Abstract
Recently, the BRAF V600E mutation was reported in all cases of hairy cell leukemia (HCL) but not in other peripheral B-cell neoplasms. We wished to confirm these results and assess BRAF status in well-characterized cases of HCL associated with poor prognosis, including the immunophenotypically defined HCL variant (HCLv) and HCL expressing the IGHV4-34 immunoglobulin rearrangement. Fifty-three classic HCL (HCLc) and 16 HCLv cases were analyzed for BRAF, including 5 HCLc and 8 HCLv expressing IGHV4-34. BRAF was mutated in 42 (79%) HCLc, but wild-type in 11 (21%) HCLc and 16 (100%) HCLv. All 13 IGHV4-34+ HCLs were wild-type. IGHV gene usage in the 11 HCLc BRAF wild-type cases included 5 IGHV4-34, 5 other, and 1 unknown. Our results suggest that HCLv and IGHV4-34+ HCLs have a different pathogenesis than HCLc and that a significant minority of other HCLc are also wild-type for BRAF V600.
Introduction
Classic hairy cell leukemia (HCLc) is a B-cell malignancy with distinctive immunophenotype, typically expressing CD20, CD22, CD25, CD11c, CD103, CD123, annexin A1, and tartrate-resistant acid phosphatase.1–3 Purine analog therapy is highly effective, with most patients achieving durable complete remissions.4,5 HCL variant (HCLv) was first identified by Cawley et al6 and recently recognized as a distinct entity by the World Health Organization.1 HCLv lacks CD25, annexin A1, and/or tartrate-resistant acid phosphatase expression, and patients respond poorly to purine analogs, with only partial response in less than 50% and relatively poor overall survival from diagnosis.7 We recently reported that HCL expressing the IGHV4-34 immunoglobulin rearrangement has a poor prognosis like HCLv, whether immunophenotypically consistent with HCLv or HCLc.8
The oncogenic v-raf murine sarcoma viral oncogene homolog B1 (BRAF) mutation is found in 6% to 7% of different cancers, including approximately 50% to 60% malignant melanoma.9–12 The 94-kDa BRAF protein encoded by the gene on chromosome 7q3413 functions as a serine/threonine kinase immediately downstream of RAS.14 BRAF mutations, most commonly c.1799T > A (V600E), can activate the mitogen-activated protein kinase pathway, leading to uncontrolled proliferation.14 Recently, the (V600E) BRAF mutation was reported in 100% of 48 patients with HCLc versus none of 195 patients with other B-cell malignancies, the latter including 16 with splenic lymphoma/leukemias or unclassifiable lymphoma/leukemias, including HCLv and red pulp small B-cell lymphoma.15 We wished to confirm these results in our HCLc cases and extend the analysis to well-characterized cases of HCLv and IGHV4-34–expressing HCL.
Methods
DNA was extracted from peripheral blood of patients being treated on or screened for HCL protocols at National Institutes of Health, approved by the Investigational Review Board of the National Cancer Institute. The diagnosis of HCLc and HCLv and molecular characterizations of IGHV rearrangements were performed as previously described.8 DNA samples were extracted using a Precision System Science automated robot (PSS USA). Twenty-four commercially available normal DNA control samples (BioChain) were used to establish baseline values.
For BRAF mutation detection, we used a previously described pyrosequencing assay, in which PCR amplification is initially performed with primers flanking the V600E mutation hotspot within exon 15 of BRAF, and is followed by targeted pyrosequencing,16 with several important modifications. Thermal cycling parameters were modified to achieve COLD-PCR conditions,17 which enrich for variant (mutant) DNA sequences during the PCR reaction. The cycling parameters are available on request. The pyrosequencing primer (5′-TAGGTGATTTTGGTCTAGCT-3′) and nucleotide dispensation order (GACACGATGATCT) were also redesigned, and pyrosequencing was performed on a PyroMark Q24 instrument (QIAGEN). The pyrogram outputs were analyzed and the COLD-PCR enhanced percentage of mutant versus wild-type alleles calculated, using the allele quantification (AQ) mode of PyroMark Version 2.0.6 software (QIAGEN). The analytical sensitivity and specificity of detecting BRAF c.1799T > A (V600E) mutation were determined using melanoma cell lines and cases with known BRAF status. The threshold AQ value for classifying HCL samples as positive for mutation was set as 3 SD above the mean (6.9%) values of 24 normal blood DNA (range, 2.5%-6.9%).
The described modifications allowed us to achieve a sensitivity of 2% to 3% tumor cells and evaluate most of the less common exon 15 mutations, in addition to the common V600E. To further prevent potential false-negative interpretations, we excluded samples with HCL constituting less than 10% of total white blood cells (WBCs) for this study, so that those included were well above the analytical sensitivity of the assay (Figure 1, data examples).
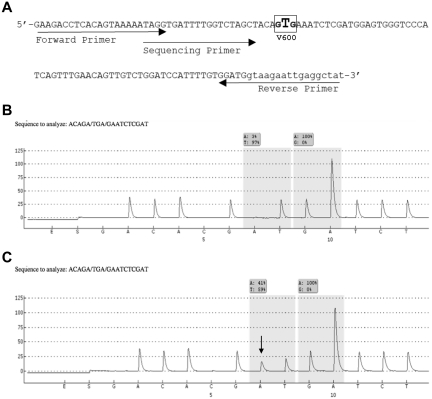
Figure 1.
BRAF primer design and representative pyrograms. (A) The modified pyrosequencing assay design from Packham et al.16 The assay detects all mutations in codon 600, including the common V600E (c.1799T > A) mutation as well as the less common variant mutations involving codons 599 through 601. (B-C) Representative pyrograms from an HCLc-expressing IGHV4-34 (case BH18; 92.6% HCL cells) revealing a wild-type result at codon 600 (B), and from an HCLc (case BH21; 77.4% HCL cells) expressing IGHV5-51 showing the c.1799T > A (V600E) mutation (C). The nucleotides on the x-axis (beginning with G) represent their dispensation order during the pyrosequencing reactions, whereas the y-axis displays arbitrary units related to peak height. Peak heights are proportional to the number of nucleotides incorporated into the growing DNA strand. The shaded boxes represent the percentage of nucleotides calculated at a particular interrogated location (AQ score). The arrow in panel C indicates the c.1799T > A mutation. The shaded box represents an AQ score of 41% A and 59% T at that position.
Results and discussion
Sixty-nine patients were selected with HCLc or HCLv and leukemic cells more than 10% of the WBCs. These included 53 (77%) HCLc and 16 (23%) HCLv, as defined by World Health Organization criteria.1 Sixty-three of the 69 cases were molecularly characterized with respect to IGHV gene usage, and most were previously reported.18 Thirteen (21%) of 63 expressed IGHV4-34, including 5 HCLc, and 8 HCLv (Table 1).
Table 1.
BRAF mutation analysis in 69 HCL and HCLv patients
| Diagnosis | IGH-VH | BRAF V600E | Wild-type | Total |
|---|---|---|---|---|
| HCLc | VH4-34+ | 0 | 5 | 5 |
| VH-other | 37 | 5 | 42 | |
| VH-unknown | 5 | 1 | 6 | |
| HCLv | VH4-34+ | 0 | 8 | 8 |
| VH-other | 0 | 8 | 8 | |
| Total | 42 | 27 | 69 |
As shown in Table 1, 42 (61%) of the 69 patients had BRAF mutations, all of the common c.1799T > A (V600E) type, and all occurring among the 53 HCLc cases. The percentage of peripheral WBCs consistent with HCL in the 42 BRAF mutant cases was 10.7% to 95% and correlated with AQ mutation values (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Twenty-seven (39%) of the 69 cases were wild-type with respect to BRAF, including all 16 HCLv but also 11 (21%) HCLc. All IGHV4-34+ cases lacked BRAF mutations, including 8 HCLv and 5 HCLc, and IGHV4-34 sequences were each more than 98% homologous to germline (unmutated). Of the remaining 6 BRAF wild-type HCLc cases, one was not typed for IGHV, whereas the remaining 5 cases expressed IGHV2-70, IGHV3-15, IGHV3-23, IGHV3-48, and IGHV4-39. Interestingly, unlike the 5 HCLc with IGHV4-34, these 5 HCLc with other IGHV rearrangements were mutated (P = .008). Of the 10 BRAF wild-type HCLc cases characterized for IGHV, median homology to germline IGHV was 99.58% for the 5 IGHV4-34+ cases versus 95.16% for the other 5 (P = .008), indicating that homology to germline was not related to BRAF mutation. Additional data, including the HCL counts, AQ values, and characteristics of each patient, can be found in supplemental Table 1.
Overall, our data confirm the results of Tiacci et al indicating that HCLc is associated with the V600E BRAF mutation.15 A more recently published study also reported 100% of 48 classic HCL samples to contain BRAF V600E.19 In contrast, however, we found BRAF to be wild-type in 11 (21%) of 53 HCLc defined phenotypically by high levels of CD25, CD103, CD123, and tartrate-resistant acid phosphatase. IGHV use in these 11 included 5 with IGHV4-34, 5 other, and 1 unknown. Of these 11 HCLc patients, 7 had bone marrow biopsies that were adequate (without myeloid cell background staining) to stain for annexin A1,2,20 including 3 IGHV4-34, 3 other IGHV, and 1 unknown IGHV, and all were positive for annexin A1. The BRAF wild-type HCLc cells constituted 19.0% to 59.0% of WBCs, well above the 3% lower limit of sensitivity.
Although our assay detects most of the unusual exon 15 BRAF mutations other than c.1799T > A, it will not detect rare exon 11 mutations, so we cannot exclude that the V600-wild-type HCLc cases may have a BRAF mutation in a different region. Nonetheless, it appears clear from our data that a significant fraction of HCLc defined phenotypically does not have the c.1799T > A (V600E) mutation. Our series was biased toward pretreated patients. Of the 69 patients, 55 were pretreated and had 1 to 8 (median, 2) prior courses of purine analog, with 30 (55%) responding to their last course for 1 to 131 (median, 14) months months. The 5 HCLc IGHV4-34− patients with wild-type BRAF were not clinically different from the BRAF mutated patients, in that 2 of the 5 were untreated, and of the other 3 with 2 or 3 prior courses of purine analog each, 2 responded for 2 and 21 months. Thus, other than patients with HCLv and/or IGHV4-34 who were always negative for BRAF V600E, additional HCLc patients lacked the BRAF mutation and could not be distinguished clinically from the others.
Our data also provide further evidence that HCLv and HCLc are distinct, as none of the 16 HCLv cases (including 8 IGHV4-34+) displayed BRAF mutations. In addition to clinical and immunophenotypic differences, molecular profiling studies also argue for a distinction in molecular pathogenesis between these morphologically similar leukemia subtypes.5
Finally, our data support the separation of IGHV4-34–expressing HCL from HCLc and provide further rationale for defining these cases on the basis of IGHV4-34 expression. It may be useful to consider IGHV4-34 HCL, with its uniformly poor responses to purine analogs, as a separate subgroup overlapping HCLv and HCLc immunophenotypically.
In conclusion, we confirm BRAF mutations in a high percentage of HCLc cases but identify a significant minority without BRAF mutations and show that HCLv and/or IGHV4-34+ HCLs uniformly lack BRAF mutations. These data support a trial in HCL using BRAF-targeted therapy or other inhibitors of the same pathway, although such a trial should be accompanied by BRAF mutation testing. Moreover, routine testing of HCL tissues, such as bone marrow for BRAF mutations, may be useful in identifying patients who have HCLv and/or IGHV4-34, which may portend a poor prognosis with single-agent purine analogs. Such patients may be optimal for clinical trials testing combined purine analog-mAb approaches.8,21–24 Moreover, the discovery of new targets in BRAF wild-type patients, which are relevant to molecular pathogenesis, may lead to new therapies for this as well as other malignancies.
Supplementary Material
Acknowledgments
The authors thank Hong Zhou, Thu-Anh Pham, and Trinh-Hoc Pham for technical assistance; Barbara Debrah for data management; and our clinical staff Sonya Duke, Rita Mincemoyer, Natasha Kormanik, and Elizabeth Maestri.
This work was supported by the intramural program of the National Cancer Institute, National Institutes of Health.
Footnotes
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: L.X., E.A., K.R.C., M.R., and R.J.K. designed research and analyzed and interpreted data; L.X., E.A., and W.N. performed research; M.S.-S. provided immunophenotypic data; and L.X., M.R., and R.J.K. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Robert J. Kreitman, National Institutes of Health, 37/5124b, 37 Convent Dr, MSC 4255, Bethesda, MD 20892; e-mail: kreitmar@mail.nih.gov.
References
- 1.Swerdlow SH, Campo E, Harris NL, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 4th Ed. Volume 2. Lyon, France: World Health Organization; 2008. [Google Scholar]
- 2.Falini B, Tiacci E, Liso A, et al. Simple diagnostic assay for hairy cell leukaemia by immunocytochemical detection of annexin A1 (ANXA1). Lancet. 2004;363(9424):1869–1870. doi: 10.1016/S0140-6736(04)16356-3. [DOI] [PubMed] [Google Scholar]
- 3.Matutes E. Immunophenotyping and differential diagnosis of hairy cell leukemia. Hematol Oncol Clin North Am. 2006;20(5):1051. doi: 10.1016/j.hoc.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 4.Goodman GR, Burian C, Koziol JA, Saven A. Extended follow-up of patients with hairy cell leukemia after treatment with cladribine. J Clin Oncol. 2003;21(5):891–896. doi: 10.1200/JCO.2003.05.093. [DOI] [PubMed] [Google Scholar]
- 5.Hockley SL, Morgan GJ, Leone PE, et al. High-resolution genomic profiling in hairy cell leukemia-variant compared with typical hairy cell leukemia. Leukemia. 2011;25(7):1189–1192. doi: 10.1038/leu.2011.47. [DOI] [PubMed] [Google Scholar]
- 6.Cawley JC, Burns GF, Hayhoe FG. A chronic lymphoproliferative disorder with distinctive features: a distinct variant of hairy-cell leukaemia. Leuk Res. 1980;4(6):547–559. doi: 10.1016/0145-2126(80)90066-1. [DOI] [PubMed] [Google Scholar]
- 7.Robak T. Hairy-cell leukemia variant: recent view on diagnosis, biology and treatment. Cancer Treat Rev. 2011;37(1):3–10. doi: 10.1016/j.ctrv.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 8.Arons E, Suntum T, Stetler-Stevenson M, Kreitman RJ. VH4-34+ hairy cell leukemia, a new variant with poor prognosis despite standard therapy. Blood. 2009;114(21):4687–4695. doi: 10.1182/blood-2009-01-201731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Badalian-Very G, Vergilio JA, Degar BA, et al. Recurrent BRAF mutations in Langerhans cell histiocytosis. Blood. 2010;116(11):1919–1923. doi: 10.1182/blood-2010-04-279083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chapman MA, Lawrence MS, Keats JJ, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471(7339):467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 12.Kimura ET, Nikiforova MN, Zhu Z, Knauf JA, Nikiforov YE, Fagin JA. High prevalence of BRAF mutations in thyroid cancer: genetic evidence for constitutive activation of the RET/PTC-RAS-BRAF signaling pathway in papillary thyroid carcinoma. Cancer Res. 2003;63(7):1454–1457. [PubMed] [Google Scholar]
- 13.Eychene A, Barnier JV, Apiou F, Dutrillaux B, Calothy G. Chromosomal assignment of two human B-raf(Rmil) proto-oncogene loci: B-raf-1 encoding the p94Braf/Rmil and B-raf-2, a processed pseudogene. Oncogene. 1992;7(8):1657–1660. [PubMed] [Google Scholar]
- 14.Vultur A, Villanueva J, Herlyn M. Targeting BRAF in advanced melanoma: a first step toward manageable disease. Clin Cancer Res. 2011;17(7):1658–1663. doi: 10.1158/1078-0432.CCR-10-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tiacci E, Trifonov V, Schiavoni G, et al. BRAF mutations in hairy-cell leukemia. N Engl J Med. 2011;364(24):2305–2315. doi: 10.1056/NEJMoa1014209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Packham D, Ward RL, Ap Lin V, Hawkins NJ, Hitchins MP. Implementation of novel pyrosequencing assays to screen for common mutations of BRAF and KRAS in a cohort of sporadic colorectal cancers. Diagn Mol Pathol. 2009;18(2):62–71. doi: 10.1097/PDM.0b013e318182af52. [DOI] [PubMed] [Google Scholar]
- 17.Li J, Wang L, Mamon H, Kulke MH, Berbeco R, Makrigiorgos GM. Replacing PCR with COLD-PCR enriches variant DNA sequences and redefines the sensitivity of genetic testing. Nat Med. 2008;14(5):579–584. doi: 10.1038/nm1708. [DOI] [PubMed] [Google Scholar]
- 18.Arons E, Roth L, Sapolsky J, Suntum T, Stetler-Stevenson M, Kreitman RJ. Evidence of canonical somatic hypermutation in hairy cell leukemia. Blood. 2011;117(18):4844–4851. doi: 10.1182/blood-2010-11-316737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boyd EM, Bench AJ, van 't Veer MB, et al. High resolution melting analysis for detection of BRAF exon 15 mutations in hairy cell leukaemia and other lymphoid malignancies. Br J Haematol. 2011;155(5):609–612. doi: 10.1111/j.1365-2141.2011.08868.x. [DOI] [PubMed] [Google Scholar]
- 20.Sherman MJ, Hanson CA, Hoyer JD. An assessment of the usefulness of immunohistochemical stains in the diagnosis of hairy cell leukemia. Am J Clin Pathol. 2011;136(3):390–399. doi: 10.1309/AJCP5GE1PSBMBZTW. [DOI] [PubMed] [Google Scholar]
- 21.Kreitman RJ, Wilson WH, Bergeron K, et al. Efficacy of the anti-CD22 recombinant immunotoxin BL22 in chemotherapy-resistant hairy-cell leukemia. N Engl J Med. 2001;345(4):241–247. doi: 10.1056/NEJM200107263450402. [DOI] [PubMed] [Google Scholar]
- 22.Kreitman RJ, Tallman MS, Robak T, et al. Phase I/II trial of anti-CD22 recombinant immunotoxin moxetumomab pasudotox (CAT-8015 or HA22) in patients with hairy cell leukemia. J Clin Oncol. doi: 10.1200/JCO.2011.38.1756. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ravandi F, O'Brien S, Jorgensen J, et al. Phase 2 study of cladribine followed by rituximab in patients with hairy cell leukemia. Blood. 2011;118(14):3818–3823. doi: 10.1182/blood-2011-04-351502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ravandi F, Jorgensen JL, O'Brien SM, et al. Eradication of minimal residual disease in hairy cell leukemia. Blood. 2006;107(12):4658–4662. doi: 10.1182/blood-2005-11-4590. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.