif] receptor 4) mRNAs. The genes for validation were selected based on the following: (a) statistical differential expression between DGFhi versus DGFlo groups and statistical differential expression between eGFRhi versus eGFRlo at 1 month posttransplantation, and (b) the genes were among the top scoring networks and pathways when using interaction networks and functional analysis for the different comparison analysis. Total RNA was subjected to reverse transcription using TaqMan® Reverse Transcription Reagents (Applied Biosystems, Foster City, CA, USA), according to the manufacturer’s protocol. Real-time PCRs were then carried out in an ABI Prism 7700 Sequence Detection System, using TaqMan® Gene Expression Assays (Applied Biosystems). Data were analyzed according to the comparative cycle threshold (Ct) method and were normalized with a housekeeping gene (GAPDH [glyceraldehyde-3-phosphate dehydrogenase]). Pearson correlation coefficient (r) was calculated to examine the relation between microarray and real-time PCR results. P < 0.05 was considered significant.
All supplementary materials are available online at www.molmed.org.
RESULTS
Study Cohort Characteristics
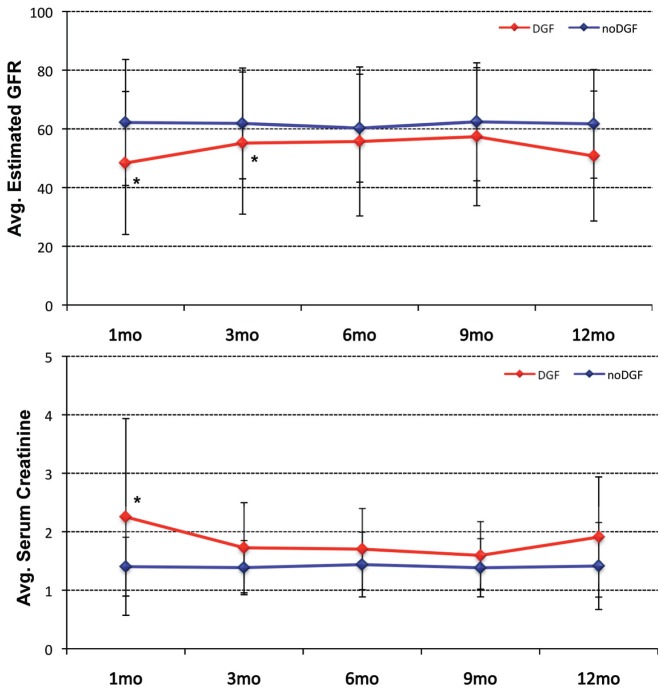
A total of 92 patients were enrolled in the study. Patients were transplanted between January 2008 and March 2010. All patients received a deceased donor kidney transplant and had a minimum follow-up of at least 12 months. Of the kidney transplant recipients included in the analysis, only one graft was lost during the first year after kidney transplant because of rapid progressive chronic allograft dysfunction with interstitial fibrosis and tubular atrophy. We first compared the two groups of DGF versus noDGF patients. The clinical definition of DGF used was the need for dialysis within the first week after kidney transplant (12). A total of 25 patients developed post–kidney transplant DGF (27%) and 67 patients did not (73%). Within the DGF group, the period of dialysis before recovery for the individual patient showed a wide range (23 ± 36 d). The clinical indications for dialysis in these 25 patients are described in Supplementary Table 1. For patients with DGF extended >3 wks, allograft biopsy was performed. In the vast majority of the performed biopsies, the histological diagnosis was acute tubular necrosis. Clinically, the two patient groups were similar regarding recipient and donor age, recipient and donor sex, last donor creatinine, cold ischemia time, warm ischemia time, incidence of acute rejection (AR) during the first year after kidney transplant or the use of pump perfusion preservation. A total of 57% of the donor kidneys underwent pump perfusion preservation, and 26.4% of the donors were extended criteria donors (including 13.2% non–heart-beating donors). The two groups only differed significantly in serum creatinine levels at 1 month post–kidney transplant and eGFR at 1-month and 1-year post–kidney transplant (Table 1 and Figure 1). The histology of the biopsies at preimplantation time is shown in Supplementary Table 2.
Table 1.
Demographic and relevant clinical information for the enrolled study patients separated according to DGF versus noDGF after transplantation.
| DGF | noDGF | DGF versus noDGF (P) | |
|---|---|---|---|
| n | 25 | 66 | |
| Recipient age | 52.6 ± 11.2 | 49.6 ± 13.8 | 0.29 |
| Recipient sex (n) [M (F)] | 17 (8) | 41 (25) | 0.395 |
| Recipient race (n) | |||
| Caucasian | 2 | 17 | <0.001 |
| African American | 22 | 45 | |
| Other | 1 | 4 | |
| Recipient hepatitis C virus status (positive, n) | 3 | 9 | 0.269 |
| CMV disease (positive, n) | 0 | 0 | N/A |
| Recipient CMV status (positive, n) | 18 | 46 | 0.144 |
| Donor age (years) | 42.8 ± 13.0 | 38.2 ± 15.0 | 0.150 |
| Donor sex (n) [M (F)] | 18 (7) | 35 (30) | 0.091 |
| Donor race (n) | |||
| Caucasian | 17 | 36 | 0.177 |
| African American | 5 | 23 | |
| Other | 2 | 1 | |
| Donor hepatitis C virus antibody (positive, n) | 3 | 8 | 0.278 |
| Donor CMV status (positive, n) | 15 | 33 | 0.139 |
| Cold ischemia time (min) | 1271 ± 530 | 1207 ± 532 | 0.607 |
| Warm ischemia time/revascularization time (min) | 29.6 ± 7.8 | 30.5 ± 7.7 | 0.647 |
| Pump perfusion preservation time (min) | 450 ± 508 | 488 ± 491 | 0.746 |
| Last donor serum creatinine (mg/dL) | 1.3 ± 0.6 | 1.0 ± 0.3 | 0.175 |
| Acute rejection episodes (n) | 4 | 90.275 | |
| HLA MM-A | 1 ± 1 | 2 ± 1 | 0.067 |
| HLA MM-B | 2 ± 1 | 2 ± 1 | 0.889 |
| HLA MM-DR | 1 ± 1 | 1 ± 1 | 0.462 |
| HLA MM total | 4 ± 2 | 4 ± 2 | 0.200 |
| Panel reactive antibodies at transplant–T cells (%) | 43.3 ± 42.9 | 40.7 ± 34.9 | 0.782 |
| Panel reactive antibodies at transplant–B cells (%) | 17.8 ± 27.6 | 22.3 ± 32.1 | 0.510 |
| Serum creatinine (mg/dL) | |||
| 1 month | 2.3 ± 1.7 | 1.4 ± 0.5 | 0.019 |
| 3 months | 1.7 ± 0.8 | 1.4 ± 0.5 | 0.052 |
| 6 months | 1.7 ± 0.7 | 1.4 ± 0.6 | 0.117 |
| 9 months | 1.6 ± 0.6 | 1.6 ± 0.7 | 0.900 |
| 12 months | 1.9 ± 1.0 | 1.4 ± 0.5 | 0.065 |
| Average eGFR (mL/min) | |||
| 1 month | 48.4 ± 24.4 | 62.2 ± 21.5 | 0.017 |
| 3 months | 55.1 ± 24.2 | 61.9 ± 18.9 | 0.229 |
| 6 months | 55.7 ± 25.4 | 60.2 ± 18.4 | 0.448 |
| 9 months | 57.3 ± 23.5 | 62.4 ± 20.1 | 0.408 |
| 12 months | 50.7 ± 22.1 | 61.7 ± 18.5 | 0.064 |
CMV, cytomegalovirus; N/A, not applicable; MM, mismatch.
Data are averages ± standard deviation (SD) unless otherwise stated. eGFR was calculated using the abbreviated Modification of Diet in Renal Disease formula. P values were calculated using Fisher exact test.
Figure 1.
Course of eGFR (top) and serum creatinine levels (bottom) for the DGF and noDGF patient groups throughout the first posttransplant year. Asterisks indicate a statistically significant difference (P ≤ 0.05) between the DGF and noDGF groups.
Table 2.
Demographic and relevant clinical information for the 91 study patients when separated on the basis of DGF versus noDGF classification in combination with eGFR values at 1 month posttransplantation.
| DGFlo | DGFhi | DGFlo versus DGFhi (P) | noDGFlo | noDGFhi | noDGFlo versus noDGFhi (P) | DGFlo versus noDGFlo (P) | DGFhi versus noDGFhi (P ) | |
|---|---|---|---|---|---|---|---|---|
| N | 13 | 12 | 16 | 50 | ||||
| Recipient age (years) | 56.2 ± 8.1 | 47.7 ± 13.1 | 0.474 | 53.3 ± 9.1 | 48.5 ± 14.7 | 0.573 | 0.928 | 0.999 |
| Recipient sex (n) [M (F)] | 10 (3) | 7 (5) | 0.286 | 11 (5) | 30 (20) | 0.375 | 0.474 | 0.582 |
| Recipient race (n) | 0.051 | 0.018 | 0.037 | |||||
| Caucasian | 1 | 1 | 7 | 10 | ||||
| African American | 12 | 10 | 8 | 37 | ||||
| Other | 0 | 1 | 1 | 3 | ||||
| Recipient hepatitis C virus status (positive) | 0 | 3 | 0 | 7 | ||||
| CMV disease (positive) | 0 | 0 | 0 | 0 | ||||
| Recipient CMV status (positive, n) | 10 | 8 | 9 | 39 | ||||
| Donor age (years) | 47.1 ± 11.7 | 38.3 ± 13.3 | 0.418 | 42.3 ± 17.7 | 37.3 ± 13.9 | 0.549 | 0.809 | 1.000 |
| Donor sex (n) [M (F)] | 9 (4) | 9 (3) | 6 (10) | 30 (20) | 0.228 | 0.092 | 0.263 | |
| Donor race (n) | 0.232 | 0.232 | 0.109 | |||||
| Caucasian | 7 | 10 | 7 | 29 | ||||
| African American | 3 | 2 | 6 | 19 | ||||
| Other | 3 | 0 | 3 | 2 | ||||
| Donor HBV antibody (positive) | 0 | 1 | 0 | 1 | ||||
| Donor hepatitis C virus antibody (positive) | 0 | 3 | 1 | 7 | ||||
| Donor CMV status (positive) | 9 | 6 | 9 | 24 | ||||
| Cold ischemia time (min) | 1,328 ± 458 | 1,208 ± 616 | 0.980 | 1,201 ± 480 | 1,209 ± 550.3 | 1.000 | 0.924 | 0.996 |
| Warm ischemia time/revascularization time (min) | 30.8 ± 8.3 | 28.3 ± 7.5 | 0.893 | 30.1 ± 6.7 | 30.6 ± 8.1 | 0.995 | 0.994 | 0.848 |
| Pump perfusion preservation time (min) | 611.5 ± 522.6 | 428.7.0 ± 511.7 | 0.904 | 500.1 ± 490.0 | 494.8 ± 496.6 | 1.000 | 0.949 | 0.999 |
| Last donor serum creatinine (mg/mL) | 1.5 ± 0.8 | 1.1 ± 0.4 | 0.223 | 0.9 ± 0.2 | 1.0 ± 0.3 | 0.941 | 0.029 | 0.999 |
| Acute rejection episodes (n) | 4 | 0 | 2 | 7 | ||||
| HLA MM-A (n) | 1 ± 1 | 1 ± 1 | 0.924 | 1 ± 1 | 2 ± 1 | 0.295 | 0.933 | 0.334 |
| HLA MM-B (n) | 2 ± 1 | 2 ± 1 | 0.871 | 1 ± 1 | 2 ± 1 | 0.541 | 0.701 | 0.811 |
| HLA MM-DR (n) | 1 ± 1 | 1 ± 1 | 1.000 | 1 ± 1 | 1 ± 1 | 0.721 | 1.000 | 0.874 |
| HLA MM total (n) | 4 ± 2 | 4 ± 2 | 1.000 | 4 ± 2 | 5 ± 1 | 0.296 | 0.997 | 0.544 |
| Panel reactive antibodies at transplant–T (%) | 40.0 ± 42.4 | 43.0 ± 44.9 | 0.997 | 33.1 ± 34.1 | 43.5 ± 35.1 | 0.791 | 0.960 | 0.989 |
| Panel reactive antibodies at transplant–B (%) | 14.7 ± 24.4 | 21.1 ± 31.1 | 0.956 | 26.0 ± 38.7 | 21.1 ± 30.1 | 0.946 | 0.767 | 1.000 |
| Serum creatinine (mg/dL) | ||||||||
| 1 month | 3.2 ± 1.9 | 1.2 ± 0.3 | <0.0001 | 1.9 ± 0.6 | 1.2 ± 0.3 | 0.013 | <0.0001 | 0.999 |
| 3 months | 2.2 ± 0.7 | 1.2 ± 0.3 | <0.0001 | 1.8 ± 0.5 | 1.3 ± 0.4 | 0.001 | 0.135 | 0.958 |
| 6 months | 2.1 ± 0.6 | 1.2 ± 0.5 | 0.001 | 1.9 ± 0.7 | 1.3 ± 0.4 | 0.001 | 0.731 | 0.983 |
| 9 months | 1.9 ± 0.4 | 1.2 ± 0.5 | 0.668 | 2.5 ± 3.3 | 1.3 ± 0.4 | 0.032 | 0.729 | 0.999 |
| 12 months | 2.5 ± 1.1 | 1.4 ± 0.6 | 0.011 | 1.6 ± 0.4 | 1.4 ± 0.8 | 0.844 | 0.048 | 1.000 |
| eGFR (mL/min) | ||||||||
| 1 month | 28.1 ± 10.6 | 70.3 ± 12.8 | <0.0001 | 38.8 ± 6.3 | 69.6 ± 19.1 | <0.0001 | 0.329 | 0.893 |
| 3 months | 38.4 ± 10.7 | 74.9 ± 20.4 | <0.0001 | 41.7 ± 9.3 | 68.0 ± 16.6 | <0.0001 | 0.942 | 0.818 |
| 6 months | 39.5 ± 11.5 | 75.1 ± 24.0 | <0.0001 | 41.1 ± 10.4 | 66.5 ± 16.0 | <0.0001 | 0.994 | 0.323 |
| 9 months | 42.4 ± 8.3 | 73.9 ± 24.0 | 0.001 | 40.5 ± 13.3 | 68.5 ± 18.4 | <0.0001 | 1.000 | 0.849 |
| 12 months | 34.3 ± 11.2 | 65.5 ± 18.8 | 0.001 | 46.3 ± 8.2 | 65.7 ± 18.4 | 0.006 | 0.386 | 0.969 |
CMV, cytomegalovirus; HBV, hepatitis B virus; MM, mismatch.
Data are averages ± SD unless otherwise stated. eGFR was calculated using the abbreviated Modification of Diet in Renal Disease formula. Statistical differences were determined by Tukey-Kramer honestly significant difference (HSD) (α = 0.05) for multiple comparison testing or by Fisher exact test.
Gene Expression Profiling of DGF Versus noDGF Grafts in Preimplantation Biopsies
One microarray failed our quality-control assessment so that preimplantation gene expression microarray data were available for 91 patients from our cohort. Gene expression microarray analysis comparing DGF (n = 25) to noDGF (n = 66) patients identified 206 probe sets (P < 0.01) differentially expressed. However, gene ontology analyses failed to identify any significantly affected biological processes or pathways overrepresented by these genes (P < 0.01). Furthermore, unsupervised hierarchical clustering did not show any clear separation between the DGF and noDGF preimplantation biopsies (not shown).
Gene expression microarray analysis comparing the two groups defined as 1 month eGFR >45 (GFRhi, N = 62) and 1 month eGFR =45 (GFRlo, N = 29) identified 208 differentially expressed probe sets (P < 0.01) (Supplementary Table 3). Expression for 80 probe sets was higher in the GFRhi group, whereas expression for 128 probe sets was higher in the GFRlo group. Core analysis was performed to interpret the data set in the context of biological processes, pathways and molecular networks. From the analysis of the differentially expressed genes, the top network associated biological functions were cellular compromise; molecular transport; inflammatory disease (network 1, score 38); and cell-to-cell signaling and interaction, cell-mediated immune response and cellular development (network 2, score 29). Moreover, the top biological functions associated with the statistical differentially expressed genes were inflammatory disease (P = 9.4E-10–9.5E-03) with overexpression of the following genes: ADA (adenosine deaminase) CD28 (T-cell-specific surface glycoprotein CD28), CD48 (CD48 antigen [B cell membrane protein]), CCL5, CD2 (T-cell surface antigen CD2), CD27 (T cell activation antigen CD27) and CXCR4, among others. Also, immune cell trafficking (P = 2.6E-08–9.5E-03) and cell-mediated immune response (P = 3.7E-08–9.3E-03) were among the top biological functions. The top canonical pathway associated with these genes was B-cell development.
Table 3.
List of differentially expressed genes with corresponding average fold changes detected between DGFlo and DGFhi as well as DGFlo and noDGFlo by microarray gene expression analysis.
| DGFlo versus DGFhi | DGFlo versus noDGFlo | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| AffyID | Entrez ID | Gene symbol | P | Average fold change | P | Average fold change |
| 200001_at | 826 | CAPNS1 | 0.00047 | 1.56 | ||
| 200814_at | 5720 | PSME1 | 0.00056 | 1.23 | ||
| 201030_x_at | 3945 | LDHB | 0.00090 | −1.21 | ||
| 201044_x_at | 1843 | DUSP1 | 0.00038 | 1.52 | ||
| 201060_x_at | 2040 | STOM | 0.00036 | 1.49 | ||
| 201137_s_at | 3115 | HLA-DPB1 | 0.00003 | 2.42 | 0.00170 | 1.84 |
| 201519_at | 9868 | TOMM70A | 0.00065 | −1.29 | ||
| 201621_at | 4681 | NBL1 | 0.00064 | 1.31 | ||
| 201666_at | 7076 | TIMP1 | 0.00081 | 2.12 | ||
| 201721_s_at | 7805 | LAPTM5 | 0.00042 | 1.97 | ||
| 201743_at | 929 | CD14 | 0.00077 | 2.38 | ||
| 201781_s_at | 9049 | AIP | 0.00078 | 1.39 | ||
| 201841_s_at | 3315 | HSPB1 | 0.00052 | 1.73 | ||
| 201988_s_at | 1389 | CREBL2 | 0.00083 | 1.36 | ||
| 202207_at | 10123 | ARL4C | 0.00031 | 2.25 | ||
| 202237_at | 4837 | NNMT | 0.00031 | 3.39 | 0.00513 | 2.39 |
| 202238_s_at | 4837 | NNMT | 0.00012 | 3.46 | 0.00013 | 3.16 |
| 202540_s_at | 3156 | HMGCR | 0.00068 | −1.63 | ||
| 202791_s_at | 9701 | SAPS2 | 0.00022 | 1.26 | ||
| 202803_s_at | 3689 | ITGB2 | 0.00056 | 1.66 | ||
| 202959_at | 4594 | MUT | 0.00067 | −1.54 | 0.00406 | −1.40 |
| 202960_s_at | 4594 | MUT | 0.00025 | −1.46 | ||
| 203392_s_at | 1487 | CTBP1 | 0.00027 | 2.35 | ||
| 204118_at | 962 | CD48 | 0.00093 | 1.53 | ||
| 204198_s_at | 864 | RUNX3 | 0.00019 | 1.25 | ||
| 204259_at | 4316 | MMP7 | 0.00024 | 2.51 | ||
| 204655_at | 6352 | CCL5 | 0.00056 | 1.74 | ||
| 204661_at | 1043 | CD52 | 0.00006 | 1.75 | 0.00422 | 1.44 |
| 205683_x_at | 64499 | TPSAB1 | 0.00056 | 1.52 | 0.00222 | 1.41 |
| 205798_at | 3575 | IL7R | 0.00009 | 1.93 | ||
| 206045_s_at | 8715 | NOL4 | 0.00013 | −1.20 | ||
| 206254_at | 1950 | EGF | 0.00048 | −2.90 | ||
| 206336_at | 6372 | CXCL6 | 0.00006 | 2.23 | 0.00114 | 1.82 |
| 206343_s_at | 3084 | NRG1 | 0.00064 | −1.76 | ||
| 206392_s_at | 5918 | RARRES1 | 0.00085 | 2.68 | ||
| 206783_at | 2249 | FGF4 | 0.00027 | −1.26 | ||
| 207134_x_at | 64499 | TPSB2 | 0.00016 | 1.56 | 0.00195 | 1.40 |
| 207174_at | 2262 | GPC5 | 0.00139 | −2.72 | 0.00037 | −2.84 |
| 207238_s_at | 5788 | PTPRC | 0.00057 | 1.75 | ||
| 208306_x_at | 3123 | HLA-DRB1 | 0.00026 | 1.62 | 0.00092 | 1.50 |
| 208675_s_at | 1650 | DDOST | 0.00054 | 1.49 | ||
| 208679_s_at | 10109 | ARPC2 | 0.00062 | 1.24 | 0.00430 | 1.18 |
| 208729_x_at | 3106 | HLA-B | 0.00052 | 1.71 | 0.00153 | 1.57 |
| 208845_at | 7419 | VDAC3 | 0.00701 | −1.27 | 0.00017 | −1.37 |
| 208894_at | 3122 | HLA-DRA | 0.00469 | 1.60 | 0.00038 | 1.74 |
| 208971_at | 7389 | UROD | 0.00098 | 1.30 | ||
| 209040_s_at | 5696 | PSMB8 | 0.00100 | 1.66 | 0.00354 | 1.52 |
| 209073_s_at | 8650 | NUMB | 0.00043 | 1.39 | ||
| 209138_x_at | 3535 | IGL@ | 0.00019 | 5.34 | ||
| 209217_s_at | 11152 | WDR45 | 0.00085 | 1.31 | ||
| 209312_x_at | 100133484 | HLA-DRB1 | 0.00025 | 1.81 | 0.00088 | 1.64 |
| 209312_x_at | 100133484 | HLA-DRB1 | 0.00088 | 1.27 | ||
| 209417_s_at | 3430 | IFI35 | 0.00076 | 1.61 | ||
| 209625_at | 5283 | PIGH | 0.00049 | −1.40 | ||
| 210072_at | 6363 | CCL19 | 0.00043 | 3.74 | ||
| 210084_x_at | 7177 | TPSAB1 | 0.00069 | 1.48 | 0.00213 | 1.39 |
| 210397_at | 1672 | DEFB1 | 0.00446 | −1.44 | 0.00079 | −1.50 |
| 210764_s_at | 3491 | CYR61 | 0.00055 | 2.00 | ||
| 210889_s_at | 2213 | FCGR2B | 0.00026 | 1.87 | ||
| 210915_x_at | 28639 | TRBC1 | 0.00004 | 1.81 | ||
| 210982_s_at | 3122 | HLA-DRA | 0.00260 | 1.64 | 0.00037 | 1.73 |
| 211395_x_at | 9103 | FCGR2C | 0.00020 | 1.52 | 0.00104 | 1.41 |
| 211430_s_at | 28396 | IGH@ | 0.00061 | 6.08 | ||
| 211645_x_at | 0 | Affy_211645_x_at | 0.00044 | 3.53 | ||
| 211654_x_at | 3119 | HLA-DQB1 | 0.00025 | 2.28 | 0.00011 | 2.26 |
| 211796_s_at | 28638 | TRBC1 | 0.00044 | 1.74 | ||
| 211911_x_at | 3106 | HLA-B | 0.00090 | 1.66 | 0.00051 | 1.65 |
| 212510_at | 23171 | GPD1L | 0.00019 | −1.51 | ||
| 212587_s_at | 5788 | PTPRC | 0.00023 | 2.44 | ||
| 212588_at | 5788 | PTPRC | 0.00063 | 2.68 | ||
| 212592_at | 3512 | IGJ | 0.00031 | 5.70 | ||
| 213193_x_at | 28639 | TRBC1 | 0.00084 | 1.39 | ||
| 213321_at | 594 | BCKDHB | 0.00006 | −1.59 | ||
| 213539_at | 915 | CD3D | 0.00047 | 1.46 | ||
| 213603_s_at | 5880 | RAC2 | 0.00012 | 1.76 | ||
| 214164_x_at | 771 | CA12 | 0.00005 | −1.38 | ||
| 214382_at | 54346 | UNC93A | 0.00090 | −1.33 | ||
| 214669_x_at | 3514 | IGKC | 0.00023 | 3.92 | ||
| 214677_x_at | 3535 | IGL@ | 0.00027 | 6.02 | ||
| 214836_x_at | 3514 | IGKC | 0.00053 | 2.76 | ||
| 215121_x_at | 28815 | IGL@ | 0.00033 | 4.51 | ||
| 215176_x_at | 100130100 | LOC100130100 | 0.00038 | 4.37 | ||
| 215193_x_at | 100133484 | HLA-DRB1 | 0.00018 | 1.86 | 0.00038 | 1.73 |
| 215379_x_at | 28815 | IGL@ | 0.00017 | 4.69 | ||
| 215646_s_at | 1462 | VCAN | 0.00099 | 2.94 | ||
| 215867_x_at | 771 | CA12 | 0.00028 | −1.34 | ||
| 215946_x_at | 91353 | IGLL3 | 0.00030 | 2.71 | ||
| 215952_s_at | 4946 | OAZ1 | 0.00097 | 1.34 | ||
| 216474_x_at | 64499 | TPSAB1 | 0.00019 | 1.60 | ||
| 216576_x_at | 28299 | IGKC | 0.00091 | 2.96 | ||
| 217022_s_at | 100126583 | IGH@ | 0.00008 | 6.43 | ||
| 217148_x_at | 3535 | IGL@ | 0.00039 | 2.30 | ||
| 217235_x_at | 28813 | IGL@ | 0.00084 | 1.72 | ||
| 217456_x_at | 3133 | HLA-E | 0.00049 | 1.47 | 0.00843 | 1.31 |
| 217478_s_at | 3108 | HLA-DMA | 0.00037 | 1.76 | 0.00270 | 1.55 |
| 217747_s_at | 6203 | RPS9 | 0.00032 | 1.38 | ||
| 217774_s_at | 51504 | HSPC152 | 0.00085 | 1.25 | ||
| 218024_at | 51660 | BRP44L | 0.00010 | −1.41 | 0.00381 | −1.27 |
| 218170_at | 51015 | ISOC1 | 0.00087 | −1.44 | ||
| 218192_at | 51447 | IP6K2 | 0.00092 | 1.24 | ||
| 218546_at | 79762 | C1orf115 | 0.00031 | −1.38 | ||
| 219076_s_at | 5827 | PXMP2 | 0.00015 | −1.90 | ||
| 219118_at | 51303 | FKBP11 | 0.00074 | 1.66 | ||
| 221523_s_at | 58528 | RRAGD | 0.00281 | −1.72 | 0.00025 | −1.88 |
| 221524_s_at | 58528 | RRAGD | 0.00092 | −1.56 | ||
| 221581_s_at | 7462 | LAT2 | 0.00083 | 1.31 | ||
| 221651_x_at | 3514 | IGK@ | 0.00036 | 3.84 | ||
| 221671_x_at | 3514 | IGK@ | 0.00030 | 3.97 | ||
| 221875_x_at | 3134 | HLA-F | 0.00075 | 1.55 | ||
| 34210_at | 1043 | CD52 | 0.00011 | 2.13 | 0.00918 | 1.59 |
| 217028_at | 7852 | CXCR4 | 0.00097 | 1.53 | ||
Only genes with a P value <0.001 in at least one of the comparisons are shown.
Combination of DGF and eGFR Values Identifies Biologically Relevant Subgroups of Patients
The comparison between DGF and noDGF did not yield any robust difference in gene expression profiles. In addition, clinically, the DGF group showed a high degree of heterogeneity regarding severity of kidney injury, reflected by the wide range in recovery time needed (Figure 2). Thus, we further segregated the DGF and the noDGF patient groups each into two subgroups according to the degree of kidney function at 1 month after kidney transplant. These four patient subgroups were as follows: DGFhi, DGF patients with 1-month eGFR >45 mL/min (rapid recovery); DGFlo, DGF patients with 1-month eGFR =45 mL/min (slow recovery); noDGFhi, noDGF patients with 1-month eGFR >45 mL/min (stable eGFR at 1 year); and noDGFlo, noDGF patients with 1-month eGFR =45 mL/min (declining eGFR at 1 year).
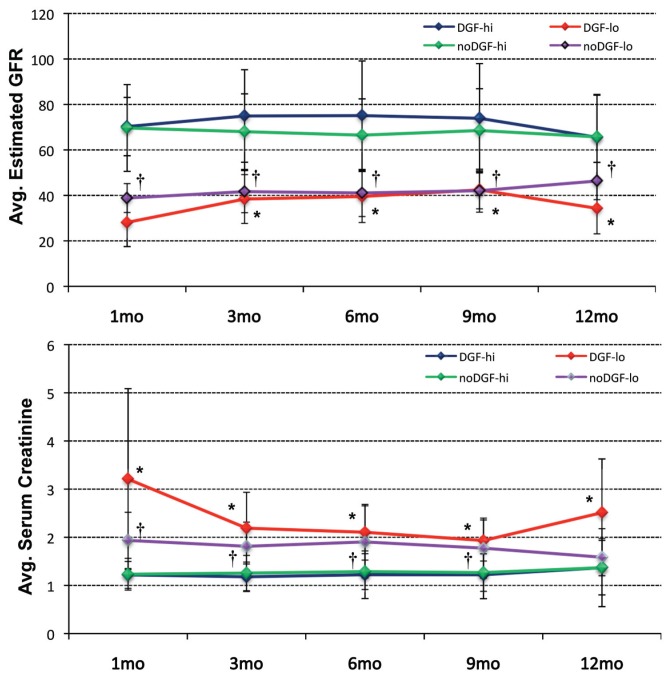
Figure 2.
Course of eGFR (top) and serum creatinine levels (bottom) for the four sub-groups of DGFhi, DGFlo, noDGFhi and noDGFlo during the first year after transplantation. Statistically significant difference (P ≤ 0.05) between the DGF and noDGF ≤45 mL/min group with its corresponding >45 mL/min counterpart are indicated by * (for DGF) or † (for noDGF). Statistical differences were identified by analysis of variance followed by Tukey-Kramer honestly significant difference (HSD) testing (α = 0.05).
Figure 2 shows that each subgroup displays a persistent and stable transplant function for at least the first year; the subgroups within the DGF as well as within the noDGF patients remain statistically different at 3, 6, 9 and 12 months (Table 2). This supports the biological validity of selecting the 1-month eGFR as the stratification variable.
The 1-month function is not associated with any of the clinical donor and recipient variables (Table 2). In addition, 1-month transplant function is independent of the occurrence of DGF. Kidneys can have a high or low eGFR in the noDGF as well as in the DGF group, and longer-term function is not associated with early hemodialysis treatment or not. Function throughout the first year is not statistically different between DGFhi and noDGFhi allografts. DGFlo versus noDGFlo kidneys again show overall no statistical difference during the first 12 posttransplant months; however, the 13 DGFlo allo-graft showed poorer early function recovery and worse 1-year function compared to the 16 noDGFlo transplants (Figure 2 and Table 2).
Gene Expression and Functional Analysis of DGF/GFR Subgroups
We next compared the gene expression profiles of the PI biopsies from the four subgroups to identify any potential differences associated with the observed segregation of samples. Four comparisons were carried out: (i) DGFlo (n = 13) versus DGFhi (n = 12), (ii) noDGFlo (n = 16) versus noDGFhi (n = 50), (iii) DGFhi versus noDGFhi, and (iv) DGFlo versus noDGFlo. Statistical analyses identified 155 probe sets differentially expressed between DGFhi and DGFlo; 79 probe sets between noDGFhi and noDGFlo; 32 probe sets between noDGFhi and DGFhi; and 130 probe sets between noDGFlo and DGFlo (P < 0.01). A total of 27 genes identified as differentially expressed among groups were also present in the GFRhi versus GFRlo gene list. A complete list of the differentially expressed probe sets can be found in Supplementary Table 4.
Functional analyses of genes differentially expressed between DGFhi and DGFlo indicate that these genes are involved in antigen processing and presentation via major histocompatibility complex (MHC) class I/II, regulation of T-cell–mediated cytotoxicity and positive thymic T-cell selection (Supplementary Table 5). Moreover, biological pathways overrepresented by the affected genes included the allograft rejection/graft versus host disease, antigen processing and presentation and cell adhesion molecules. Example genes identified in these categories included PSMB8, FCGR2B, HLA-G, HLA-F, HLA-E, HLA-DRB1, HLA-DRA, HLA-DPB1, HLA-DPA1, HLA-DQB1, HLA-DQA1, HLA-B, HLA-DMA, IL7R, PTPRC, CD3D, CXCL6, CCL19, CCL5 and LAT2 (Table 3). Between noDGFhi and noDGFlo, intracellular transport and protein localization were identified as significant.
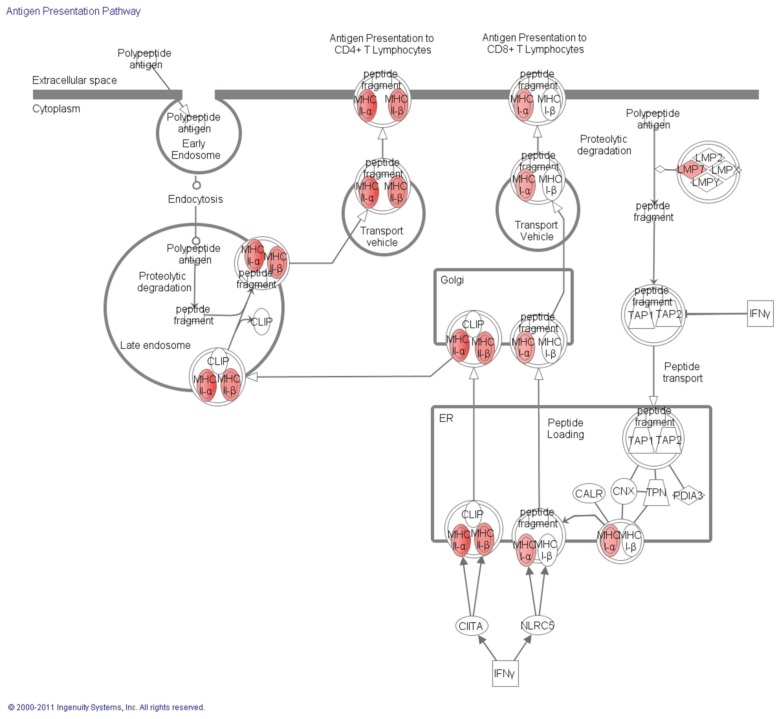
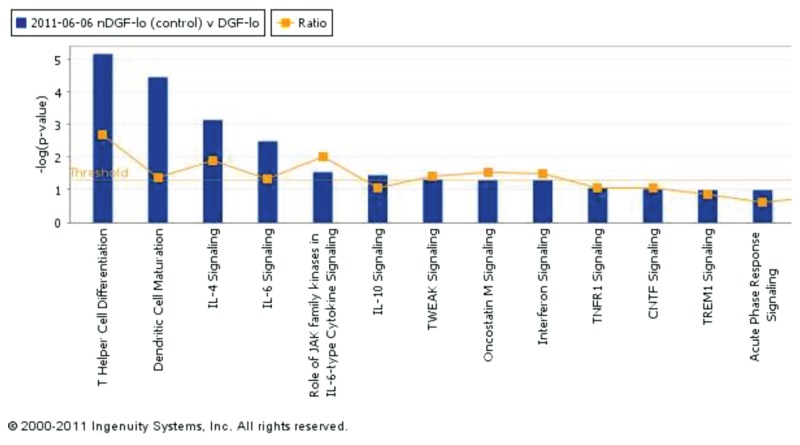
Finally, functional analyses between noDGFlo and DGFlo showed that differentially expressed genes between these groups, similar to the comparison between DGFlo and DGFhi, are involved in antigen processing and presentation via MHC class I/II (Figure 3) and regulation of T-cell–mediated cytotoxicity, whereas biological pathways overrepresented by the affected genes included allograft rejection/graft versus host disease, antigen processing and presentation and cell adhesion molecules. Genes identified included HLA-G, HLA-E, HLA-DRB1, HLA-DRA, HLA-DPB1, HLA-DPA1, HLA-DQB1, HLA-DQA1, HLA-B, HLA-C, HLA-DMA, PSMB8, PSME1, HSP90AB1, and PRDX1 (see Table 3). The top-scoring network of interactions among the probe sets identified as significantly differentially expressed when comparing noDGFlo versus DGFlo is shown in Supplementary Figure 1. Also, from the analysis of the canonical pathways, cytokine signaling involved in this comparison between groups is shown the Figure 4.
Figure 3.
Graphic representation from canonical pathways using ingenuity pathways analysis of the antigen presentation signaling showing (in red) all the differentially expressed genes between noDGFlo versus DGFlo that were overexpressed in noDGFlo. IFNγ, interfon-γ. LMP, proteasome (prosome, macropain) subunit, beta type, 9 (PSMB9); TAP, transporter associated with antigen processing; CLIP, class II–associated invariant chain peptides; CIITA, class II, major histocompatibility complex, transactivator; NLRC, NLR family; CALR, calreticulin; CNX, calnexin; TPN, TAP binding protein; PDIA, protein disulfide isomerase family A.
Figure 4.
Canonical pathways identified from the analysis of the differentially expressed genes between noDGFlo versus DGFlo. Canonical pathways are displayed along the x axis; the y axis displays the –log of P value, which is calculated by the Fisher exact test right-tailed. The orange points represent the ratio of the number of genes in a given pathway that meet cutoff criteria divided by the total number of genes in that pathway. TWEAK, TNF-like weak inducer of apoptosis; TNFR1, tumor necrosis factor receptor superfamily, member 1A; CNTF, ciliary neurotrophic factor; TREM1, triggering receptor expressed on myeloid cells 1.
Microarray results were validated for a number of genes using the remaining stock RNA from samples used in the microarray study. All genes tested showed a similar trend to that observed in the microarray data with a Pearson correlation of 0.928 (Supplementary Figure 2).
Markers Validation Using RT-qPCR
Five genes were also validated in an independent group of patients including DGFhi (n = 15) and DGFlo (n = 20) (validation set). Supplementary Figure 3 shows that the significance of the identified genes was validated in the independent set of patients.
DISCUSSION
To identify robust biomarkers capable of predicting graft outcome, a better understanding of the mechanisms leading to DGF and its impact on long-term function is necessary, in particular as more and more marginal organs are accepted to increase the donor pool (2123). The heterogeneity in the causes of DGF; the multitude of donor, recipient and procedural factors influencing early graft function; and the variety of definitions used for DGF reflect its limitations as a robust clinical end point and the failure to find reliable biomarkers of kidney quality (4,6,7). In the present report, we evaluated in 91 consecutive deceased donor kidney transplants clinical variables and gene expression profiles in relation to the development of DGF as well as transplant function throughout the first year.
The occurrence of DGF was neither associated with any of the tested clinical variables, including ischemia times, pump perfusion, donor and recipient sex, age and race or incidence of rejections, nor with kidney function beyond 3 months after kidney transplant. In addition, despite 206 probe sets differentially expressed in the preimplantation biopsies of kidneys that developed DGF versus those that did not, the functional analyses failed to identify any overrepresented biological pathways from this list of probe sets separating DGF from noDGF. These findings corroborate the clinical experience that the DGF classification is subjective, lacks biological relevance for longer-term outcome and is as such not reflecting organ quality and a poor reference point to identify reliable biomarkers (24).
However, the classifictaion of patients in groups according with the eGFR at 1 month posttransplantation (GFRhi versus GFRlo) showed an important set of genes associated with inflammation and immune response differentially expressed between groups.
To better capture the “quality” of the kidneys and dissect the intrinsic heterogeneity when using the DGF definition, we further subclassified the patient cohort according to 1-month eGFR values. This however identified four subgroups of patients with distinct and persistent clinical 1-year transplant function. Patients in the DGFlo and noDGFlo group remained statistically different from patients in their counterpart group throughout the first year after transplantation. Furthermore, the DGFhi and noDGFhi groups were not statistically different post–kidney transplant, whereas the DGFlo and noDGFlo groups were only statistically different from each other in serum creatinine at 1 month posttransplantation. This analysis shows that grafts with DGF and fast recovery (DGFhi) behave clinically similar, at least for the first year, to grafts from the noDGF group.
These findings suggest that the occurrence of DGF in kidneys with good quality has little or no clinical impact on long-term survival of the graft. This stratification further identified subsets of kidneys that have a poorer posttransplant function (DGFlo and noDGFlo), independent of whether they had experienced DGF or not. The clinical variables available before transplant did not identify these groups of kidneys with better or poorer function during the first post-transplant year. Gene expression profiling obtained from preimplantation biopsies identified the subset of grafts (DGFlo, n = 13) that developed DGF and continued to have a low GFR through-out the first year. Patients in this DGFlo group showed increased expression of a number of HLA and immune-related genes when compared with the other groups. Immune response genes have been shown to be overexpressed with renal ischemia in implantation biopsies (9,11). However, these studies included a mix of living and deceased donor biopsies in their profile analysis. Donor factors have been estimated to account for 35–45% of the variability in early allograft function (25). There is a high likelihood that the molecular mechanisms associated with ischemia-reperfusion injury differ depending on the donor type, as suggested by the independent segregation of the samples in the unsupervised cluster analysis presented in the previously described articles. Moreover, one study identified 132 transcripts including coagulation and complement genes that separated deceased and living donor kidneys (8). In concordance, Mueller et al. (10) identified 3,718 probe sets differentially expressed at an adjusted P value of <0.01 from the comparison between deceased versus living donors kidneys, showing high differences between groups also after reperfusion.
It is intriguing to find immune activation already before implantation in kidneys that will undergo DGF and will maintain a poor 1-year graft function (DGFlo group). The biopsy time point excludes any impact of recipient factors on these transcriptome findings. The major difference between the two low GFR groups is the donor serum creatinine (1.5 versus 0.9 in the DGFlo versus noDGFlo groups, respectively; P = 0.034). Hence, the signature of immune activation might be reflecting increased injury to an older tissue with impaired repair capacity resulting in significantly poorer 1-month (serum creatinine 3.2 versus 1.9 or 1.2 mg/dL in DGFlo versus noDGFlo versus DGFhi, respectively) and 12-month graft function (serum creatinine 2.5 versus 1.6 versus 1.4 mg/dL in DGFlo versus noDGFlo versus DGFhi, respectively).
These findings were further corroborated by an analysis on the basis of the 1-month GFR alone. Kidneys with lower GFR again expressed genes associated with inflammation and immune response, overlapping with the identified genes and pathways associated with poorer outcomes in the analyses of the four subgroups.
Overall, these findings suggest that a subset of allografts may have increased expression of immune-related genes before transplantation. It is possible that early immune activation within the grafts and possible prolonged upregulation of these and other immune-related genes may lead to long-term detrimental effects, an issue that needs to be further investigated.
It remains unclear why there is segregation between patients within the noDGF group. From the analysis of the differentially expressed genes in this group, most of these genes were down-regulated in the noDGFlo group and were found to be involved in intracellular localization and cellular transport. These gene changes may provide a biological explanation to what has been described in the literature as “slow graft function” (26–28). Finally, few differentially expressed probe sets were identified between the DGFhi and noDGFhi sub-groups, indicating that transcriptionally these two groups of patients are very similar.
It is clear from the data presented (see Figure 2) that patients with 1-month eGFR values =45 mL/min have a tendency to remain as such by 1 year post-transplantation and that eGFR may be in fact a more accurate clinical marker of short- and longer-term function than DGF (29,30). Further follow-up of these patients will reveal whether this tendency continues beyond the 1-year posttransplantation mark and whether the graft’s long-term function has been significantly affected by development of DGF or the persistent lower graft function.
In the present analyses, gene expression changes effectively separated the DGFlo group from the other three groups of patients. Upregulated genes differentially expressed in this group included various genes involved in T-cell activation (DDOST, IL7R, CXCR4, ITGB2, PTPRC, HLA-G, HLA-DQA1, CD48 and HLA-DMA) and/or chemotaxis (CXCL6, CCL19, CCL5, CXCR4, ITGB2, RAC2 and VCAN).
The gene expression signals identified in this subset of patients (DGFlo) suggest the presence of active donor antigen-presenting cells with the potential of attracting and activating recipient T cells. Additionally, some of the gene signals identified may also suggest the presence of activated T cells already present on the graft at preimplantation. These kidney grafts may represent a set of real “poor-quality” grafts, reflected in the poorer medium-term graft function observed at 12 months after transplant. If such is the case, then these donor organs would require early identification, ideally before implantation, to identify kidney transplant recipients at risk of lower long-term allograft function. Further investigation of these donor organs may eventually help improve organ allocation and posttransplant management.
Kainz et al. (31) recently showed in a randomized control trial that steroid pretreatment of the deceased organ donor suppressed inflammation in the transplant organ but did not reduce the rate or duration of DGF. This study sought to elucidate those factors that caused DGF in the steroid-treated subjects. Genome-wide gene expression profiles were used from 20 steroid-pretreated donor organs and were analyzed on the level of regulatory protein–protein interaction networks (32). A total of 63 significantly down-regulated sequences associated with DGF that could be functionally categorized according to Protein Analysis Through Evolutionary Relationships ontologies into two main biological processes (transport and metabolism) were identified. The recognized genes suggested hypoxia as the cause of DGF, which cannot be compensated by steroid treatment.
In summary, organ quality and clinical performance does not appear to be accurately captured by the DGF/noDGF classification. Stratification of patients on the basis of a combination of 1-month eGFR and DGF classification appears to have more predictive value and be more relevant to medium-term (12-month) graft performance than DGF classification alone. Comparative GE profiling of preimplantation biopsies using this classification has identified a subgroup of DGF kidney transplant recipients with elevated expression of immune-related genes. A better understanding of molecular pathways affected early on in the graft (that is, at preimplantation) may signal high risk for graft dysfunction, giving the chance for potential therapeutic approaches to be taken.
Supplemental Data
ACKNOWLEDGMENTS
The research results included in this report were supported by a National Institute of Diabetes and Digestive and Kidney Diseases grant (R01DK080074).
VR Mas, RF Mueller and DG Maluf participated in the research design. MJ Scian, JL Suh, KG David, AL King and TWB Gehr performed research. VR Mas, MJ Scian, KJ Archer and TF Mueller performed data analysis. VR Mas, MJ Scian, KJ Archer, MP Posner, TF Mueller and DG Maluf prepared the manuscript.
Footnotes
Online address: http://www.molmed.org
DISCLOSURE
The authors declare that they have no competing interests as defined by Molecular Medicine, or other interests that might be perceived to influence the results and discussion reported in this paper.
REFERENCES
- 1.Veroux M, Corona D, Veroux P. Kidney transplantation: future challenges. Minerva Chir. 2009;64:75–100. [PubMed] [Google Scholar]
- 2.Knoll G. Trends in kidney transplantation over the past decade. Drugs. 2008;68(Suppl 1):3–10. doi: 10.2165/00003495-200868001-00002. [DOI] [PubMed] [Google Scholar]
- 3.Schold JD, Kaplan B. The elephant in the room: failings of current clinical endpoints in kidney transplantation. Am J Transplant. 2010;10:1163–6. doi: 10.1111/j.1600-6143.2010.03104.x. [DOI] [PubMed] [Google Scholar]
- 4.Mueller TF, Solez K, Mas V. Assessment of kidney organ quality and prediction of outcome at time of transplantation. Semin Immunopathol. 2011;33:185–99. doi: 10.1007/s00281-011-0248-x. [DOI] [PubMed] [Google Scholar]
- 5.Mas VR, Mueller TF, Archer KJ, Maluf DG. Identifying biomarkers as diagnostic tools in kidney transplantation. Expert Rev Mol Diagn. 2011;11:183–96. doi: 10.1586/erm.10.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yarlagadda SG, et al. Marked variation in the definition and diagnosis of delayed graft function: a systematic review. Nephrol Dial Transplant. 2008;23:2995–3003. doi: 10.1093/ndt/gfn158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moore J, et al. Assessing and comparing rival definitions of delayed renal allograft function for predicting subsequent graft failure. Transplantation. 2010;90:1113–6. doi: 10.1097/TP.0b013e3181f86966. [DOI] [PubMed] [Google Scholar]
- 8.Hauser P, et al. Genome-wide gene-expression patterns of donor kidney biopsies distinguish primary allograft function. Lab Invest. 2004;84:353–61. doi: 10.1038/labinvest.3700037. [DOI] [PubMed] [Google Scholar]
- 9.Kainz A, et al. Alterations in gene expression in cadaveric vs. live donor kidneys suggest impaired tubular counterbalance of oxidative stress at implantation. Am J Transplant. 2004;4:1595–6004. doi: 10.1111/j.1600-6143.2004.00554.x. [DOI] [PubMed] [Google Scholar]
- 10.Mueller TF, et al. The transcriptome of the implant biopsy identifies donor kidneys at increased risk of delayed graft function. Am J Transplant. 2008;8:78–85. doi: 10.1111/j.1600-6143.2007.02032.x. [DOI] [PubMed] [Google Scholar]
- 11.Melk A, et al. Transcriptional analysis of the molecular basis of human kidney aging using cDNA microarray profiling. Kidney Int. 2005;68:2667–79. doi: 10.1111/j.1523-1755.2005.00738.x. [DOI] [PubMed] [Google Scholar]
- 12.Mas VR, et al. Gene expression patterns in deceased donor kidneys developing delayed graft function after kidney transplantation. Transplantation. 2008;85:626–35. doi: 10.1097/TP.0b013e318165491f. [DOI] [PubMed] [Google Scholar]
- 13.Levey AS, et al. A more accurate method to estimate glomerular filtration rate from serum creatinine: a new prediction equation. Modification of Diet in Renal Disease Study Group. Ann Intern Med. 1999;130:461–70. doi: 10.7326/0003-4819-130-6-199903160-00002. [DOI] [PubMed] [Google Scholar]
- 14.Kainz A, et al. Gene-expression profiles and age of donor kidney biopsies obtained before transplantation distinguish medium term graft function. Transplantation. 2007;83:1048–54. doi: 10.1097/01.tp.0000259960.56786.ec. [DOI] [PubMed] [Google Scholar]
- 15.Archer KJ, Dumur CI, Joel SE, Ramakrishnan V. Assessing quality of hybridized RNA in Affymetrix GeneChip experiments using mixed-effects models. Biostatistics. 2006;7:198–212. doi: 10.1093/biostatistics/kxj001. [DOI] [PubMed] [Google Scholar]
- 16.Archer KJ, Guennel T. An application for assessing quality of RNA hybridized to Affymetrix GeneChips. Bioinformatics. 2006;22:2699–701. doi: 10.1093/bioinformatics/btl459. [DOI] [PubMed] [Google Scholar]
- 17.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.R Development Core Team R. A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: p. 2007. [Google Scholar]
- 19.Collini A, et al. Long-term outcome of renal transplantation from marginal donors. Transplant Proc. 2006;38:3398–99. doi: 10.1016/j.transproceed.2006.10.055. [DOI] [PubMed] [Google Scholar]
- 20.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37(Web Server issue):W305. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fraser SM, et al. Acceptable outcome after kidney transplantation using “expanded criteria donor” grafts. Transplantation. 2010;89:88–96. doi: 10.1097/TP.0b013e3181c343a5. [DOI] [PubMed] [Google Scholar]
- 22.Ciancio G, et al. Favorable outcomes with machine perfusion and longer pump times in kidney transplantation: a single-center, observational study. Transplantation. 2010;90:882–90. doi: 10.1097/TP.0b013e3181f2c962. [DOI] [PubMed] [Google Scholar]
- 23.Mühlberger I, Perco P, Fechete R, Mayer B, Oberbauer R. Biomarkers in renal transplantation ischemia reperfusion injury. Transplantation. 2009;88(Suppl 3):S14–9. doi: 10.1097/TP.0b013e3181af65b5. [DOI] [PubMed] [Google Scholar]
- 24.Yarlagadda SG, Klein CL, Jani A. Long-term renal outcomes after delayed graft function. Adv Chronic Kidney Dis. 2008;15:248–56. doi: 10.1053/j.ackd.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 25.Tyson M, et al. Early graft function after laparoscopically procured living donor kidney transplantation. J Urol. 2010;184:1434–9. doi: 10.1016/j.juro.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 26.Suri D, Meyer TW. Influence of donor factors on early function of graft kidneys. J Am Soc Nephrol. 1999;10:1317–23. doi: 10.1681/ASN.V1061317. [DOI] [PubMed] [Google Scholar]
- 27.Ciancio G, et al. Favorable outcomes with machine perfusion and longer pump times in kidney transplantation: a single-center, observational study. Transplantation. 2010;90:882–90. doi: 10.1097/TP.0b013e3181f2c962. [DOI] [PubMed] [Google Scholar]
- 28.Tyson M, et al. Early graft function after laparoscopically procured living donor kidney transplantation. J Urol. 2010;184:1434–9. doi: 10.1016/j.juro.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 29.Hawley CM, et al. Estimated donor glomerular filtration rate is the most important donor characteristic predicting graft function in recipients of kidneys from live donors. Transpl Int. 2007;20:64–72. doi: 10.1111/j.1432-2277.2006.00400.x. [DOI] [PubMed] [Google Scholar]
- 30.Johnston O, et al. Reduced graft function (with or without dialysis) vs. immediate graft function: a comparison of long-term renal allograft survival. Nephrol Dial Transplant. 2006;21:2270–4. doi: 10.1093/ndt/gfl103. [DOI] [PubMed] [Google Scholar]
- 31.Kainz A, et al. Steroid pretreatment of organ donors to prevent postischemic renal allograft failure: a randomized, controlled trial. Ann Intern Med. 2010;153:222–30. doi: 10.7326/0003-4819-153-4-201008170-00003. [DOI] [PubMed] [Google Scholar]
- 32.Wilflingseder J, et al. Impaired metabolism in donor kidney grafts after steroid pretreatment. Transpl Int. 2010;23:796–804. doi: 10.1111/j.1432-2277.2010.01053.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.