To the Editor: During March and early April 2009, a new swine-origin influenza A (H1N1) virus emerged in Mexico and the United States; this virus subsequently spread across the globe by human-to-human transmission at an unprecedented rate. Pandemic (H1N1) 2009 virus also affected pigs. On May 2, 2009, the Canadian Food Inspection Agency notified the World Organisation for Animal Health that the novel influenza A virus had been confirmed on a pig farm in Alberta, Canada. Infection of pigs with pandemic (H1N1) 2009 virus has been observed in multiple countries (1). In this study, we report transmission of pandemic (H1N1) 2009 virus from humans to pigs in the People’s Republic of China.
During August 2009–April 2010, swine influenza virus surveillance was conducted in the provinces of central and eastern China, including Henan, Hubei, Hunan, Jiangxi, and Anhui. A total of 1,021 samples, comprising tracheal mucus swabs and lungs, from pigs on 30 farms, were collected, and dozens of swine influenza viruses, H1N1, H1N2, H3N8, H9, and H10 subtypes, were isolated. Eight isolates were subtype H1N1, including 4 novel pandemic (H1N1) 2009 viruses: A/swine/Nanchang/3/2010 (H1N1) (GenBank accession nos. JF275917–24), A/swine/Nanchang/5/2010 (H1N1) (GenBank accession nos. JF275933–40) and A/swine/Nanchang/6/2010 (H1N1) (GenBank accession nos. JF275941–48), which were isolated from tracheal mucus, and A/swine/Nanchang/F9/2010 (H1N1) (GenBank accession nos. JF275925–32), isolated from the lung. Pigs from which the novel viruses were isolated showed mild respiratory signs, including depression, cough, and transient increase in body temperature. Compared with the sequence of A/California/04/2009 (H1N1), genomic sequencing of the 4 pandemic viruses showed 15 common point mutations, such as polymerase basic protein 2, T588I; polymerase acidic protein, A70V, P224S, D547E; hemagglutinin, P100S, D103E, S145P, T214A, S220T, I338V; nucleocapsid protein, V100I, H289Y; neuraminidase, V106I, N248D; and nonstructural protein, I123V.
Recent studies have shown that the novel pandemic (H1N1) 2009 human influenza viruses were almost avirulent for mice (50% mouse lethal dose >106 PFU for A/CA/04/09 [2,3]). In this study, mice were anesthetized with ketamine/xylazine, as described (4) and 50 μL of phosphate-buffered saline containing the indicated doses (105 50% egg infectious dose) of the 4 viruses were instilled into anesthetized mice through nostrils. Interestingly, the 4 pandemic (H1N1) 2009 viruses isolated from pigs could cause systemic infection on mice. All mice had extensive loss of body weight, and some of the infected mice died within 2 weeks postinfection (data not shown).
To detect whether the 4 isolated pandemic (H1N1) 2009 viruses could cause clinical diseases in pigs and what kinds of pathologic damage could be induced in infected pigs, pigs were intratracheally challenged with the 4 pandemic (H1N1) 2009 viruses at a dose of 107 50% egg infectious dose and monitored for clinical signs and pathologic changes. Clinical signs in pigs were mild, and no deaths occurred. Except for slightly labored breathing, no infected pigs showed dominant signs, such as cough, nasal discharge, facial edema, and dyspnea. Body temperatures of infected pigs started to increase at 2 days postinfection (dpi), peaked ≈3 dpi at 41.5°C, and returned to the initial temperature by 5 dpi.
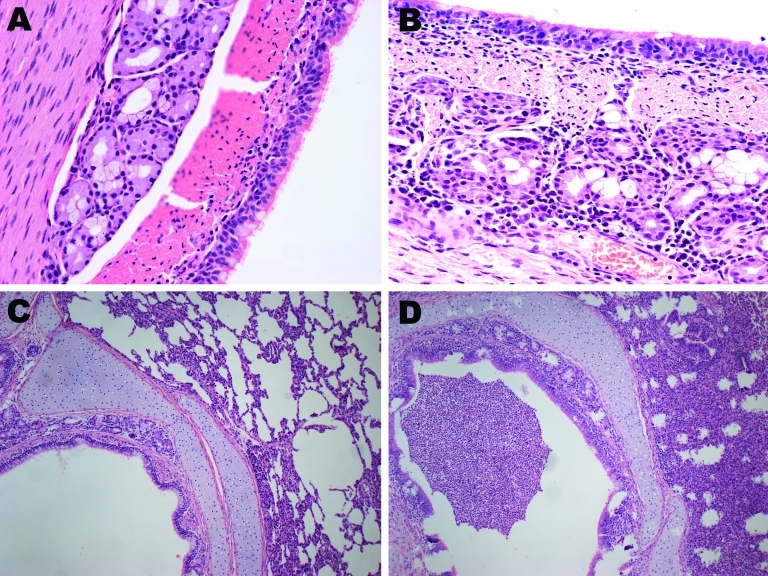
We observed necrosis of the tracheal wall and pneumonia foci in the 4 pandemic (H1N1) 2009 virus–infected pigs. Hematoxylin and eosin–stained sections of the trachea from the infected pigs showed mild necrotizing tracheitis. The necrotic epithelial cells were present in the lumen, and a mixed inflammatory cell infiltrate was present throughout (Figure). In the lung, we also observed alveolar septal edema and interstitial inflammatory cell infiltrates, as well as histologic changes, including alveolar epithelial hyperplasia, a mixed inflammatory infiltrate, and interstitium broadening (Figure). Additionally, immunohistochemical analysis for the distribution of viral antigens showed positive staining in bronchial epithelial cells and alveolar pneumocytes.
Figure.
Hematoxylin and eosin–stained trachea and lung controls and samples from pigs infected with pandemic (H1N1) 2009 virus. A) Control trachea sample; B) mixed inflammatory cell infiltrate present throughout trachea sample from infected pig. C) Control lung sample; D) mixed inflammatory infiltrate and interstitium broadening in lung sample from infected pig. Original magnifications: panels A and B, ×40; panels C and D, ×10.
Previous studies showed that pandemic (H1N1) 2009 may have become established in swine populations in Canada, Norway, and Hong Kong (1,5–8). The human-to-pig transmission of pandemic (H1N1) 2009 may substantially affect virus evolution and subsequent epidemiology. Although the pandemic was mild, the virus could develop further reassortment in swine and gain virulence. On the other hand, subtype H5N1 and H9N2 viruses have become established in pigs, so the introduction of pandemic (H1N1) 2009 virus to pigs has provided the possibility for the incorporation of avian virus genes into mammalian-adapted viruses. That transmission could occur from humans to pigs and vice versa is especially troublesome. Given the possible production of novel viruses of potential threat to public health, we should emphasize influenza surveillance in pigs and establishment of the genetic basis of the viral genome for rapidly identifying such reassortment events.
Acknowledgments
This study was supported by China National Basic Research Program (China “973” Program 2011CB505004 and 2010CB534002), National Natural Science Foundation of China (no. 31072154 and 30800832) and National Transgenic Major Program (2009ZX08009-141B).
Footnotes
These authors contributed equally to this article.
References
- 1.Pasma T, Joseph T. Pandemic (H1N1) 2009 infection in swine herds, Manitoba, Canada. Emerg Infect Dis. 2010;16:706–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Itoh Y, Shinya K, Kiso M, Watanabe T, Sakoda Y, Hatta M, et al. In vitro and in vivo characterization of new swine origin H1N1 influenza viruses. Nature. 2009;460:1021–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maines TR, Jayaraman A, Belser JA, Wadford DA, Pappas C, Zeng H, et al. Transmission and pathogenesis of swine-origin 2009 A (H1N1) influenza viruses in ferrets and mice. Science. 2009;325:484–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tompkins SM, Lo CY, Tumpey TM, Epstein SL. Protection against lethal influenza virus challenge by RNA interference in vivo. Proc Natl Acad Sci U S A. 2004;101:8682–6. 10.1073/pnas.0402630101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hofshagen M, Gjerset B, Er C, Tarpai A, Brun E, Dannevig B, et al. Pandemic influenza A (H1N1)v: human to pig transmission in Norway? Euro Surveill. 2009;14:pii:19406. [DOI] [PubMed]
- 6.Howden KJ, Brockhoff EJ, Caya FD, McLeod LJ, Lavoie M, Ing JD, et al. An investigation into human pandemic influenza virus (H1N1) 2009 on an Alberta swine farm. Can Vet J. 2009;50:1153–61. [PMC free article] [PubMed] [Google Scholar]
- 7.Weingartl HM, Berhane Y, Hisanaga T, Neufeld J, Kehler H, Emburry-Hyatt C, et al. Genetic and pathobiologic characterization of pandemic H1N1 2009 influenza viruses from a naturally infected swine herd. J Virol. 2010;84:2245–56. 10.1128/JVI.02118-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vijaykrishna D, Poon LL, Zhu HC, Ma SK, Li OT, Cheung CL, et al. Reassortment of pandemic H1N1/2009 influenza A virus in swine. Science. 2010;328:1529. 10.1126/science.1189132 [DOI] [PMC free article] [PubMed] [Google Scholar]