Abstract
The mammalian intestine is home to ~100 trillion bacteria that perform important metabolic functions for their hosts. The proximity of vast numbers of bacteria to host intestinal tissues raises the question of how symbiotic host-bacterial relationships are maintained without eliciting potentially harmful immune responses. Here we show that RegIIIγ, a secreted antibacterial lectin, was essential for maintaining a ~50 μm zone that physically separates the microbiota from the small intestinal epithelial surface. Loss of host-bacterial segregation in RegIIIγ−/− mice was coupled to increased bacterial colonization of the intestinal epithelial surface and enhanced activation of intestinal adaptive immune responses by the microbiota. Together, our findings reveal that RegIIIγ is a fundamental immune mechanism that promotes host-bacterial mutualism by regulating the spatial relationships between microbiota and host.
Complex communities of intestinal bacteria inhabit the mammalian intestine, making essential contributions to host metabolism (1). It remains unclear how these large and diverse bacterial populations are maintained at homeostasis without negative health consequences such as chronic immune activation and inflammation. Much of our current understanding of intestinal homeostasis is built around the idea that the microbiota continuously interact with the intestinal immune system, and that microbiota species composition dictates the balance between pro-inflammatory and tolerogenic immune responses (2–4). However, studies of the spatial relationships between microbiota and host point to the existence of a more general strategy for maintaining homeostasis. These studies show that the physical separation of microbiota from the intestinal surface is critical for limiting immune activation and maintaining mutualistic host-bacterial associations (5). Consistent with these findings, perturbed spatial relationships between microbiota and host correlate with disease states such as inflammatory bowel disease (7). Nevertheless, little is known about the immune mechanisms that promote host-bacterial segregation and maintain homeostatic spatial relationships between host and microbiota.
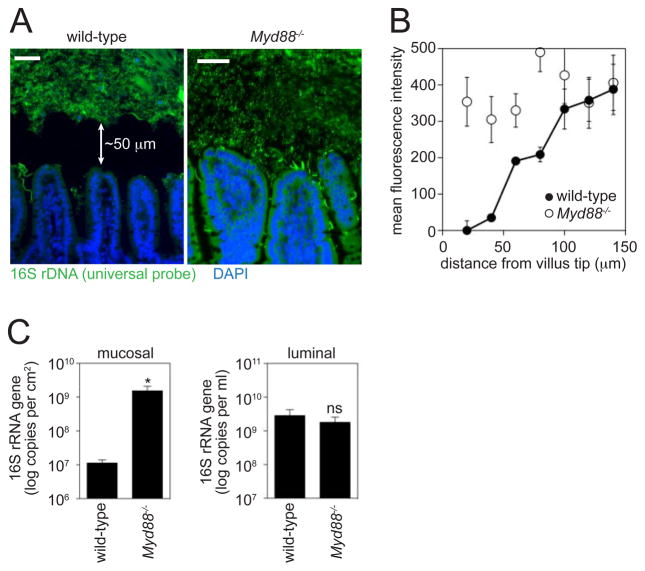
To discover immune mechanisms that restrict host-bacterial contact, we first characterized the location of the microbiota relative to host tissue in the mouse small intestine. In the colon, the physical separation of host and microbiota depends in part on the viscous gel-like mucus layer that covers the epithelium and inhibits bacterial access to the mucosal surface (5). In light of these studies, we prepared small intestines with Carnoy’s fixative (6), which preserves the mucus layer and maintains the native spatial relationships between bacteria and host tissue (7). When bacterial location was determined by fluorescence in situ hybridization (FISH) of a universal 16S rRNA gene probe, we observed that the majority of bacteria were not in direct contact with the intestinal surface. Rather, the microbiota were located ~50 μm from the villus tip (Fig. 1A,B, fig. S1), similar to the microbiota localization in the colon (5).
Fig. 1.
MyD88 promotes physical separation of the microbiota and the small intestinal surface. (A) Visualization of microbiota localization relative to the small intestinal mucosal surface by FISH. Sections were hybridized to a probe that recognizes the 16S rRNA genes of all bacteria (green), and counterstained with DAPI to visualize nuclei (blue). Scale bars=50 μm. Arrows indicate the distance from the villus tip to the microbiota. Mice were co-housed littermates from intercrossed Myd88+/− mice. Sections are representative of >10 groups of littermates. (B) Quantification of fluorescence intensity extending from the villus tip into the lumen (N=5 mice/genotype). (C) Mucosa-associated and luminal bacteria were quantified by Q-PCR determination of 16S rRNA gene copy number in the terminal ileum. N=5 mice/genotype. Data are from three groups of littermates. *, p<0.05; Error bars, ±SEM; ns, not significant.
Toll-like receptors (TLRs) are pattern recognition receptors that detect conserved molecular signatures of microorganisms and initiate immune responses. To explore the role of innate immunity in maintaining host-microbiota segregation, we studied mice lacking MyD88, a signaling adaptor for several TLRs. FISH analysis showed a marked increase in the number of bacteria in direct contact with the small intestinal epithelial surface in Myd88−/− mice as compared to co-housed wild-type littermates (Fig. 1A,B). We further measured bacterial loads in a culture-independent manner by quantitative PCR (Q-PCR) determination of total 16S rRNA gene copy number. When we quantified bacteria recovered from the intestinal surface, we detected ~100-fold higher mucosal bacterial loads in Myd88−/− mice as compared to wild-type littermates (Fig. 1C), consistent with the FISH analysis. In contrast, luminal bacterial loads were not significantly different between Myd88−/− and wild-type littermates (Fig. 1C), indicating that MyD88 does not regulate overall numbers of colonizing bacteria. Moreover, pyrosequencing of 16S rRNA genes in the terminal ileal lumen revealed no significant differences in luminal bacterial community composition between Myd88−/− and wild-type mice (fig. S2A,B). The increase in mucosa-associated bacteria was unlikely to result from a diminished mucus barrier, as Myd88−/− mice did not exhibit reduced mucus protein expression or secretion (fig. S3A,B). These results suggest that MyD88-dependent innate immune effectors limit the numbers of mucosal surface bacteria, maintaining a distinct physical separation between microbiota and host in the small intestine.
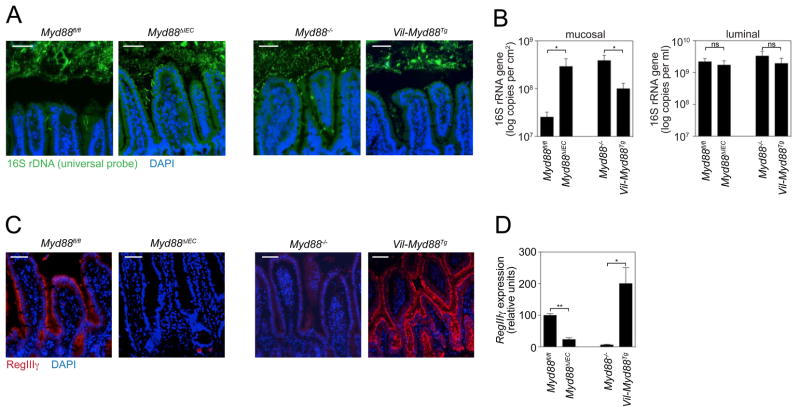
As a first step in identifying the MyD88-dependent mechanisms that promote host-microbial segregation, we investigated their cellular origin. We generated mice with an epithelial cell-specific deletion of Myd88 (Myd88 ΔIEC) by crossing mice carrying the intestinal epithelial cell (IEC)-restricted villin-Cre transgene (8) with mice harboring a loxP-flanked Myd88 allele (Myd88fl/fl)(9, 10). Myd88 ΔIEC mice lost the clear segregation between the microbiota and the mucosal surface that was readily apparent in their co-housed Myd88fl/fl littermates (Fig. 2A). This defect correlated with increased mucosal bacterial numbers as measured by Q-PCR (Fig. 2B). However, there were no statistically significant differences in luminal bacterial loads (Fig. 2B), consistent with our findings in whole-body Myd88−/− mice (Fig. 1C). Thus, epithelial cell Myd88 is required to limit bacterial colonization of the mucosal surface in the small intestine, but does not regulate overall numbers of colonizing bacteria.
Fig. 2.
Epithelial cell MyD88 is necessary and sufficient to limit bacterial association with the small intestinal surface. Myd88 ΔIEC and Vil-Myd88Tg mice were each analyzed alongside co-housed littermates (Myd88fl/fl and Myd88−/−, respectively). (A) FISH analysis of microbiota localization in terminal ileum using a universal bacterial 16S rRNA gene probe. Images are representative of >10 littermate groups. (B) Mucosa-associated and luminal bacteria were quantified by Q-PCR determination of 16S rRNA gene copy number in the terminal ileum. N=5 mice/genotype; data are from three littermate groups. (C) RegIIIγ was detected in distal small intestine with anti-RegIIIγ antiserum (13) (red). Nuclei were counterstained with DAPI (blue). Images are representative of three littermate groups. (D) Q-PCR quantification of RegIIIγ transcripts in terminal ileum (N=5 mice/genotype from three littermate groups). Scale bars=50 μm; *, p<0.05; **, p<0.01; Error bars, ±SEM; ns, not significant.
To determine whether epithelial cell Myd88 was also sufficient to maintain physical separation of host and microbiota, we first generated transgenic mice that expressed Myd88 under the control of the IEC-specific villin promoter. We then crossed this line onto the Myd88−/− background to produce mice with IEC-restricted Myd88 expression (Vil-MyD88Tg)(fig. S4). Vil-Myd88Tg mice exhibited restoration of the distinct zone separating the microbiota from host tissue (Fig. 2A), and had fewer mucosa-associated bacteria as compared to co-housed Myd88−/− littermates (Fig. 2B). There were no significant differences in overall numbers of luminal bacteria (Fig. 2B), supporting the idea that epithelial MyD88 does not impact luminal bacterial loads. We noted that the numbers of mucosa-associated bacteria in Vil-Myd88Tg mice were significantly greater (p=0.03) than in control Myd88fl/fl mice (Fig 2B). This could suggest a role for non-epithelial MyD88 signaling in limiting mucosa-associated bacteria. However, we cannot rule out the possibility that this difference arises from differences in microbiota composition rather than host genotype, as the comparison is between non-littermates that have not been co-housed. Overall, our findings show that epithelial cell Myd88 is both necessary and sufficient to limit bacterial colonization of the mucosal surface, establishing that the maintenance of homeostatic host-bacterial spatial relationships is an epithelial cell-intrinsic function.
A plausible mechanism by which intestinal epithelial cells could limit bacterial-mucosal contact is through the production of secreted antibacterial proteins. Consistent with this idea, MyD88 controls the expression of several key antimicrobial proteins in Paneth cells, a specialized epithelial lineage (11, 12). This group includes RegIIIγ, a directly bactericidal C-type lectin that specifically targets Gram-positive bacteria (13). RegIIIγ is expressed most prominently in the distal small intestine (ileum), where it is produced by multiple epithelial lineages including enterocytes and Paneth cells (11, 13). On the basis of these findings, we hypothesized that RegIIIγ might be one epithelial cell-derived MyD88-dependent factor that limits bacterial colonization of the mucosal surface. As an initial test of this idea we asked whether RegIIIγ expression correlates with the physical segregation of microbiota and host. Myd88 ΔIEC mice showed reduced expression of RegIIIγ mRNA and protein relative to Myd88fl/fl littermates, whereas Vil-Myd88Tg mice showed increased RegIIIγ expression relative to Myd88−/− littermates (Fig. 2C,D). Thus, epithelial cell-intrinsic MyD88 signaling controls RegIIIγ expression throughout the intestinal epithelium, and this expression correlates with the physical separation of the microbiota from the host mucosal surface.
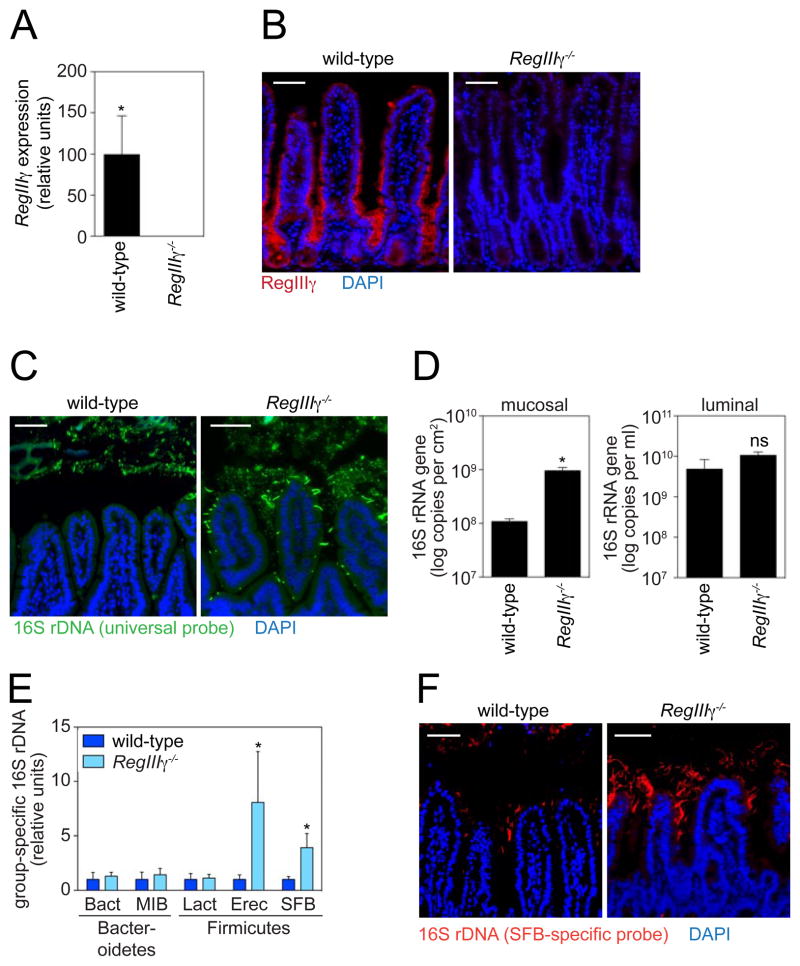
To directly assess the role of RegIIIγ in maintaining host-microbial homeostasis, we generated RegIIIγ−/− mice by gene targeting. We deleted a 2.6 kb region that encompasses the six RegIIIγ exons and then removed the neomycin cassette from the targeted allele (fig. S5A,B). RegIIIγ−/− mice were born at normal Mendelian ratios but were defective in production of RegIIIγ transcript and protein (Fig. 3A,B). The mice appeared healthy and showed no signs of enteropathy by histology (fig. S5C) or intestinal permeability measurement (fig. S5D). However, FISH analysis revealed perturbed spatial relationships between the microbiota and the host mucosal surface in RegIIIγ−/− mice, which exhibited a marked increase in numbers of mucosa-associated bacteria relative to co-housed wild-type littermates (Fig. 3C,D). In contrast, we did not detect significant differences in luminal bacterial numbers (Fig. 3D), consistent with our findings in Myd88−/− mice (Fig. 1C). We detected no defects in microbiota localization in the colons of RegIIIγ−/− mice (fig. S6A), which accords with the low colonic expression of RegIIIγ relative to small intestine (fig. S6B,C). Thus, RegIIIγ is a key MyD88-dependent mechanism that limits bacterial colonization of the small intestinal mucosal surface and maintains homeostatic spatial relationships between microbiota and host.
Fig. 3.
RegIIIγ limits mucosal surface association by Gram-positive bacteria. (A) Q-PCR analysis of RegIIIγ transcripts in terminal ileum of wild-type and RegIIIγ−/− mice. N=3 mice/group. (B) RegIIIγ was detected in distal small intestine using a polyclonal antibody raised against a peptide unique to RegIIIγ (red). (C) FISH analysis of wild-type and RegIIIγ−/− mice using a universal bacterial 16S rRNA gene probe (green). Mice were co-housed littermates from intercrossed RegIIIγ+/− mice. Images are representative of five littermate groups. (D) Quantification of total ileal mucosa-associated and luminal bacteria by Q-PCR determination of 16S rRNA gene copy number. N=5–7 mice/genotype from five littermate groups. (E) Q-PCR quantification of specific bacterial groups. Bacteria were recovered from the ileal mucosal surface. Values for each bacterial group are expressed relative to the 16S rDNA levels in wild-type mice. N=5–7 mice/genotype from five littermate groups. (F) Comparison of co-housed wild-type and RegIIIγ−/− littermates by FISH using an SFB-specific probe (red). Images are representative of five littermate groups. All tissues were counterstained with DAPI (blue). Scale bars=50 μm; *, p<0.05; Error bars, ±SEM; ns, not significant.
Because the antibacterial activity of RegIIIγ is restricted to Gram-positive bacteria in vitro (13), we predicted that RegIIIγ−/− mice would exhibit a preferential increase in the abundance of mucosa-associated Gram-positive bacteria. We therefore compared the abundance of specific bacterial groups in RegIIIγ−/− mice and their co-housed wild-type littermates by Q-PCR with 16S rRNA gene primers targeting specific bacterial groups. These included members of the Gram-negative Bacteroidetes phylum (Bacteroides and Mouse Intestinal Bacteroides [MIB] groups), and members of the Gram-positive Firmicutes phylum (Lactobacillus, Eubacterium rectale, and segmented filamentous bacteria [SFB] groups). Whereas RegIIIγ−/− mice harbored similar numbers of mucosa-associated bacteria belonging to the Gram-negative Bacteroidetes phylum, they showed increased numbers of bacteria belonging to the Gram-positive E. rectale and SFB groups (Fig. 3E). FISH analysis with an SFB-specific 16S rRNA gene probe corroborated the expansion of this bacterial group at the mucosal surface (Fig. 3F). In contrast, we detected no significant differences in abundances of key Gram-positive or Gram-negative groups when comparing luminal bacterial communities by Q-PCR (fig. S7). Furthermore, the luminal communities were similar at the phylum level when compared by pyrosequencing of 16S rRNA genes in the terminal ileal lumen (fig. S2A,B). These results establish that RegIIIγ limits associations between Gram-positive members of the microbiota and the small intestinal epithelial surface. Further, they reveal that RegIIIγ activity is restricted to the mucosal surface niche and has little impact on luminal bacteria, at least under co-housed conditions. The possibility remains that following separate caging of wild-type and RegIIIγ−/− mice, community changes at the mucosal surface could be propagated into the lumen over time.
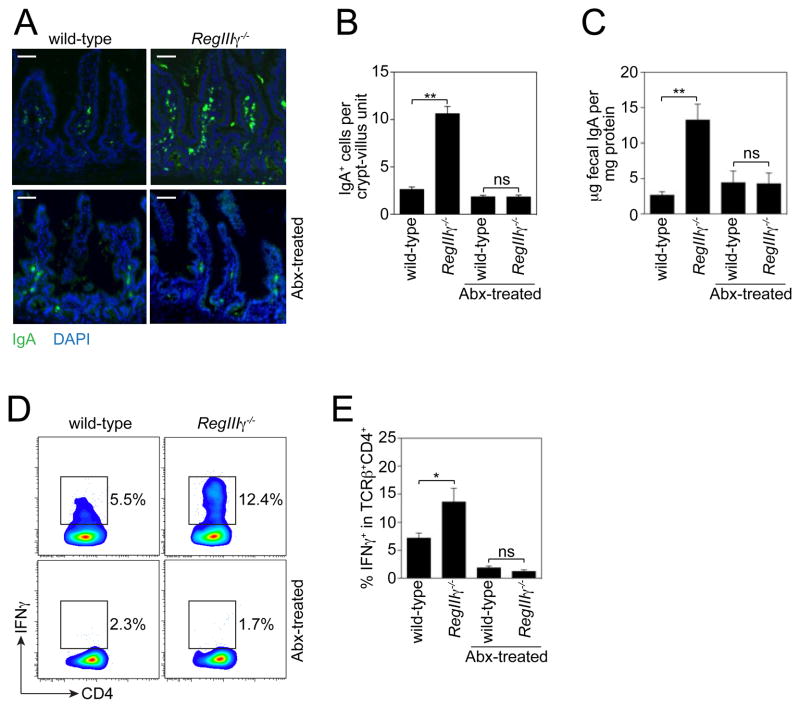
We next asked whether RegIIIγ −/− mice exhibit enhanced activation of adaptive immune responses as a consequence of the increased numbers of mucosa-associated bacteria. Immunoglobulin A (IgA) is a critical adaptive immune effector that is secreted into the intestinal lumen in large quantities, restricting bacterial penetration of mucosal tissues (14, 15). Immunofluorescence analysis revealed higher numbers of IgA+ cells in the small intestinal lamina propria of RegIIIγ−/− mice as compared to their co-housed wild-type littermates (Fig. 4A,B). This correlated with ~5-fold higher fecal IgA in RegIIIγ−/− mice (Fig. 4C). RegIIIγ−/− mice also exhibited increased frequencies of lamina propria CD4+ T helper (Th) cells that produced IFNγ (Th1 cells) (Fig. 4D,E). In contrast, we did not detect significant differences in the proportion of CD4+ T cells producing Foxp3 (Treg cells) or IL-17 (Th17 cells) (fig. S8A–C). This was initially surprising in light of reports that SFB induce Th17 cells in the lamina propria of the small intestine (2). However, we note that overall numbers of colonizing SFB did not differ markedly between co-housed RegIIIγ−/− and wild-type littermates (fig. S2A and S7), suggesting that Th17 cell frequencies may be more sensitive to overall SFB levels in the intestinal lumen than to numbers of mucosa-adherent SFB.
Fig. 4.
RegIIIγ−/− mice show increased activation of adaptive immunity. (A) Immunofluorescence detection of IgA+ cells in terminal ileum of RegIIIγ−/− mice and co-housed wildtype littermates. Images are representative of five littermate groups. (B) IgA+ cells were quantified by counting 10 well-oriented crypt-villus units in 10 mice/genotype from five littermate groups. (C) Total fecal IgA was determined by ELISA. N=5–7 mice per non-antibiotic-treated group from five littermate groups; N=3 mice per antibiotic-treated (Abx) group from two littermate groups. (D) Flow cytometric analysis of Th1 CD4+ T cell frequencies in RegIIIγ−/− mice. Small intestinal lamina propria cells were isolated from co-housed RegIIIγ−/− and wild-type littermates. Cells were gated on T cell receptor β chain (TCRβ) and CD4 (fig. S8A), and IFNγ+ cells were quantified as a percentage of this population. The IFNγ gate was determined based on an isotype control. Representative plots are shown. (E) IFNγ+ cells as a percentage of the TCRβ+CD4+ cell population. N=6 mice/group from three groups of littermates; representative of three independent trials. Scale bars=50 μm; *, p<0.05; **, p<0.01; Error bars, ±SEM; ns, not significant.
Finally, we established that the enhanced activation of adaptive immunity in RegIIIγ−/− mice was microbiota-dependent by depleting intestinal bacteria with broad-spectrum antibiotics. Both the enhanced IgA response and the Th1 response in RegIIIγ−/− mice disappeared with antibiotic treatment (Fig. 4A–E). This argues that the effects of RegIIIγ on adaptive immunity do not arise from direct effects of RegIIIγ on the adaptive immune system, but are instead a consequence of RegIIIγ’s impact on microbiota localization. Altogether, these results demonstrate that RegIIIγ restricts bacterial colonization of the intestinal epithelial surface and consequently limits activation of adaptive immune responses by the microbiota.
In conclusion, our findings provide essential insight into the immune mechanisms that promote host-bacterial mutualism by regulating the physical location of the microbiota. We propose that epithelial MyD88 and RegIIIγ are key components of an essential regulatory feedback loop that senses bacterial population density at or near the intestinal surface, with RegIIIγ expression activated as a mechanism to restrict numbers of surface-associated Gram-positive bacteria and limit activation of adaptive immunity. The mucosal niche-specific activity of RegIIIγ could be due to restricted diffusion of RegIIIγ through the mucus barrier, the result of specific binding interactions between RegIIIγ and mucus glycoproteins, or because of a requirement for environmental conditions that are specific to the mucosal surface niche. The mucosal surface specificity of RegIIIγ contrasts with the in vivo activities of the α-defensins, antibacterial proteins that are constitutively expressed (16) and regulate overall microbial community structure in the small intestine (17). We speculate that these niche-specific antibacterial functions could arise from size differences between the two protein families: α-defensins are considerably smaller (2–3 kDa) than RegIIIγ (15 kDa), and therefore may more readily diffuse through the mucus layer. The mucosal surface-specific function of RegIIIγ is consistent with the observation of enhanced Reg family expression in the mucosa of IBD patients exhibiting increased numbers of mucosal adherent bacteria (18). Further, RegIIIγ’s niche-specific antibacterial function may be critical for its role in limiting infection by antibiotic resistant Gram-positive bacteria, such as vancomycin-resistant enterococci (19). More generally, our findings raise the question of whether there are distinct antibacterial factors that maintain spatial homeostasis with Gram-negative bacteria.
Supplementary Material
Acknowledgments
We thank A. DeFranco for the Myd88fl/fl mice, R. Medzhitov for the FLAG-MyD88 transgene, and A. Mobley for assistance with flow cytometry. This work was supported by NIH R01 DK070855 (LVH), a Crohn’s and Colitis Foundation of America Fellowship Award (SV), the Human Microbiome Project Data Analysis and Coordination Center (OK), a Packard Fellowship in Science and Engineering (REL), a Hartwell Foundation Investigator Award (REL), a Beckman Young Investigator award (RL), a Burroughs Wellcome Foundation New Investigators in the Pathogenesis of Infectious Diseases Award (LVH), and the Howard Hughes Medical Institute (LVH). The data reported in this paper are tabulated in the main text and Supporting Online Material and the sequence data have been deposited in RG-RAST under project accession number qiime:135.
References and Notes
- 1.Turnbaugh PJ, et al. Nature. 2006;444:1027. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 2.Ivanov, et al. Cell. 2009;139:485. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atarashi K, et al. Science. 2011;331:337. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Round JL, et al. Science. 2011;332:974. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johansson ME, et al. Proc Natl Acad Sci USA. 2008;105:15064. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Materials and Methods are available as supporting material on Science online.
- 7.Swidsinski A, Weber J, Loening-Baucke V, Hale LP, Lochs H. J Clin Microbiol. 2005;43:3380. doi: 10.1128/JCM.43.7.3380-3389.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Madison BB, et al. J Biol Chem. 2002;277:33275. doi: 10.1074/jbc.M204935200. [DOI] [PubMed] [Google Scholar]
- 9.Hou B, Reizis B, DeFranco AL. Immunity. 2008;29:272. doi: 10.1016/j.immuni.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ismail AS, et al. Proc Natl Acad Sci USA. 2011;108:8743. doi: 10.1073/pnas.1019574108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Proc Natl Acad Sci USA. 2008;105:20858. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brandl K, Plitas G, Schnabl B, Dematteo RP, Pamer EG. J Exp Med. 2007;204:1891. doi: 10.1084/jem.20070563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Science. 2006;313:1126. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Macpherson AJ, et al. Science. 2000;288:2222. doi: 10.1126/science.288.5474.2222. [DOI] [PubMed] [Google Scholar]
- 15.Hapfelmeier S, et al. Science. 2010;328:1705. doi: 10.1126/science.1188454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Putsep K, et al. J Biol Chem. 2000;275:40478. doi: 10.1074/jbc.M007816200. [DOI] [PubMed] [Google Scholar]
- 17.Salzman NH, et al. Nat Immunol. 2010;11:76. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ogawa H, et al. Inflamm Bowel Dis. 2003;9:162. doi: 10.1097/00054725-200305000-00003. [DOI] [PubMed] [Google Scholar]
- 19.Brandl K, et al. Nature. 2008;455:804. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koren O, et al. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4592. doi: 10.1073/pnas.1011383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caporaso JG, et al. Nat Methods. 2010;7:335. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vijay-Kumar, et al. Science. 2010;328:228. [Google Scholar]
- 23.Szentkuti L, Enss ML. Comp Biochem Physiol A Mol Integr Physiol. 1998;119:379. doi: 10.1016/s1095-6433(97)00434-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.