Abstract
During the 2007 equine influenza outbreak in Australia, respiratory disease in dogs in close contact with infected horses was noted; influenza (H3N8) virus infection was confirmed. Nucleotide sequence of the virus from dogs was identical to that from horses. No evidence of dog-to-dog transmission or virus persistence in dogs was found.
Keywords: Canine influenza, equine influenza, dogs, horses, respiratory disease, influenza, viruses, dispatch
Respiratory disease in dogs caused by type A influenza virus was first noted in racing greyhounds in Florida in January 2004 (1). This subtype H3N8 virus has a presumptive but unidentified equine origin. The geographic extent of infection in racing greyhounds and in pet dogs suggest that this virus has become enzootic to the United States (1,2).
In the United Kingdom, pneumonia in dogs and influenza (H3N8) virus have been retrospectively linked, and subtype H3N8 infections have been identified serologically in dogs likely to have been in close contact with horses during the 2003 outbreak of equine influenza (3,4). A 78-bp segment of the hemagglutinin (HA) gene identified in dogs with pneumonia had complete homology with local equine strains (3). Unlike the situation in the United States, no evidence of continuing circulation of an influenza virus of equine origin in the canine population has been found in the United Kingdom.
In Australia, in late 2007, an outbreak of equine influenza virus (EIV) infection occurred in horses. During this outbreak, respiratory disease was noted in dogs of various ages and breeds that were kept near infected horses. Investigations were undertaken to exclude influenza virus infection.
The Study
The first reported case was in a dog near a large stable; the dog became inappetant and lethargic and had had a slight nasal discharge and a persistent cough for several days. Over the next 2–3 weeks, dogs in or near stables with infected horses, including dogs whose owners were handling infected horses or dogs (n = 6) that were only housed with infected dogs, were examined. Samples were also collected from dogs kept with horses at 5 other locations 20–60 km from the first case. Of the 40 dogs, examined, 10 had clinical signs consistent with influenza (anorexia, lethargy, and, for some, a harsh cough that persisted for several weeks). All affected dogs recovered.
Nasal swabs and serum were collected from each of the 40 dogs; 23 were seropositive according to influenza type A blocking ELISA (5) and hemagglutinin inhibition (HI) assay (5) using A/equine/Sydney/2007 virus as antigen (Table). HI titers were 16–256 (geometric mean 122). Results were discordant for 5 dogs: for 2, HI titer was high but ELISA results were negative; for 3, ELISA results were positive but HI titer was negative. These discrepancies may have been resolved had later sampling been possible. Convalescent-phase serum samples were collected 14–16 days later from 26 of the dogs; seroconversion was noted for 4 of the 5 dogs with discordant ELISA and HI results. Testing of 19 dogs 2 years later showed no change in HI titer, although ELISA results were negative for each. Each seropositive dog had been in close proximity to EIV-infected horses but not always in direct contact. No evidence of lateral transmission was found for dogs that did not have contact with horses.
Table. Clinical signs and serologic findings for 40 dogs exposed to equine influenza virus, Australia, October 2007*.
| Dog | Breed | Age/sex | Clinical signs | Day of sample collection |
PCR | Titer |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ELISA |
HI assay |
||||||||||||
| 1st | 2nd | 1st | 2nd | 3rd† | 1st | 2nd | 3rd† | ||||||
| 1 | Cattle dog x | 6 mo/F | Cough | 5 | NS | Neg | 70 | NA | NA | 64 | NA | NA | |
| 2 | Whippet | 6 mo/F | Cough, inappetance, lethargy, nasal discharge | 5 | 25 | Neg | 79 | 69 | Neg | 128 | 128 | 128 | |
| 3 | Rottweiler | UK | Cough, lethargy | 10 | 25 | Neg | Neg | 65 | Neg | Neg | Neg | Neg | |
| 4 | Dalmation | 3 y/MN | Cough, inappetance, lethargy | 11 | NS | Neg | 74 | NA | NA | 64 | NA | NA | |
| 5 | Kelpi x | 9.5 y/FN | Cough, lethargy | 12 | 26 | Neg | 66 | 74 | NA | 64 | 128 | NA | |
| 6 | Border collie | 5 y/MN | Inappetance, lethargy | 13 | 27 | Neg | 60 | 75 | Neg | 64 | 32 | 64 | |
| 7 | Cattle dog | 4.5 y/M | Cough, lethargy | 13 | 27 | Neg | 64 | 57 | Neg | 256 | 128 | 128 | |
| 8 | German shepherd | 9 y/M | Inappetance, lethargy | 14 | 30 | Neg | Neg | Neg | NA | Neg | Neg | NA | |
| 9 | Jack Russell | 9 y/MN | Cough | 14 | 30 | Neg | Neg | 65 | NA | Neg | Neg | NA | |
| 10 | Lowchen | 2 y/MN | Cough, inappetance, lethargy, nasal discharge | 26 | NS | Neg | Neg | NA | Neg | Neg | NA | Neg | |
| 11 | Cattle dog x | 18 mo/F | None | 10 | 26 | Neg | 77 | 75 | NA | 64 | 64 | NA | |
| 12 | Fox terrier x poodle | 15 y/FN | None; lived with dog 4 | 11 | NS | Neg | Neg | Neg | NA | Neg | Neg | NA | |
| 13 | Kelpie x | 4 y/MN | None | 12 | 26 | Neg | 64 | 71 | Neg | 128 | 64 | 128 | |
| 14 | German shepherd | 1.5 y/MN | None | 12 | 26 | Neg | 71 | 67 | Neg | 128 | 64 | 64 | |
| 15 | Kelpie x labrador | 10 y/FN | None | 12 | 26 | Neg | 71 | 80 | Neg | 256 | 32 | Neg | |
| 16 | Cattle x kelpie | 3 y/MN | None | 12 | 27 | Neg | 64 | 61 | Neg | 128 | 32 | 64 | |
| 17 | Unknown | UK | None | 12 | 27 | Neg | 66 | 66 | Neg | 256 | 64 | 32 | |
| 18 | Border collie | 1 y/M | None | 12 | 27 | Neg | 78 | 59 | Neg | 128 | 64 | 128 | |
| 19 | Jack Russell x | 2.5 y/MN | None | 12 | 26 | Neg | 49 | 71 | NA | 32 | 16 | NA | |
| 20 | Greyhound | 18 mo/F | None | 12 | 26 | Neg | Neg | Neg | NA | 256 | 128 | NA | |
| 21 | Greyhound | 2 y. M | None | 12 | 26 | Neg | Neg | 55 | NA | Neg | Neg | NA | |
| 22 | Greyhound | 5 y/M | None | 12 | 25 | Neg | Neg | Neg | NA | Neg | Neg | NA | |
| 23 | Greyhound | 18 mo/F | None | 12 | 25 | Neg | Neg | Neg | NA | Neg | Neg | NA | |
| 24 | Greyhound | 18 m/F | None | 12 | 25 | Neg | Neg | Neg | NA | Neg | Neg | NA | |
| 25 | Cattle x kelpie | 4 y/FN | None | 12 | 26 | Neg | 51 | 73 | Neg | 128 | 64 | 64 | |
| 26 | Poodle | 5 mo/M | None, lived with dog 7 | 13 | 27 | Neg | Neg | Neg | Neg | 8 | Neg | Neg | |
| 27 | Jack Russell | 5 y/F | None, lived with dog 5 | 13 | 27 | Neg | Neg | Neg | NA | Neg | Neg | NA | |
| 28 | Cattle x hunterway | 4.5 y/M | None | 14 | 30 | Pos | Neg | 50 | NA | Neg | 64 | NA | |
| 29 | Border collie | 4 y/FN | None | 15 | NS | Neg | Neg | NA | NA | Neg | NA | NA | |
| 30 | Cattle | 13 y/MN | None | 15 | NS | Neg | Neg | NA | NA | >32 | NA | NA | |
| 31 | Jack Russell | UK | None | 15 | NS | Neg | 76 | NA | NA | 64 | NA | NA | |
| 32 | Rottweiler | UK | None | 15 | NS | Neg | 76 | NA | Inc | 128 | NA | 128 | |
| 33 | Fox terrier | 4 y/MN | None | 15 | NS | Neg | 78 | NA | NA | 32 | NA | NA | |
| 34 | Labrador | 13 y/FN | None | 15 | NS | Neg | 51 | NA | NA | 64 | NA | NA | |
| 35 | x | UK | None | 18 | NS | Neg | Neg | NA | Neg | Neg | NA | Neg | |
| 36 | x | UK | None | 18 | NS | Neg | 73 | NA | Neg | 64 | NA | 64 | |
| 37 | x | UK | None | 18 | NS | Neg | Neg | NA | Neg | Neg | NA | Neg | |
| 38 | x | UK | None | 18 | NS | Neg | Neg | NA | NA | Neg | NA | NA | |
| 39 | x | UK | None | 18 | NS | Neg | 67 | NA | Neg | 256 | NA | 128 | |
| 40 | Poodle x | 9 y/UK | None, lived with dog 10 | 26 | NS | Neg | Neg | NA | Neg | Neg | NA | Neg | |
*HI, hemagglutination inhibition; x, cross-breed; NS, not sampled; neg, negative; NA, not applicable; pos, positive; UK, unknown; MN, male neutered; FN, female neutered. †Samples collected ≈2 y after second sample.
Nasal swabs from 1 clinically healthy dog had a positive result in an influenza A real-time reverse transcription–PCR assay (5) on 2 consecutive days. The dog remained clinically healthy and was seropositive (titer 64) on day 16 after the first positive swab was collected. Attempts to isolate virus from these swabs were unsuccessful.
Nucleic acid sequencing was conducted for the HA, neuraminidase (NA), and matrix (M) genes amplified by PCR from the RNA purified from 2 samples from this dog (A/canine/Sydney/6525/2007 and A/canine/Sydney/6692/2007) and from a nasal swab from an infected horse (A/equine/Sydney/6085/2007) in the same stable (GenBank accession nos. GU045761–GU045769). Sequences were aligned with representative sequences from GenBank by using Clustal W (www.clustal.org/) before phylogenetic trees with bootstrapping were generated (n = 1,000; random seed n = 111) with MegAlign (Lasergene; DNAStar, Madison, WI, USA). Complete nucleotide homology was found for each of the HA, NA, and M gene sequences from the 2 dogs and the sequence from the infected horse in the same stable (A/equine/Sydney/6085/2007).
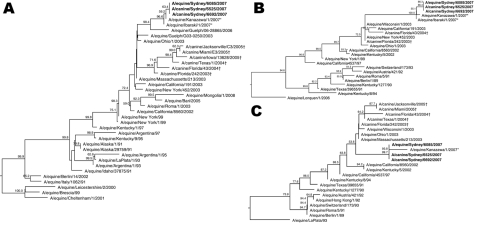
When influenza subtype H3N8 sequences from horses and dogs were compared with other subtype H3N8 sequences in GenBank, the HA, NA, and M sequences were most similar to strains A/equine/Kanazawa/1/2007 and A/equine/Ibaraki/1/2007, which were isolated during the 2007 equine influenza outbreak in Japan (Figure). The HA, NA, and M gene sequences from the dogs in Australia were positioned on separate clades of the phylogenetic trees, as opposed to those from subtype H3N8 viruses from dogs in the United States, which all grouped closely together (Figure).
Figure.
Phylogenetic trees of influenza subtype H3N8 viruses showing analyses conducted on A) hemagglutinin genes, B) neuraminidase genes, and C) matrix genes. Sequences from dogs are from the same animal on successive days. Boldface indicates viruses identified in dogs and horses in Australia, 2007, asterisks (*) indicate viruses from horses in Japan, and daggers (†) indicate viruses from dogs in the United States. Bootstrap values >50 are indicated at branch nodes. Bootstrap trials = 1,000; seed = 111.
Conclusions
Researchers in Japan have described transmission of EIV from 3 experimentally infected horses to 3 dogs individually housed with each horse (6). Their findings were mostly consistent with ours, but there were some differences. Both studies showed direct linkage between active influenza virus infection in dogs and horses. Because some naturally infected dogs were only in the vicinity of stables and not in direct contact with horses, we believe that EIV may be readily transmitted from horses to dogs in close proximity. The mechanism of spread remains unclear, although in the United Kingdom aerosol transmission was believed to be a major means of spread to dogs (4). Studies conducted during the equine influenza outbreak in Australia indicate that the levels of virus excretion from horses not previously exposed to the virus can be extremely high (A.J. Read et al., unpub. data). Although humans readily spread virus from horse to horse, either directly during handling or by fomite transmission, human transmission of EIV to dogs that were not in the immediate vicinity of infected horses was not found. Similarly, dog-to-dog transmission was not found when infected dogs were transported and kept with other dogs in urban locations where there was no opportunity for contact with horses.
Although clinical signs were not observed for any of the dogs in Japan, >35% of the naturally infected dogs in Australia exhibited clinical signs, some quite severe and protracted. Nevertheless, virus was rarely detected in nasal secretions of the dogs in Australia, and there was no evidence of horizontal transmission to other dogs. The lack of clinical signs in experimentally infected dogs may be because of the small numbers of dogs or because of inoculum attenuation after passage in embryonated chicken eggs. That the experimentally infected dogs in Japan also had lower HI titers than did naturally infected dogs may be relevant.
Finally, when 19 of the dogs in Australia were tested 2 years after infection and without opportunity for reexposure, with only 1 exception, the HI antibody titers had not changed. This finding supports the interpretation that antibodies detected in dogs in the United Kingdom (3,4) had been acquired during the equine influenza outbreak several years earlier.
The nucleotide gene sequences encoding the 2 surface proteins (HA and NA) and the M protein from the infected dog in Australia matched those from the horse with which it had contact and did not have any of the nucleotide changes that have been identified in viruses from dogs in the United States (2). Such changes may be critical to, or a consequence of, the adaptation of EIVs to dogs and may play a role in enhancing the infectivity of these viruses for dogs because there is no evidence of continuing circulation of virus in dogs in Australia.
Acknowledgments
We are indebted to the staff of the Virology Laboratory for technical support and to the NSW Department of Primary Industries for funding. The Melbourne WHO Collaborating Centre for Reference and Research in Influenza is supported by the Australian Government Department of Health and Ageing.
Biography
Dr Kirkland is a veterinary virologist and head of the virology laboratory at the Elizabeth Macarthur Agricultural Institute. His research interests include the epidemiology and pathogenesis of viral diseases of animals, development of rapid diagnostic assays, and new and emerging diseases.
Footnotes
Suggested citation for this article: Kirkland PD, Finlaison DS, Crispe E, Hurt AC. Influenza virus transmission from horses to dogs, Australia. Emerg Infect Dis [serial on the Internet]. 2010 Apr [date cited]. http://dx.doi.org/10.3201/eid1604.091489
References
- 1.Crawford PC, Dubovi EJ, Castleman WL, Stephenson I, Gibbs EP, Chen L, et al. Transmission of equine influenza virus to dogs. Science. 2005;310:482–5. 10.1126/science.1117950 [DOI] [PubMed] [Google Scholar]
- 2.Payungporn S, Crawford PC, Kouo TS, Chen L, Pompey J, Castleman WL, et al. Influenza A virus (H3N8) in dogs with respiratory disease, Florida. Emerg Infect Dis. 2008;14:902–8. 10.3201/eid1406.071270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daly JM, Blunden AS, MacRae S, Miller J, Bowman SJ, Kolodziejek J, et al. Transmission of equine influenza to English foxhounds. Emerg Infect Dis. 2008;14:461–4. 10.3201/eid1403.070643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Newton R, Cooke A, Elton D, Bryant N, Rash A, Bowman S, et al. Canine influenza: cross-species transmission from horses. Vet Rec. 2007;161:142–3. [DOI] [PubMed] [Google Scholar]
- 5.Selleck PW, Kirkland PD. 2009. Avian influenza. In: Australian and New Zealand standard diagnostic procedures for animal diseases, Sub-Committee on Animal Health Laboratory Standards for Animal Health Committee, Australia [cited 2010 Feb 14]. http://www.scahls.org.au
- 6.Yamanaka T, Nemoto M, Tsujimura K, Kondo T, Matsumura T. Interspecies transmission of equine influenza virus (H3N8) to dogs by close contact with experimentally infected horses. Vet Microbiol. 2009;139:351–5. 10.1016/j.vetmic.2009.06.015 [DOI] [PubMed] [Google Scholar]