Clostridium difficile infections (CDIs) have increased in incidence and severity within the past decade in North America and Europe (1), in large part because of the emergence of the hypervirulent North American pulsed-field type 1 (NAP1/027/III) strains (2–5). Recently, interest has increased in the ribotype 078 strain. A 2007 North American study showed that ribotype 078 strains predominated in swine and cattle (83%–94% prevalence), but were rare in a group of hospitalized persons (4% prevalence) (6). However, in studies from Europe and the United States, 078/V strains were found at a prevalence ranging from 3% to 11% (7–9). In a subsequent study by the US group, analysis of the toxinotype V strains from humans and food animals showed that 83% of strains were either NAP7 or NAP8 (10). A Dutch group has recently shown that 078/V strains increased from 3% to 13% during February 2005–2008 and can be considered hypervirulent (11). Our study aimed to determine the incidence rate of infections attributed to hypervirulent NAP7/078/V and NAP8/078/V strains of C. difficile in hospitals in Canada.
The Study
The Canadian Nosocomial Infection Surveillance Program is a collaborative effort between the Canadian Hospital Epidemiology Committee, a subcommittee of the Association of Medical Microbiology and Infectious Disease Canada, the Centre for Infectious Disease Prevention and Control, and the National Microbiology Laboratory of the Public Health Agency of Canada. The Canadian Nosocomial Infection Surveillance Program conducted prospective surveillance including collection of stool specimens from patients showing the presence of CDI during November 2004–April 2005 and during March and April in 2007 and 2008.
An infection was considered healthcare-associated CDI if the patient’s symptoms occurred at least 72 hours after hospital admission or if the symptoms resulted in readmission of a patient who had been hospitalized within the 2 months before the symptom onset date and who was not a resident in a long-term care facility or nursing home (12). An infection was considered community-onset CDI if the healthcare-associated definition was not met. Outcomes 30 days postinfection were recorded to capture severe cases, which were defined as infections in patients admitted to an intensive care unit, in patients who had undergone colectomy, or in patients who had died (12). Deaths were assessed by the Canadian Hospital Epidemiology Committee member and categorized into 3 groups: 1) death directly attributable to CDI, 2) death indirectly related to CDI by exacerbation of an existing disease condition, or 3) death not a result of CDI. The assessment was made from information obtained from medical charts, nurse logs, laboratory reports, and consultation with nursing and medical staff.
All stool specimens were cultured for C. difficile, and isolates were analyzed by PCR and pulsed-field gel electrophoresis (PFGE) at the National Microbiology Laboratory. PFGE, ribotyping, and toxinotyping were performed as described (10,11). MICs were determined by agar dilution or Etest. The primers used for PCR and sequencing are listed in Table 1. Macrorestriction patterns were analyzed with BioNumerics V4.5 (Applied Maths, Sint-Martens-Latem, Belgium).
Table 1. Primers used in study of hospitalized patients with Clostridium difficile infection, Canada, 2004–2008.
| Primer | Sequence (5′ → 3′) | Specificity |
|---|---|---|
| tcd3 | TGCAATTATAAAAACATCTTTAAAC | tcdC PaLoc negative regulator |
| tcd4 | TATATCTAATAAAAGGGAGATTG | |
| cdtB-F1 | TGGACAGGAAGAATAATTCCTTC | cdtB binary toxin subunit B |
| cdtB-R1 | TGCAACTAACGGATCTCTTGC | |
| E5 | CTCAAAACTTTTTAACGAGTG | ermB erythromycin/clindamycin resistance |
| E6 | CCTCCCGTTAAATAATAGATA | |
| GyrAF | TTGAAATAGCGGAAGAAATGA | gyrA DNA gyrase subunit A |
| GyrAR | TTGCAGCTGTAGGGAAATC | |
| GyrBF | GAAGGTCAAACTAAAACAAA | gyrB DNA gyrase subunit B |
| GyrBR | GGGCTCCATCTACATCG |
Fifteen NAP7 and 4 NAP8 patterns were identified from isolates obtained from 2,794 patients (overall prevalence 0.68%). Table 2 lists the patients and epidemiologic information, and the Figure shows the corresponding genomic fingerprint patterns. During the study period, the incidence rate increased as follows: 8/1,785 (0.5%) in 2004–2005; 5/638 (0.8%) in 2007; and 6/371 (1.6%) in 2008. Of the 19 patients identified, 14 were men with an average age of 70.8 years (not including 1 pediatric case), and 4 were women with an average age of 52.2 years; the overall average age was 61.5 years (Table 2). CDI was considered as community onset in 7 (37%) of 19 cases, and severe CDI was manifested in 3 (15.8%) case-patients (1 was healthcare-associated CDI and 2 were community-onset CDI). At 30 days postinfection for CDI, 26.3% of all patients had died, 1 death a direct result of CDI (5.3%), and 1 indirectly related; 10.6% of total deaths were attributable to CDI.
Table 2. Epidemiologic information from hospitalized patients with Clostridium difficile infection, Canada, 2004–2008*.
| Year and patient ID | Province | Age, y/sex | Source | Severe CDI† | Outcome‡ |
|---|---|---|---|---|---|
| 2004–2005 | |||||
| O1-0059 | Ontario | 62/M | Healthcare-associated | No | Discharged |
| O2-0053 | Ontario | 35/M | Community-onset | No | Died-not attrib |
| O3-0042 | Ontario | 64/F | Community-onset | No | Discharged |
| Q1-0028 | Quebec | 66/M | Healthcare-associated | Yes | Died-attrib |
| H1-0040 | Nova Scotia | 70/M | Healthcare-associated | No | Discharged |
| S1-0054 | Saskatchewan | 72/M | Community-onset | No | Discharged |
| S1-0063 | Saskatchewan | 82/M | Community-onset | Yes | Discharged |
| O7-0121 |
Ontario |
74/M |
Healthcare-associated |
No |
Survived-hosp |
| 2007 | |||||
| O1-7-0011 | Ontario | 87/M | Community-onset | No | Survived-hosp |
| O4-7-0011 | Ontario | 82/M | Community-onset | Yes | Died-contrib |
| Q1-7-0017 | Quebec | 40/F | Healthcare-associated | No | Discharged |
| O8B-7-0002 | Ontario | 65/M | Healthcare-associated | No | Died-not attrib |
| Q5-7-0013 |
Quebec |
71/M |
Healthcare-associated |
No |
Discharged |
| 2008 | |||||
| B1-8-0052 | British Columbia | 44/F | Healthcare-associated | No | Discharged |
| B1-8-0059 | British Columbia | 73/M | Healthcare-associated | No | Discharged |
| A3-8-0022 | Alberta | 38/F | Community-onset | No | Discharged |
| O2B-8-0015 | Ontario | 75/F | Community-onset | No | Survived-hosp |
| Q1-8-0008 | Quebec | 81/M | Healthcare-associated | No | Died-not attrib |
| O5-8-0001 | Ontario | 2/M | Healthcare-associated | No | Discharged |
*ID, identification; CDI, Clostridium difficile infection; Died-not attrib, death not attributable to CDI; Died-attrib, death directly attributable to CDI; Survived-hosp, patient survived but was still in a hospital at endpoint; Died-contrib, CDI indirectly contributed to death. †Required admission to intensive care unit due to CDI, received a colectomy, or died. ‡At 30 days after diagnosis of CDI.
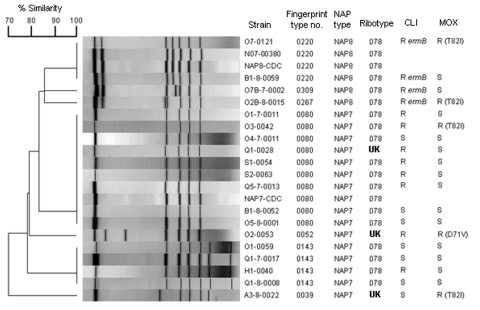
Figure.
Dendrogram analysis of macrorestriction patterns (SmaI) of the NAP7 and NAP8 Clostridium difficile strains isolated from the patients listed in Table 2. C. difficile N07-00380 is a ribotype 078 control strain. C. difficile NAP7-CDC and NAP8-CDC control strains are toxinotype V. Isolates exhibiting high-level clindamycin resistance (>256 μg/mL) and harboring ermB are indicated. The amino acid change found in the gyrA protein is shown for the moxifloxacin-resistant strain antimicrobial drug–resistance mechanisms.
Sequence analysis of the tcdC gene showed that all strains carried a C184T transition that introduces a stop codon leading to a presumptive truncated protein of 61 residues, and a 39-bp deletion located downstream of the alternative stop codon. This tcdC variant has been previously described for toxinotype V strains (13). Sixteen of the isolates were ribotype 078 and 3 isolates had unknown ribotypes. All 2004/2005 and 2007 isolates were toxinotype V. The 2008 isolates were not toxinotyped. All 19 strains were susceptible to metronidazole and vancomycin. Seven isolates were susceptible to clindamycin (MIC <8 μg/mL) and 12 were resistant (6 had MICs = 8, 2 had MICs = 16, and 4 had MICs >256). Only the 4 latter strains carried ermB and all were NAP8. Fourteen isolates that were susceptible to moxifloxacin (MIC <8 μg/mL) had identical gyrA and gyrB quinolone-resistance–determining regions (QRDR) sequences to the genes in C. difficile 630 (GenBank accession no. AM180355). Five moxifloxacin-resistant isolates (MIC > 8 μg/mL) had no mutations in the gyrB QRDR but each had 1 mutation in the gyrA QRDR. One with MIC = 8 had an Asp71Val mutation; 3 with MIC = 16 and 1 with MIC >32 had a Thr82Ile mutation. These mutations have been previously described in moxifloxacin-resistant C. difficile (14).
Conclusions
C. difficile NAP7 and NAP8/078/V strains are relatively rare in hospitalized patients with CDI in Canada, in contrast to their prevalence in Europe and the United States (7–11). However, incidence rates have tripled from 0.5% in 2004 to 1.6% in 2008 (p = 0.22). There was a high association with a community onset, although dataset was too small to statistically confirm that increased cases were more likely to be community onset; 2 (40%) of 5 deaths were attributable to CDI. Although the number of strains studied here was small, data are consistent with other studies that indicate a community association for NAP7 and NAP8/078/V strains (9–11). The prevalence of these strains in Canada may be higher than suggested here if they are a common cause of community-associated CDI, as studies have indicated (10,11). The role of animals in acquisition of NAP7 and NAP8/078/V strains was not evaluated because animal and food contact data were not available.
Molecular typing of C. difficile is typically performed by using ribotyping in Europe and PFGE/macrorestriction analysis in North America; both groups may use toxinotyping, which strictly looks at PaLoc variation. We showed a high correlation between NAP7, NAP8, ribotype 078, and toxinotype V strains by the 3 typing methods, which enabled results of separate studies to be compared. Furthermore, tcdC analysis provides an additional diagnostic tool for these strains because the gene has a 39-bp deletion and a C184T-transition in all isolates we studied.
Continued surveillance is warranted in humans, animals, and retail meat to determine whether NAP7 and 8/078/V strains will continue to emerge in patients hospitalized in Canada and to determine whether the sources of these infections are related to animals or food. Surveillance is especially important given that these strains appear to be hypervirulent as has been reported for NAP1/027/III strains (11).
Acknowledgments
We gratefully thank Krista Wilkinson for critical reading of the manuscript. Expert technical assistance was provided by Romeo Hizon, Tim Du, and Stuart McCorrister. C. difficile isolates representing NAP7 and NAP8 were kindly provided by B. Limbago (Centers for Disease Control and Prevention, Atlanta, GA, USA).
Dr Mulvey is chief of the Antimicrobial Resistance and Nosocomial Infections Section of the National Microbiology Laboratory of the Public Health Agency of Canada. His research interests include the molecular epidemiology of antimicrobial-resistant bacterial pathogens.
Footnotes
Suggested citation for this article: Mulvey MR, Boyd DA, Gravel D, Hutchinson J, Kelly S, McGeer A, et al. Hypervirulent Clostridium difficile strains in hospitalized patients, Canada. Emerg Infect Dis [serial on the Internet]. 2010 Apr [date cited]. http://dx.doi.org/10.3201/eid1604.091152
Parts of this study were presented at the 48th Interscience Conference on Antimicrobial Agents and Chemotherapy/46th Infectious Disease Society of America meeting in Washington DC, USA, October 25–28, 2008.
Members of the Canadian Nosocomial Infection Surveillance Program who participated in the surveillance for C. difficile infection: Elizabeth Bryce, Vancouver General Hospital, Vancouver, British Columbia; John Conly, Foothills Medical Centre, Calgary, Alberta; John Embil, Health Sciences Centre, Winnipeg, Manitoba; Joanne Embree, Health Sciences Centre, Winnipeg; Sarah Forgie, Stollery Children’s Hospital, Edmonton, Alberta; Charles Frenette, McGill University Health Centre, Montreal, Quebec; Camille Lemieux, University Health Network, Toronto, Ontario; Elizabeth Henderson, Peter Lougheed Centre, Calgary; Michael John, London Health Sciences Centre, London, Ontario; Lynn Johnston, QEII Elizabeth Health Sciences Centre, Halifax, Nova Scotia; Pamela Kibsey, Victoria General Hospital, Victoria, British Columbia; Joanne Langley, IWK Health Centre, Halifax; Mark Loeb, Hamilton Health Sciences Corporation, Hamilton, Ontario; Anne Matlow, Hospital for Sick Children, Toronto; Sophie Michaud, CHUS-Hôpital Fleurimont, Sherbrooke, Quebec; Marianne Ofner, Centre for Communicable Diseases and Infection Control, Public Health Agency of Canada, Ottawa, Ontario; Virginia Roth, The Ottawa Hospital, Ottawa; Eva Thomas, Children’s and Women’s Health Center, Vancouver; William Thompson, South East Regional Health Authority, Moncton, New Brunswick; Nathalie Turgeon, Hôtel-Dieu de Québec du CHUQ, Quebec, Quebec; Mary Vearncombe, Sunnybrook Health Sciences Centre, Toronto; Karl Weiss, Maisonneuve-Rosemont Hospital, Montreal; Alice Wong, Royal University Hospital, Saskatoon, Saskatchewan; Dick Zoutman, Kingston General Hospital, Kingston, Ontario.
To determine the incidence rate of infections with North American pulsed-field types 7 and 8 (NAP7/NAP8) strains of Clostrodium difficile, ribotype 078, and toxinotype V strains, we examined data collected for the Canadian Nosocomial Infections Surveillance Program (CNISP) CDI surveillance project during 2004–2008. Incidence of human infections increased from 0.5% in 2004/2005 to 1.6% in 2008.
References
- 1.Kelly CP, LaMont JT. Clostridium difficile—more difficult than ever. N Engl J Med. 2008;359:1932–40. 10.1056/NEJMra0707500 [DOI] [PubMed] [Google Scholar]
- 2.McDonald LC, Killgore GE, Thompson A, Owens RC Jr, Kazakova SV, Sambol SP, et al. An epidemic, toxin gene-variant strain of Clostridium difficile. N Engl J Med. 2005;353:2433–41. 10.1056/NEJMoa051590 [DOI] [PubMed] [Google Scholar]
- 3.Loo VG, Poirier L, Miller MA, Oughton M, Libman MD, Michaud S, et al. A predominantly clonal multi-institutional outbreak of Clostridium difficile–associated diarrhea with high morbidity and mortality. N Engl J Med. 2005;353:2442–9. 10.1056/NEJMoa051639 [DOI] [PubMed] [Google Scholar]
- 4.Warny M, Pepin J, Fang A, Killgore G, Thompson A, Brazier J, et al. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet. 2005;366:1079–84. 10.1016/S0140-6736(05)67420-X [DOI] [PubMed] [Google Scholar]
- 5.MacCannell DR, Louie TJ, Gregson DB, Laverdiere M, Labbe A-C, Laing F, et al. Molecular analysis of Clostridium difficile ribotype 027 isolates from eastern and western Canada. J Clin Microbiol. 2006;44:2147–52. 10.1128/JCM.02563-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keel K, Brazier S, Post KW, Weese S, Songer JG. Prevalence of PCR ribotypes among Clostridium difficile isolates from pigs, calves, and other species. J Clin Microbiol. 2007;45:1963–4. 10.1128/JCM.00224-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goorhuis A, Debast SB, van Leengoed LAMG, Harmanus C, Notermans DW, Bergwerff AA, et al. Clostridium difficile PCR ribotype 078: an emerging strain in humans and in pigs? J Clin Microbiol. 2008;46:1157–8. 10.1128/JCM.01536-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rupnik M, Widmer A, Zimmermann O, Eckert C, Barbut F. Clostridium difficile toxinotype V, ribotype 078, in animals and humans. J Clin Microbiol. 2008;46:1963–4. 10.1128/JCM.00598-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Limbago B. Long CM, Thompson AD, Killgore GE, Hannett G, Havill N, et al. Isolation and characterization of Clostridium difficile responsible for community-associated disease. In: Abstracts of the Second International Clostridium difficile Symposium, Maribor, Slovenia, June 6–9, 2007. [cited 2009 Jun 10]. http://clostridia.net/ICDS.htm
- 10.Jhung MA, Thompson AD, Killgore GE, Zukowski WE, Songer G, Warny M, et al. Toxinotype V Clostrdium difficile in humans and food animals. Emerg Infect Dis. 2008;14:1039–45. 10.3201/eid1407.071641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goorhuis A, Bakker D, Corver J, Debast SB, Harmanus C, Notermans DW, et al. Emergence of Clostridium difficile infection due to a new hypervirulent strain, polymerase chain reaction ribotype 078. Clin Infect Dis. 2008;47:1162–70. 10.1086/592257 [DOI] [PubMed] [Google Scholar]
- 12.Gravel D, Miller M, Simor A, Taylor G, Gardam M, McGeer A, et al. Health-care associated Clostridium difficile infection in adults admitted to acute care hospitals in Canada: a Canadian Nosocomial Infection Surveillance Program study. Clin Infect Dis. 2009;48:568–76. 10.1086/596703 [DOI] [PubMed] [Google Scholar]
- 13.Spigaglia P, Mastrantonio P. Molecular analysis of the pathogenicity locus and polymorphism in the putative negative regulator of toxin production (TcdC) among Clostridium difficile clinical isolates. J Clin Microbiol. 2002;40:3470–5. 10.1128/JCM.40.9.3470-3475.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dridi L, Tankovic A, Burghoffer B, Barbut F, Petit J-C. gyrA and gyrB mutations are implicated in cross-resistance to ciprofloxacin and moxifloxacin in Clostridium difficile. Antimicrob Agents Chemother. 2002;46:3418–21. 10.1128/AAC.46.11.3418-3421.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]