Abstract
Synthetic triterpenoids are multitarget compounds exhibiting promise as preventative and therapeutic agents for cancer. Their proposed mechanism of action is by forming Michael adducts with reactive nucleophilic groups on target proteins. Our previous work demonstrates that the 2-cyano-3,12-dioxooleana-1,9-dien-28-oic acid (CDDO) and its derivatives promote B-lymphoid cell apoptosis through a mitochondria-mediated pathway linked to mitochondrial protein aggregation. As one function of the Lon protease is to eliminate abnormal mitochondrial proteins, we hypothesized that CDDO-induced protein aggregation and lymphoma apoptosis occur by inactivating this enzyme. Here, we show that CDDO and its derivatives directly and selectively inhibit Lon. CDDO blocks Lon-mediated proteolysis in biochemical and cellular assays, but does not inhibit the 20S proteasome. Furthermore, a biotinylated-CDDO conjugate modifies mitochondrial Lon. A striking common phenotype of CDDO-treated lymphoma cells and Lon-knockdown cells is the accumulation of electron-dense aggregates within mitochondria. We also show that Lon protein levels are substantially elevated in malignant lymphoma cells, compared with resting or activated B cells. Finally, we demonstrate that Lon knockdown leads to lymphoma cell death. Together, these findings suggest that Lon inhibition plays a contributory role in CDDO-induced lymphoma cell death, and support the concept that mitochondrial Lon is a novel anticancer drug target.
Introduction
The malignant transformation of normal cells into cancer cells is driven principally by enhanced oncogenic protein function and/or inactivation of tumor suppressors. To promote this transformation process, tumor cells undergo an extensive reprogramming of normal growth and survival pathways that are mediated by nononcogenic proteins. The identification of nononcogenic proteins that are essential for the survival and proliferation of cancer cells provides potential new drug targets for anticancer therapeutics. Nononcogenic proteins participating in the cell stress response have emerged as a unique and important class of viable targets. Recent work demonstrates that pharmacologic inhibition or down-regulation of the master transcriptional regulator of the cell stress response-heat shock factor 1 (HSF1),1 as well as of the molecular chaperones HSP70 or HSP90, selectively inhibit tumor development.2,3 Remarkably, the inhibition or down-regulation of these essential heat-shock response proteins effectively limits cancer cell growth without substantially compromising normal cell survival.1,2,4
Luo and colleagues6 have expanded on the classic hallmarks of cancer originally proposed by Hanahan and Weinberg5 to include several common stress phenotypes of tumorigenesis. The neoplastic transformation of cancer cells gives rise to diverse oncogenic stressors such as DNA damage and mitotic stress, as well as to metabolic, proteotoxic, and oxidative stress. Cancer cells thus depend on conserved defense mechanisms to survive such oncogenic stresses, and they rely on these antistress systems to a greater extent than normal cells, which are not chronically subjected to such elevated stress. It is proposed that the dependency of tumor cells on stress-response pathways can be exploited therapeutically, either by augmenting oncogenic stress (“stress overload”) or by blocking stress response systems, thereby increasing tumor sensitivity to stress (“stress sensitization”).6
The ATP-dependent Lon protease is highly conserved from bacteria (La protease) to mammalian mitochondria and peroxisomes, and operates in protein quality control and stress response pathways by selectively degrading misfolded, misassembled, or damaged proteins.7,8 Mitochondrial Lon supports cell viability during hypoxic, proteotoxic, and endoplasmic reticulum (ER) stress,9–11 which are common stress phenotypes of cancer cells. During hypoxia, Lon is up-regulated by the hypoxia inducible factor-1α (HIF1-α).10 Lon participates in remodeling the cytochrome c oxidase holoenzyme (COX) by degrading the Cox4-1 subunit, thereby permitting the assembly of an alternate subunit Cox4-2, which confers enzyme activity optimized for low oxygen, thus adapting cancer cells to a hypoxic environment.10 Lon is also up-regulated by the ER stress response, which is activated by misfolded proteins in the ER lumen.11 In contrast to normal cells, many tumor cells exhibit higher levels of ER stress associated with chromosome instability, genotoxic, and hypoxic stress. One potential reason why unfolded proteins in the ER up-regulate the Lon protease in mitochondria is that the ER stress response signals a block in cytosolic protein synthesis,12 thereby decreasing protein import into mitochondria, leading to the stoichiometric imbalance of proteins that promotes misfolding, misassembly, and aggregation. Lon-mediated degradation probably functions to relieve the load of abnormal mitochondrial proteins caused by ER stress. In addition, Lon has been shown to specifically degrade mildly oxidized proteins within mitochondria.13–15 Thus, the up-regulation of Lon may be critical for cancer cell survival by preventing mitochondrial proteotoxicity elicited by oxidative, hypoxic, and ER stress.
The synthetic oleanane triterpenoids (SOs) such as 2-cyano-3, 12-dioxooleana-1,9-dien-28-oic acid (CDDO), are multifunctional electrophilic agents that in a dose-dependent manner are either antitumorigenic, anti-inflammatory, or cytoprotective.16 The proposed mechanism underlying the anticancer effects of SOs is by the formation of Michael adducts between SOs and reactive nucleophiles, such as free thiols on target proteins.17–20 The wide spectrum of SO effects is explained, in part, by their concentration-dependent induction of oxidative stress, which alters various redox-sensitive proteins and signaling networks. SOs show unique promise in the prevention and treatment of cancer,16 and may also act synergistically with other anticancer agents to induce stress overload and/or stress sensitization. Destabilization of mitochondria is among the diverse effects of SO. Specifically, the methyl ester derivative of CDDO (CDDO-Me), at cytotoxic doses, leads to depletion of mitochondrial glutathione, increased production of reactive oxygen species (ROS) and permeabilization of the mitochondrial inner membrane.21–24 In addition, our previous work shows that CDDO and its derivatives mediate apoptosis in lymphoma cells through a novel mitochondria-mediated mechanism.25 CDDO leads to mitochondrial protein thiol modification and the generation of mitochondrial protein aggregates. These aggregates are associated with opening of the mitochondrial permeability transition pore and the production of mitochondrial superoxide, which initiates apoptosis. Based on these findings, we hypothesized that one possible mechanism of CDDO-induced mitochondrial protein aggregation and lymphoma cell death is through inhibition of the mitochondrial Lon protease.
In this study, we show that mitochondrial Lon is a cellular target of CDDO and its derivatives, which are compounds that we and others have previously demonstrated are cytotoxic for human lymphoma cells.24–28 We show that Lon is substantially up-regulated in human malignant B-cell lymphoma patient samples and cell lines compared with resting or activated normal donor peripheral blood B cells. We also show that CDDO and its derivatives directly and selectively inhibit Lon-mediated proteolysis. Although these compounds block mitochondrial Lon, they exhibit little inhibitory effect on the 20S proteasome. Furthermore, our results suggest that the underlying mechanism of Lon inhibition is by the formation of adducts between CDDO and Lon. We also show that Granta mantle cell lymphoma (MCL) cells exhibit electron-dense inclusion bodies specifically in mitochondria but not in other cellular compartments when exposed to doses of CDDO that lead to Lon-CDDO adducts and lymphoma cell death. This same phenotype is observed in a colon carcinoma cell line knocked-down for Lon. Lastly, we show that genetically knocking down Lon protein expression in Granta cells results in cell death. In light of our previous findings that CDDO-induced apoptosis of lymphoma cells promotes mitochondrial protein aggregation,25 we propose that Lon inhibition contributes to CDDO-induced lymphoma cell death, and that mitochondrial Lon is a novel potential target for anticancer therapeutics.
Methods
Cell lines and culture conditions
Lymphoma cell lines and patient samples were cultured and processed as described in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). All samples were obtained under a University of Rochester institutional review board–approved protocol. The doxycycline (DOX)–inducible Lon-knockdown cell line (LonRNAi) and the parental cell line LS174T, have been previously described.29 HeLa ρ0 cells were generated according to published procedures.30
Reagents and antibodies
CDDO and CDDO-Me were kindly provided by Dr Michael Sporn (Dartmouth University, NH). Custom-made antibodies recognizing Lon were previously described.29,31 Commercially obtained antibodies were to BiP (BD Biosciences), ClpP (Abcam), p53 (Calbiochem), TFAM (Aviva), actin and his-tag (Santa Cruz Biotechnology), caspase-3 and -8 (BD Transduction Labs).
Peptidase activity assays
Human Lon was purified as previously described,32 with some modifications (supplemental Figure 1). The 20S proteasome was obtained commercially (Boston Biochem). The peptidase activities of Lon and the 20S proteasome were performed in quadruplicate (20 μL) in 384-well plates. Purified Lon (800nM monomer), 20S proteasome (4nM complex), and no enzyme controls were preincubated for 1 hour at 37°C in the absence or presence of CDDO in reaction buffer (150mM NaCl, 50mM Hepes (N-2-hydroxyethylpiperazine-N'-2-ethanesulfonic acid) pH 8.0, 10mM MgCl2) with DMSO controlled to 3.4%. Peptidase reactions were initiated with the addition of the fluorescent dipeptide substrate rhodamine 110, bis-(CBZ-L-alanyl-L-alanine amide; AA2-Rh110, Anaspec; 6μM) and ATP (2mM), and incubated for 3 hours at 37°C after which fluorescence was measured at excitation/emission = 485/535 nm using a Perkin Elmer Victor3 V. The relative fluorescence units (RFUs) of the background (no enzyme control) measurements were subtracted, and the resultant values were normalized to percent activity of the no drug reactions. Data were fit to 4-parameter dose-response curves using GraphPad Prism 5 software, and the error bars represent the standard deviation (SD) of 4 replicate reactions. Three identical experiments were performed, and representative figures are shown.
Reactions containing Lon and the native mitochondrial protein substrate TFAM were carried out as previously described.33 Briefly, the no enzyme control and purified Lon (300nM monomer) were preincubated for 1 hour at 37°C with various concentrations of CDDO in reaction buffer with DMSO controlled to 2%. After the additions of ATP (2mM) and his-tagged TFAM substrate (60nM), reactions were incubated for 1 hour at 37°C and analyzed by immunoblotting.
CDDO adduct formation
Cultured Granta cells (5 × 106) were treated with 2.5 μM biotinylated or nonbiotinylated-CDDO at 37°C for 1 hour22; cell extracts were prepared and then incubated with streptavidin (SA)–sepharose at 4°C for 90 minutes. SA-sepharose was pelleted and washed twice with phosphate-buffered saline (PBS). The CDDO-biotin interacting proteins pulled down by SA-sepharose were analyzed by immunoblotting. For purified Lon and cell extracts SA-sepharose was reacted with either biotinylated or nonbiotinylated-CDDO (2.5μM) in PBS containing 1% NP40 at room temperature for 90 minutes. SA-sepharose was washed twice with PBS then incubated with purified Lon (2 μg), or cell extracts (20 μg) at 4°C for 90 minutes. The CDDO-biotin interacting proteins pulled down by SA-sepharose were washed extensively with PBS, resuspended in reducing sample buffer and analyzed by immunoblotting.
Electron microscopy
Granta cells (1 × 106/mL) were cultured in the presence or absence of CDDO (2.5μM) at 37°C for 24 hours. LonRNAi and LS174T cells were cultured with or without DOX (50 ng/mL) for 2.5 weeks, to knockdown Lon or as a control, respectively. Granta cells grown in suspension, and LonRNAi or LS174T cells grown on slides, were fixed at 4°C overnight, in 2.5% glutaraldehyde/4.0% paraformaldehyde in 0.1 M phosphate buffer. The samples were processed as described in supplemental Methods, and examined using a Hitachi 7650 transmission electron microscope with an attached Gatan Erlangshen 11 megapixel digital camera.
Lentiviral transduction
On the day of transduction, Granta 519 cells (2 × 105) were seeded in 2 mL of complete RPMI medium (10% FBS, 10mM Hepes, pH7.4, 200mM Glutamax 100mM pyruvate, and 10 000 U/mL penicillin-streptomycin) in a 12-well plate and incubated for 30 minutes at 37°C at 5% CO2. Polybrene (8 μg/mL) and the respective lentiviral particles (1 × 106, multiplicity of infection [MOI] of 5) for the expression of control or Lon shRNA, or copGFP (Santa Cruz Biotechnology) were added to the cells and gently mixed. The plate was centrifuged at 1000g for 90 minutes at room temperature. One milliliter of medium was replaced with 2 mL of fresh complete RPMI with minimal disruption of the cells. The plate was incubated at 37°C. At the various time points, the cells were counted to determine the number of viable cells (trypan blue excluding), or dead and sick cells (trypan blue permeant). Fresh medium (2 mL) was replaced every 48 hours. At the indicated time points, cells were harvested and processed for immunoblotting.
Results
Lon protein levels are up-regulated in human B-cell lymphoma cells
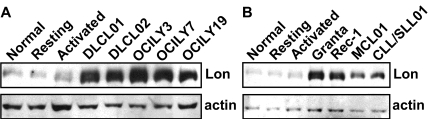
Based on our previous work, which shows that CDDO and its derivatives induce apoptosis in lymphoma cells by a mechanism linked to mitochondrial protein aggregation, we speculated that these compounds target the mitochondrial Lon protease. To confirm that Lon is indeed expressed in lymphoma cells, we examined its protein levels in patient samples and lymphoma cell lines, as well as in normal peripheral blood B cells. Figure 1 shows that Lon was consistently up-regulated in both patient biopsy specimens and lymphoma cell lines compared with resting or activated normal peripheral blood B cells. A substantial increase in Lon was observed in malignant diffuse large cell lymphoma (DLCL) cells from a patient splenectomy specimen (DLCL01) and DLCL cells from a malignant pleural effusion (DLCL02; Figure 1A) as well as in the DLCL cell lines OCI Ly-3, -7, and -19 (Figure 1A). Lon was similarly up-regulated in Granta and Rec-1 MCL cell lines, MCL cells from a patient lymph node biopsy (MCL01) and in chronic lymphocytic leukemia (CLL)/small lymphocytic lymphoma (SLL) cells from a patient lymph node biopsy (CLL/SLL01; Figure 1B). By contrast, Lon expression was markedly lower in freshly isolated normal donor peripheral blood B cells that were positively selected using CD19+ magnetic beads (normal), as well as in normal B cells that had been cultured for 72 hours with activation of the toll-like receptor 9 (TLR9) using CpG oligonucleotides, and the B-cell receptor using anti-IgM antibodies (activated), or left untreated (resting; Figure 1A-B). These data show that Lon, as a potential target of CDDO, is significantly up-regulated in lymphoma patient samples and lymphoma cell lines, compared with normal resting or activated peripheral blood B cells.
Figure 1.
Lon is up-regulated in patient biopsy specimens and lymphoma cell lines compared with resting or activated normal peripheral blood B cells. Protein extracts (20 μg) obtained from cells as described in “Methods” and supplemental Methods, were immunoblotted with antibodies to Lon or actin. (A-B) Extracts from freshly isolated normal donor peripheral blood B cells that were positively selected using CD19+ magnetic beads (normal), as well as in normal B cells that had been cultured for 72 hours with activation of the TLR9 using CpG oligonucleotides, and the B-cell receptor using anti-IgM (activated), or untreated (resting). (A) Extracts from malignant DLCL cells from a patient splenectomy specimen (DLCL01) and DLCL cells from a malignant pleural effusion (DLCL-02), as well as in the diffuse large cell lymphoma (DLCL) cell lines OCI Ly-3, -7 and -19. (B) Extracts from Granta and Rec-1 MCL cell lines, MCL cells from a patient lymph node biopsy (MCL01) and in chronic lymphocytic leukemia (CLL)/small lymphocytic lymphoma (SLL) cells from a patient lymph node biopsy (CLL/SLL01).
CDDO and its derivatives inhibit the protease activity of purified Lon but not the 20S proteasome
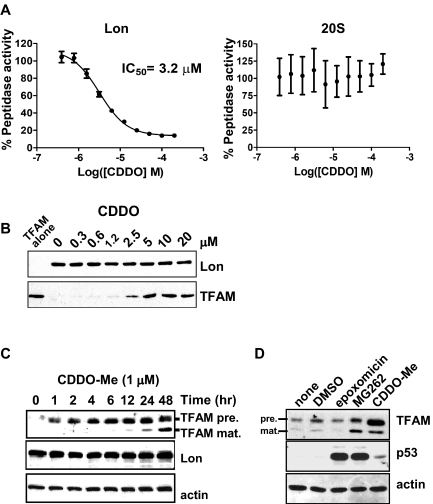
To determine whether CDDO directly inhibits Lon, degradation assays were performed using purified protease and peptide or protein substrates. We used a fluorescently labeled reporter substrate (Z-Ala-Ala)2-Rh110 (AA2-Rh110) consisting of nonnucleophilic alanine dipeptides flanking rhodamine 110, which cannot form adducts with CDDO. Lon-dependent cleavage of AA2-Rh110 resulted in increased relative fluorescence. However, when Lon was preincubated with CDDO before the addition of the peptide reporter, its peptidase activity was inhibited in a dose-dependent manner (Figure 2A). CDDO inhibited Lon with an IC50 of 3.2μM, with a 95% confidence interval between 2.8 to 3.7μM. As CDDO was apparently a slow binding inhibitor, its preincubation with Lon was required to measure effective inhibition. The dimethylsulfoxide (DMSO) vehicle had no inhibitory effect on Lon activity (supplemental Figure 2). Results showed that CDDO as well as its imidazole or methyl ester derivatives CDDO-Im and CDDO-Me, inhibited Lon in a dose-dependent manner with the derivatives showing greater inhibition than CDDO (S.H.B. and C.K.S, unpublished observations, July 21, 2006).
Figure 2.
CDDO inhibits purified Lon but not the purified 20S proteasome. (A) Human Lon (800nM monomer) or the 20S proteasome (4nM) were preincubated for 1 hour at 37°C in the presence or absence of CDDO at the indicated concentrations, after which ATP (2 mM) and the fluorogenic dipeptide, AA2-Rh110 (6μM) were added and incubated for 3 hours at 37°C before fluorescence values were measured. (B) Purified Lon (300nM monomer) was pre-incubated in the presence or absence of CDDO at the indicated concentrations for 1 hour at 37°C, after which ATP (2mM) and TFAM (60nM) were added for 1 hour at 37°C. Samples were immunoblotted for TFAM and Lon. (C-D) Cell extracts (20 μg) from HeLa ρ0 cells treated with or without CDDO-Me (1μM; C); or, with or without epoxomicin (5μM), MG262 (5μM), CDDO-Me (1μM) or DMSO vehicle for 24 hours (D). Samples were immunoblotted with antibodies recognizing TFAM, Lon, actin, or p53.
The selectivity of CDDO was examined using the cytosolic 20S proteasome, which is the target of the drug bortezomib used clinically to treat patients with MCL and multiple myeloma.34 In the absence of CDDO, the purified proteolytic 20S component of the proteasome efficiently cleaved the AA2-Rh110 reporter peptide. However, CDDO had no inhibitory effect on the 20S proteasome even at the highest concentration tested (200μM; Figure 2A). By contrast, the 20S-mediated cleavage of AA2-Rh110 was efficiently blocked by the established proteasome inhibitor MG262 with an IC50 of 0.7-1.0 nM (supplemental Figure 3).
We also tested the effect of CDDO on Lon-mediated degradation of a purified endogenous mitochondrial protein substrate. Mitochondrial transcription factor A (TFAM) is a substrate of Drosophila,35 as well as human mitochondrial Lon (B.L., J.L., X. Nie, M.L., Y. I. Morozov, D. F. Bogenhagen, D. Temiakov, and C.K.S., manuscript submitted). Purified Lon rapidly degraded purified TFAM with a half-life less than 30 minutes (supplemental Figure 4). Lon-mediated degradation of TFAM was completely blocked by CDDO at 2.5μM (Figure 2B). Taken together, these results demonstrate that CDDO directly and selectively inhibits Lon-mediated proteolysis of both heterologous and native protein substrates.
Selective in situ inhibition of Lon but not the proteasome by CDDO and its derivatives
To determine the in situ effects of CDDO-Me on mitochondrial Lon and the 26S proteasome, we examined the expression levels of their respective protein substrates in HeLa cells. We have demonstrated that Lon degrades TFAM when it is not bound to mitochondrial DNA (mtDNA), either because mtDNA is absent from cells,36,37 or, because DNA-binding by TFAM is impaired (B.L., J.L., X. Nie, M.L., Y. I. Morozov, D. F. Bogenhagen, D. Temiakov, and C.K.S., manuscript submitted). In HeLa cells depleted of mitochondrial DNA (HeLa ρ0 cells), the steady state levels of TFAM were barely detectable (Figure 2C, time = 0 hours). However, incubation with CDDO-Me (1μM) during a 48-hour time course led to increased levels of both the precursor and mature forms of TFAM, whereas no change in Lon expression was observed during this time course (Figure 2C). Thus, the increased protein levels of TFAM resulted from CDDO-dependent inhibition of Lon and not from decreased Lon expression.
The effect of CDDO-Me on the 26S proteasome was also assessed in HeLa ρ0 cells by examining the protein levels of p53. HeLa are cervical carcinoma cells that express the human papillomavirus (HPV) E6 protein, which promotes the rapid degradation of p53 by the 26S proteasome. In extracts from untreated HeLa ρ0 cells, p53 was barely detectable by immunoblotting. On treatment with the proteasome inhibitors epoxomicin or MG262 (5μM) for 24 hours, p53 levels were substantially increased (Figure 2D). By contrast, p53 was only minimally increased on treatment with CDDO-Me (1μM) for 24 hours. However, TFAM levels were up-regulated in the same extracts from CDDO-Me–treated cells, to a level consistent with that observed in the time course (Figure 2C). In addition, our previous work and that of others has shown that Lon is selectively inhibited by the proteasome inhibitors MG262 or MG132, but not epoxomicin.38,39 As expected, MG262 (5μM, Figure 2D) and MG132 (5μM, data not shown) led to increased TFAM, whereas epoxomicin did not (5μM, Figure 2D). Notably, CDDO-Me was as effective as MG262 in stabilizing TFAM. Collectively, these results show that CDDO-Me selectively blocks Lon-mediated proteolysis both in vitro and in situ, and has little effect on the 26S proteasome.
CDDO interacts directly with purified and cellular Lon
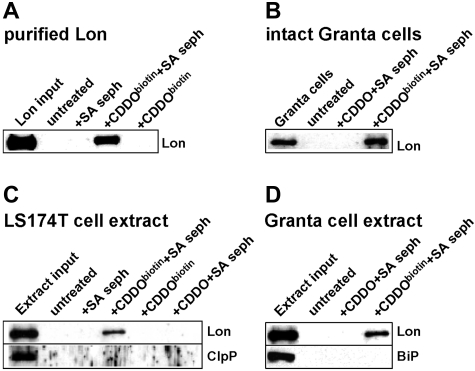
The proposed mechanism by which CDDO and its derivatives inactivate target proteins is by the formation of Michael adducts with reactive nucleophilic groups such as free thiols. To test whether CDDO forms adducts with Lon, we used a biotinylated conjugate of CDDO (CDDObiotin), which has comparable activity to its parent compound albeit with reduced potency,26,40 and which colocalizes with mitochondria.22 CDDO or CDDObiotin were first reacted with SA-sepharose, and then combined with purified Lon. The SA-sepharose was pulled down, washed extensively and then immunoblotted for Lon. Results show that Lon is specifically pulled down only in complexes containing CDDObiotin and SA-sepharose, thereby demonstrating a direct interaction between CDDO and the Lon protease (Figure 3A).
Figure 3.
CDDO directly interacts with purified Lon and forms a complex with Lon in cultured cells. (A) CDDObiotin (2.5μM) was reacted with purified Lon (2 μγ) in PBS containing 1% NP40 for 90 minutes at room temperature, after which CDDObiotin was pulled down with SA-sepharose (SA-seph). (B) Granta cells were treated with or without CDDO or CDDObiotin for 1 hour at 37°C, followed by extraction of cellular proteins and incubation with SA-seph as indicated for 90 minutes at 4°C; (C-D) LS174T or Granta cells extracts (20 μg), respectively, were reacted with or without CDDO or CDDObiotin for 90 minutes at 4°C, followed by SA-seph as in panel B. Pull-down reactions were immunoblotted with antibodies to Lon, ClpP, or BiP. Untreated mock samples contained input but without CDDObiotin and SA-seph.
To determine whether CDDO binds Lon in situ, Granta cells were incubated with or without CDDO or CDDObiotin (2.5μM) for 1 hour, and then washed extensively to remove CDDObiotin in the culture medium. The cells were lysed, incubated with SA-sepharose to pull down biotin-containing proteins and the biotin-streptavidin complexes were immunoblotted for Lon (Figure 3B). Lon was pulled down specifically from cells treated with CDDO-biotin but not unbiotinylated CDDO. Similar experiments were performed using Granta and LS174T cell extracts that were incubated with or without CDDO or CDDObiotin. We observed that CDDObiotin pulls down Lon, but not the ER stress protein BiP, nor ClpP, another ATP-dependent protease within the mitochondrial matrix (Figure 3C-D). Together, these results demonstrate that CDDO interacts directly and specifically with mitochondrial Lon.
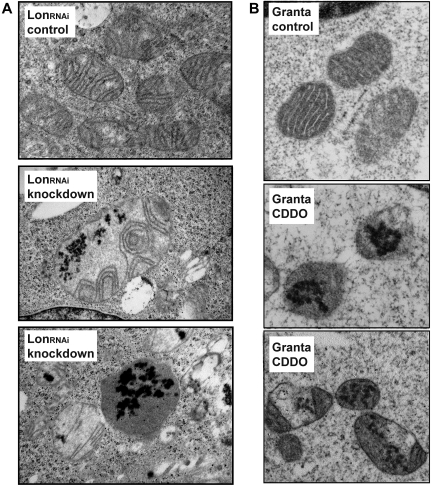
Mitochondrial inclusion bodies are a common phenotype of Lon-knockdown cells and CDDO-treated lymphoma cells
Our published work demonstrates that a Lon knockout in the budding yeast Saccharomyces cerevisiae leads to loss of ATP-dependent proteolysis within the mitochondrial matrix resulting in the accumulation of mitochondrial aggregates as shown by electron microscopy (EM).41 A similar morphologic defect was observed in the human LS174T colon carcinoma cell line LonRNAi carrying a stably integrated DOX-inducible short hairpin RNA (shRNA) leading to the knockdown of Lon29 (Figure 4A). LonRNAi knockdown for 2.5 weeks with DOX led to a substantial depletion of Lon (supplemental Figure 5). EM analysis of LonRNAi knockdown cells showed extensive electron dense aggregates within mitochondria (Figure 4A), but not in other cellular compartments (compare supplemental Figure 6A-B). No inclusions were observed in control cells cultured in the absence of DOX (Figure 4A), or in cells carrying a control shRNA cultured with DOX (data not shown). In light of these findings, we predicted that CDDO-mediated inhibition of Lon in lymphoma cells would result in a morphologic defect similar to the LonRNAi knockdown. To test this prediction, EM analysis was performed using Granta cells that had been treated with or without CDDO (2.5μM) for 24 hours, which are conditions that elicited cell death as demonstrated by PARP cleavage and activation of caspases 3 and 8 (supplemental Figure 7A-B). We observed the accumulation of electron dense aggregates only in the mitochondria of CDDO-treated Granta cells, which were reminiscent of those seen in LonRNAi knockdown mitochondria (Figure 4B bottom panels). No aggregates were present in any other compartment of CDDO-treated Granta cells (compare supplemental Figure 8A-B), nor were such inclusion bodies observed in mitochondria from untreated Granta cells (Figure 4B top panel), or in DMSO vehicle-treated cells (data not shown). We showed that the presence of mitochondrial inclusions were not an epiphenomenon of cell death in general, as mitochondrial inclusions were absent in Granta cells treated with cytotoxic doses of doxorubicin (supplemental Figures 9-10).
Figure 4.
CDDO and Lon knockdown induce the accumulation of electron dense inclusions within mitochondria. (A) Mitochondria from LonRNAi cells, expressing Lon (control) or knocked down for Lon by doxycycline (DOX) treatment for 2.5 weeks. (B) Mitochondria from Granta cells cultured in the absence (control) or presence of CDDO (2.5μM) for 24 hours.
Lon knockdown elicits lymphoma cell death
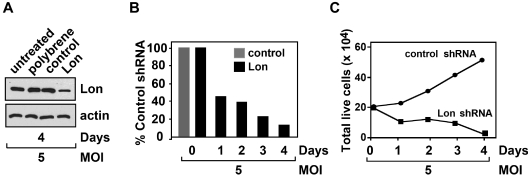
To determine directly the effect of down-regulating Lon on lymphoma cell survival and proliferation, Granta cells were transduced with lentiviral particles containing Lon or control shRNAs. Transduction efficiencies of ∼ 90% were achieved using a MOI of 5 as determined using a lentivirus-delivered green fluorescent protein construct (supplemental Figure 11). As shown in Figure 5A, Lon protein was significantly reduced in the Lon shRNA transduced cells after 4 days compared with control shRNA transduced cells, untreated or polybrene-treated Granta cells. We determined the effect of Lon knockdown on Granta cell growth and survival by scoring the number of viable cells that excluded trypan blue. Lon knockdown elicited progressive cell death over time with only ∼ 10% of cells surviving after the 4-day time course compared with the control shRNA cells (Figure 5B). The cell death observed for Lon knockdown Granta cells was in marked contrast to the control shRNA transduced cells, which continued to proliferate with a doubling-time of ∼ 3 days. Our data therefore showed that the genetic knockdown of Lon results in lymphoma cell death. These results together with the observations that CDDO inhibits Lon-mediated proteolysis and interacts with purified Lon and mitochondrial Lon in cultured Granta cells, strongly supports the assertion that CDDO-induced lymphoma cell death is mediated, in part, by inactivating Lon.
Figure 5.
Lon knockdown in Granta cells leads to cell death. Granta cells (2 × 105) were transduced with lentivirus particles (MOI 5) for the expression of control or Lon shRNAs. (A) Four days after transduction, cell extracts from untreated, polybrene-treated Granta cells, control shRNA, and Lon shRNA transduced Granta cells were immunoblotted with antibodies recognizing Lon or actin. (B-C) During a 4-day time course, cell viability was scored using trypan blue dye exclusion. The values are represented as the percentage of Lon shRNA viable cells relative to control, which was 100% (B). The total number of viable cells at the individual time-points was plotted over the 4-day time course (C). Results are representative of at least 3 independent experiments.
Discussion
Molecular chaperones of the heat shock protein (HSP) family as well as the ATP-dependent 26S proteasome play critical roles in facilitating tumor growth and survival, and have thus emerged as nononcogenic protein targets for cancer therapy.1–3 Overexpression of these stress response proteins is observed in various cancers and linked, in some cases, to poor prognosis and resistance to chemotherapeutic treatment.42,43 As the mitochondrial ATP-dependent Lon protease plays a similar role, assisting cells to survive and adapt to various proteotoxic stresses that are linked to oncogenesis (eg, hypoxic, oxidative, and ER stress),10,11,15 we speculated that Lon might also be a potential anticancer drug target.
Our previous work showed that the synthetic triterpenoid CDDO and its derivatives promote B-lymphoid cell apoptosis through a novel mitochondria-mediated pathway associated with mitochondrial protein aggregation.25 Based on these findings, we hypothesized that one mechanism of CDDO-induced mitochondrial protein aggregation and lymphoma cell death is by inhibiting Lon. We first set out to determine whether Lon is overexpressed in malignant B-lymphoid cells. Our results demonstrate that Lon is indeed overexpressed in human B-cell lymphoma cell lines and patient derived samples compared with normal resting or activated peripheral blood B cells (Figure 1). This is consistent with previous results showing an association between increased levels of Lon and malignant transformation. For example, human mammary epithelial cells overexpressing the proto-oncogene ErbB2 show significantly higher levels of Lon compared with normal mammary epithelial cells.44 Similarly, Lon mRNA is up-regulated in a mouse epidermal cell line transformed by epidermal growth factor (EGF), in contrast to control cells, and is also overexpressed in irreversibly transformed mouse epidermal cells compared with parental cells.44 In addition, Lon is overexpressed in rat Zajdela hepatoma tumor cells.45
Our findings strongly suggest that Lon is a physiologic target of CDDO, and the first specific mitochondrial protein target of this compound to be identified. We demonstrate that CDDO and its derivatives selectively and directly block the protease activity of Lon. CDDO or CDDO-Me inhibited Lon-mediated proteolysis in situ (Figure 2C-D) at a concentration that also induced apoptosis in Granta cells (supplemental Figure 7), Ramos or OCI-Ly19 lymphoma cells (1μM).25 A similar concentration of CDDO-Me also blocked Lon-mediated turnover of TFAM in cultured cells (Figure 2C-D). Strikingly, a common phenotype of CDDO-treated Granta cells and LonRNAi LS174T colon carcinoma cells is the accumulation of electron dense aggregates within the mitochondrial matrix. By contrast, such inclusions are not present in other cellular compartments demonstrating a unique effect of CDDO on mitochondrial homeostasis. The electron dense bodies induced by CDDO treatment or Lon-depletion probably represent aggregated misfolded, misassembled, or damaged proteins. Our results are consistent with previous findings that antisense depletion of Lon in human fibroblasts leads to similar mitochondrial inclusions that are associated with apoptotic cell death.46
We show that knockdown of Lon in Granta cells promotes cell death, which is consistent with our hypothesis that Lon inhibition contributes to CDDO-mediated apoptosis. To our knowledge this is the first demonstration that the pharmacologic or genetic down-regulation of a mitochondrial ATP-dependent protease is a potential therapeutic strategy for lymphoid malignancies. In contrast to the effects of CDDO and genetically knocking down Lon in Granta MCL cells, neither CDDO treatment nor Lon-knockdown in LS174T colon carcinoma cells resulted in cell death (data not shown). This is perhaps explained by cancer cell-specific sensitivities to cell death associated with mitochondrial proteotoxicity. Further experiments are required to determine the requirement of Lon-dependent degradation in cancer cell metabolism and survival.
Lon and other mitochondrial quality control proteins may represent a new and unique class of anticancer drug targets. In this study, CDDO and its derivatives selectively inhibited Lon but not the 20S or 26S proteasome, demonstrating that the anti-inflammatory and anticancer activities of these compounds are unlikely to be mediated directly or primarily through proteasome inhibition. Our findings that Lon is specifically pulled down with CDDO-biotin in biochemical and cell culture experiments, suggests a direct interaction by adduct formation between free thiol groups within Lon and Michael acceptor moieties of the triterpenoids (Figure 3). CDDObiotin selectively interacts with Lon, but not with BiP- an ER stress protein, which is linked to tumor progression and angiogenesis,47 nor with ClpP- the proteolytic component of the ATP-dependent protease ClpXP within the mitochondrial matrix (Figure 3). Further experiments are required to determine the mechanism by which CDDO inhibits Lon, for example, by inactivating one or both of its catalytic functions, or by promoting conformational defects or disassembly of the holoenzyme.
A fundamental question is—how can compounds that interfere with essential cellular functions such as mitochondrial ATP-dependent proteolysis, be effective anticancer agents without harming normal cells? One view is that cancer cells are more vulnerable than normal cells to anticancer agents that increase cellular stress. Indeed, proteasome inhibitors, histone deacetylase inhibitors, and redox active agents48 have been shown to increase oxidative stress in tumor cells, which in general already have augmented levels of ROS.49 It is therefore speculated that such drugs push tumor cells over the threshold for tolerating increased oxidative stress, and are thus selectively more toxic for tumor cells compared with normal cells. That the Lon protease plays a fundamentally critical role in antioxidant stress response by degrading toxic aggregates of oxidized mitochondrial proteins,15 supports the premise that inhibitors of Lon and possibly of other mitochondrial quality control proteins may have therapeutic utility either alone or in combination with other anticancer agents. In this regard, we previously showed that CDDO exhibits an additive effect on cell death when administered in combination with bortezomib (aka PS-341).25 This suggests that subjecting cancer cells to bimodal proteotoxic overload by inhibiting mitochondrial as well as cytosolic ATP-dependent proteases may provide a chemotherapeutic strategy for augmenting cancer cell death. Indeed, recent efforts have focused on developing new anticancer therapeutics by targeting drugs to mitochondria.50 Thus, it is anticipated that the development of specific high affinity inhibitors of Lon and other mitochondrial proteases and chaperones will aid in determining their full potential as drug targets in treating cancer and possibly other diseases.
Supplementary Material
Acknowledgments
The authors thank Dr Michael B. Sporn for providing CDDO and its derivatives. They are grateful for the expertise of Karen L. de Mesy Bentley, University of Rochester Electron Microscope Research Core.
This work was supported by a National Institutes of Health, National Cancer Institute SPORE in Lymphoma (1P50CA130805), the Leukemia & Lymphoma Society and the Lymphoma Research Foundation (S.H.B.); the National Institutes of Health (R01GM61095, R01GM084039, R21NS067668) and the Foundation of University of Medicine and Dentistry of New Jersey (C.K.S.).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: S.H.B. and C.K.S. designed, analyzed, and interpreted research and wrote the paper; S.V. optimized and performed the Granta lentivirus transduction experiments; M.L. performed the lentivirus transduction and cell counting experiments; J.L. designed and performed the CDDO inhibition experiments using purified Lon, TFAM, and the 20S proteasome, carried out the statistical analysis, and contributed to writing the paper; B.L. performed the immunoblotting of HeLa ρ0 cells treated with CDDO; S.P.H. designed research; K.M.M. performed the CDDO-biotin experiments; H.M.M. and J.S. performed research; M.A. provided the biotinylated-CDDO used in this study; and P.S.B. provided unpublished data and insightful discussions.
Conflict-of-interest disclosure: S.H.B. has been a consultant for Millenium Pharmaceuticals. M.A. is a shareholder and a consultant for Reata Pharmaceuticals and holds a patent for CDDO-Me, which is in clinical development. The remaining authors declare no competing financial interests.
The current affiliation for B.L. is Institute of Biophysics, School of Laboratory Medicine and Life Sciences, Wenzhou Medical College, Wenzhou, Zhejiang, China.
Correspondence: Carolyn K. Suzuki, New Jersey Medical School, University of Medicine and Dentistry of New Jersey, Dept of Biochemistry and Molecular Biology, 185 S Orange Ave, E661, Newark, NJ 07103; e-mail: suzukick@umdnj.edu; or Steven H. Bernstein, James P. Wilmot Cancer Center, University of Rochester Medical Center, 601 Elmwood Ave, Rochester, NY 14642; e-mail: steven_bernstein@urmc.rochester.edu.
References
- 1.Dai C, Whitesell L, Rogers AB, Lindquist S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 2007;130(6):1005–1018. doi: 10.1016/j.cell.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leu JI, Pimkina J, Frank A, Murphy ME, George DL. A small molecule inhibitor of inducible heat shock protein 70. Mol Cell. 2009;36(1):15–27. doi: 10.1016/j.molcel.2009.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trepel J, Mollapour M, Giaccone G, Neckers L. Targeting the dynamic HSP90 complex in cancer. Nat Rev Cancer. 2010;10(8):537–549. doi: 10.1038/nrc2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banerji U, O'Donnell A, Scurr M, et al. Phase I pharmacokinetic and pharmacodynamic study of 17-allylamino, 17-demethoxygeldanamycin in patients with advanced malignancies. J Clin Oncol. 2005;23(18):4152–4161. doi: 10.1200/JCO.2005.00.612. [DOI] [PubMed] [Google Scholar]
- 5.Hanahan D, Weinberg R. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 6.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136(5):823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Venkatesh S, Lee J, Singh K, Lee I, Suzuki CK. Multitasking in the mitochondrion by the ATP-dependent Lon protease. Biochim Biophys Acta. 2012;1823(1):56–66. doi: 10.1016/j.bbamcr.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee I, Suzuki CK. Functional mechanics of the ATP-dependent Lon protease- lessons from endogenous protein and synthetic peptide substrates. Biochim Biophys Acta. 2008;178(5):727–735. doi: 10.1016/j.bbapap.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bota DA, Davies KJ. Protein degradation in mitochondria: implications for oxidative stress, aging and disease: a novel etiological classification of mitochondrial proteolytic disorders. Mitochondrion. 2001;1(1):33–49. doi: 10.1016/s1567-7249(01)00005-8. [DOI] [PubMed] [Google Scholar]
- 10.Fukuda R, Zhang H, Kim JW, Shimoda L, Dang CV, Semenza GL. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell. 2007;129(1):111–122. doi: 10.1016/j.cell.2007.01.047. [DOI] [PubMed] [Google Scholar]
- 11.Hori O, Icinoda F, Tamatani T, et al. Transmission of cell stress from endoplasmic reticulum to mitochondria: enhanced expression of Lon protease. J Cell Biol. 2002;157(7):1151–1160. doi: 10.1083/jcb.200108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koumenis C. ER stress, hypoxia tolerance and tumor progression. Curr Mol Med. 2006;6(1):55–69. doi: 10.2174/156652406775574604. [DOI] [PubMed] [Google Scholar]
- 13.Bayot A, Gareil M, Rogowska-Wrzesinska A, Roepstorff P, Friguet B, Bulteau AL. Identification of novel oxidized protein substrates and physiological partners of the mitochondrial ATP-dependent Lon-like protease Pim1. J Biol Chem. 2010;285(15):11445–11457. doi: 10.1074/jbc.M109.065425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bender T, Leidhold C, Ruppert T, Franken S, Voos W. The role of protein quality control in mitochondrial protein homeostasis under oxidative stress. Proteomics. 2010;10(7):1426–1443. doi: 10.1002/pmic.200800619. [DOI] [PubMed] [Google Scholar]
- 15.Bota DA, Davies KJA. Lon protease preferentially degrades oxidized mitochondrial aconitase by an ATP-stimulated mechanism. Nat Cell Biol. 2002;4(9):674–680. doi: 10.1038/ncb836. [DOI] [PubMed] [Google Scholar]
- 16.Liby KT, Yore MM, Sporn MB. Triterpenoids and rexinoids as multifunctional agents for the prevention and treatment of cancer. Nat Rev Cancer. 2007;7(5):357–369. doi: 10.1038/nrc2129. [DOI] [PubMed] [Google Scholar]
- 17.Ahmad R, Raina D, Meyer C, Kharbanda S, Kufe D. Triterpenoid CDDO-Me blocks the NF-kappaB pathway by direct inhibition of IKKbeta on Cys-179. J Biol Chem. 2006;281(47):35764–35769. doi: 10.1074/jbc.M607160200. [DOI] [PubMed] [Google Scholar]
- 18.Ahmad R, Raina D, Meyer C, Kufe D. Triterpenoid CDDO-methyl ester inhibits the Janus-activated kinase-1 (JAK1)–>signal transducer and activator of transcription-3 (STAT3) pathway by direct inhibition of JAK1 and STAT3. Cancer Res. 2008;68(8):2920–2926. doi: 10.1158/0008-5472.CAN-07-3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Porter WW, Suh N, et al. A synthetic triterpenoid, 2-cyano-3,12-dioxooleana-1,9-dien-28-oic acid (CDDO), is a ligand for the peroxisome proliferator-activated receptor gamma. Mol Endocrinol. 2000;14(10):1550–1556. doi: 10.1210/mend.14.10.0545. [DOI] [PubMed] [Google Scholar]
- 20.Couch RD, Browning RG, Honda T, et al. Studies on the reactivity of CDDO, a promising new chemopreventive and chemotherapeutic agent: implications for a molecular mechanism of action. Bioorg Med Chem Lett. 2005;15(9):2215–2219. doi: 10.1016/j.bmcl.2005.03.031. [DOI] [PubMed] [Google Scholar]
- 21.Samudio I, Konopleva M, Hail N, Jr, et al. 2-Cyano-3,12-dioxooleana-1,9-dien-28-imidazolide (CDDO-Im) directly targets mitochondrial glutathione to induce apoptosis in pancreatic cancer. J Biol Chem. 2005;280(43):36273–36282. doi: 10.1074/jbc.M507518200. [DOI] [PubMed] [Google Scholar]
- 22.Samudio I, Konopleva M, Pelicano H, et al. A novel mechanism of action of methyl-2-cyano-3,12 dioxoolean-1,9 diene-28-oate: direct permeabilization of the inner mitochondrial membrane to inhibit electron transport and induce apoptosis. Mol Pharmacol. 2006;69(4):1182–1193. doi: 10.1124/mol.105.018051. [DOI] [PubMed] [Google Scholar]
- 23.Samudio I, Kurinna S, Ruvolo P, et al. Inhibition of mitochondrial metabolism by methyl-2-cyano-3,12-dioxooleana-1,9-diene-28-oate induces apoptotic or autophagic cell death in chronic myeloid leukemia cells. Mol Cancer Ther. 2008;7(5):1130–1139. doi: 10.1158/1535-7163.MCT-07-0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ikeda T, Sporn M, Honda T, Gribble GW, Kufe D. The novel triterpenoid CDDO and its derivatives induce apoptosis by disruption of intracellular redox balance. Cancer Res. 2003;63(17):5551–5558. [PubMed] [Google Scholar]
- 25.Brookes P, Morse K, Ray D, et al. The triterpenoid CDDO and its derivatives elicit human lymphoid cell apoptosis through a novel pathway involving the unregulated mitochondrial permeability transition pore. Cancer Res. 2007;67(4):1793–1802. doi: 10.1158/0008-5472.CAN-06-2678. [DOI] [PubMed] [Google Scholar]
- 26.Couch RD, Ganem NJ, Zhou M, et al. 2-cyano-3,12-dioxooleana-1,9(11)-diene-28-oic acid disrupts microtubule polymerization: a possible mechanism contributing to apoptosis. Mol Pharmacol. 2006;69(4):1158–1165. doi: 10.1124/mol.105.018572. [DOI] [PubMed] [Google Scholar]
- 27.Kress CL, Konopleva M, Martinez-Garcia V, et al. Triterpenoids display single agent antitumor activity in a transgenic mouse model of chronic lymphocytic leukemia and small B cell lymphoma. PLoS One. 2007;2(6):e559. doi: 10.1371/journal.pone.0000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ray DM, Morse KM, Hilchey SP, et al. The novel triterpenoid 2-cyano-3,12-dioxooleana-1,9-dien-28-oic acid (CDDO) induces apoptosis of human diffuse large B-cell lymphoma cells through a peroxisome proliferator-activated receptor gamma-independent pathway. Exp Hematol. 2006;34(9):1201–1210. doi: 10.1016/j.exphem.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 29.Lu B, Yadav S, Shah PG, et al. Roles for the human ATP-dependent Lon protease in mitochondrial DNA maintenance. J Biol Chem. 2007;282(24):17363–17374. doi: 10.1074/jbc.M611540200. [DOI] [PubMed] [Google Scholar]
- 30.King MP, Attardi G. Isolation of human cell lines lacking mitochondrial DNA. Methods Enzymol. 1996;264:304–313. doi: 10.1016/s0076-6879(96)64029-4. [DOI] [PubMed] [Google Scholar]
- 31.Lu B, Liu T, Crosby JA, Thomas-Wohlever J, Lee I, Suzuki CK. The ATP-dependent Lon protease of Mus musculus is a DNA-binding protein that is functionally conserved between yeast and mammals. Gene. 2003;306:45–55. doi: 10.1016/s0378-1119(03)00403-7. [DOI] [PubMed] [Google Scholar]
- 32.Liu T, Lu B, Lee I, Ondrovicova G, Kutejova E, Suzuki CK. DNA and RNA binding by the mitochondrial Lon protease is regulated by nucleotide and protein substrate. J Biol Chem. 2004;279(14):13902–13910. doi: 10.1074/jbc.M309642200. [DOI] [PubMed] [Google Scholar]
- 33.Ondrovicova G, Liu T, Singh K, et al. Cleavage Site Selection within a Folded Substrate by the ATP-dependent Lon Protease. J Biol Chem. 2005;280(26):25103–25110. doi: 10.1074/jbc.M502796200. [DOI] [PubMed] [Google Scholar]
- 34.Diefenbach CS, O'Connor OA. Mantle cell lymphoma in relapse: the role of emerging new drugs. Curr Opin Oncol. 2010;22(5):419–423. doi: 10.1097/CCO.0b013e32833d58f2. [DOI] [PubMed] [Google Scholar]
- 35.Matsushima Y, Goto Y, Kaguni LS. Mitochondrial Lon protease regulates mitochondrial DNA copy number and transcription by selective degradation of mitochondrial transcription factor A (TFAM). Proc Natl Acad Sci U S A. 2010;107(43):18410–18415. doi: 10.1073/pnas.1008924107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larsson NG, Oldfors A, Holme E, Clayton DA. Low levels of mitochondrial transcription factor A in mitochondrial DNA depletion. Biochem Biophys Res Commun. 1994;200(3):1374–1381. doi: 10.1006/bbrc.1994.1603. [DOI] [PubMed] [Google Scholar]
- 37.Seidel-Rogol BL, Shadel GS. Modulation of mitochondrial transcription in response to mtDNA depletion and repletion in HeLa cells. Nucleic Acids Res. 2002;30(9):1929–1934. doi: 10.1093/nar/30.9.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frase H, Hudak J, Lee I. Identification of the proteasome inhibitor MG262 as a potent ATP-dependent inhibitor of the salmonella enterica serovar typhimurium Lon protease. Biochemistry. 2006;45(27):8264–8274. doi: 10.1021/bi060542e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Granot Z, Kobiler O, Melamed-Book N, et al. Turnover of mitochondrial steroidogenic acute regulatory (StAR) protein by Lon protease: the unexpected effect of proteasome inhibitors. Mol Endocrinol. 2007;21(9):2164–2177. doi: 10.1210/me.2005-0458. [DOI] [PubMed] [Google Scholar]
- 40.Honda T, Janosik T, Honda Y, et al. Design, synthesis, and biological evaluation of biotin conjugates of 2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oic acid for the isolation of the protein targets. J Med Chem. 2004;47(20):4923–4932. doi: 10.1021/jm049727e. [DOI] [PubMed] [Google Scholar]
- 41.Suzuki CK, Suda K, Wang N, Schatz G. Requirement for the yeast gene LON in intramitochondrial proteolysis and maintenance of respiration. Science. 1994;264:273–276. 891. doi: 10.1126/science.8146662. [DOI] [PubMed] [Google Scholar]
- 42.Simpson NE, Lambert WM, Watkins R, et al. High levels of Hsp90 cochaperone p23 promote tumor progression and poor prognosis in breast cancer by increasing lymph node metastases and drug resistance. Cancer Res. 2010;70(21):8446–8456. doi: 10.1158/0008-5472.CAN-10-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Virrey JJ, Dong D, Stiles C, et al. Stress chaperone GRP78/BiP confers chemoresistance to tumor-associated endothelial cells. Mol Cancer Res. 2008;6(8):1268–1275. doi: 10.1158/1541-7786.MCR-08-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu Y, Wang M, Lin H, Huang C, Shi X, Luo J. Epidermal growth factor up-regulates the transcription of mouse lon homology ATP-dependent protease through extracellular signal-regulated protein kinase- and phosphatidylinositol-3-kinase-dependent pathways. Exp Cell Res. 2002;280(1):97–106. doi: 10.1006/excr.2002.5621. [DOI] [PubMed] [Google Scholar]
- 45.Luciakova K, Sokolikova B, Chloupkova M, Nelson BD. Enhanced mitochondrial biogenesis is associated with increased expression of the mitochondrial ATP-dependent Lon protease. FEBS Lett. 1999;444:186–188. doi: 10.1016/s0014-5793(99)00058-7. [DOI] [PubMed] [Google Scholar]
- 46.Bota DA, Ngo JK, Davies KJ. Down-regulation of the human Lon protease impairs mitochondrial structure and function and causes cell death. Free Radic Biol Med. 2005;38(5):665–677. doi: 10.1016/j.freeradbiomed.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 47.Dong D, Ni M, Li J, et al. Critical role of the stress chaperone GRP78/BiP in tumor proliferation, survival, and tumor angiogenesis in transgene-induced mammary tumor development. Cancer Res. 2008;68(2):498–505. doi: 10.1158/0008-5472.CAN-07-2950. [DOI] [PubMed] [Google Scholar]
- 48.Tew KD, Townsend DM. Redox platforms in cancer drug discovery and development. Curr Opin Chem Biol. 2010;15(1):156–161. doi: 10.1016/j.cbpa.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trachootham D, Alexandre J, Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat Rev Drug Discov. 2009;8(7):579–591. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- 50.Biasutto L, Dong LF, Zoratti M, Neuzil J. Mitochondrially targeted anticancer agents. Mitochondrion. 2010;10(6):670–681. doi: 10.1016/j.mito.2010.06.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.