Translation termination in eukaryotic cells is determined by proteins Sup35 (eRF3) and Sup45 (eRF1) [1], which interact with a large number of partners [2]. In yeast Saccharomyces cerevisiae, protein Sup35 can form an aggregating epigenetically inherited conformer (prion) [PSI+] [3]. This prion is carried through the cytoplasm and causes disturbances in translation termination, which are phenotypically identified as the dominant omnipotent nonsense suppression. [PSI+] variants with different properties (nonsense suppression efficiency and transmission stability in mitosis) can be obtained in the same yeast strain. The presence of prion [PSI+] leads to lethality in the haploid yeast strain carrying mutations in the gene encoding another termination factor, Sup45 [4]. We have shown that the combination in the diploid strain of some mutant alleles of the SUP45 gene in the heterozygous state with prion [PSI+] entails the death of the hybrid [5]. The synthetic lethality of prion [PSI+] and mutant allele of the sup45 gene depends both on the type of mutant allele and the prion variant. Variant [PSI+], which is a strong suppressor (“strong” [PSI+], or [PSI+]S), causes synthetic lethality with all nonsense mutations and some missense mutations sup45 in the heterozygote. Our data indicate that the lethality of hybrids is correlated with a decreased activity of the Sup45 protein in the cell in case of sup45 mutations. This paper describes a test system that allows identification of proteins that affect the stability of prion [PSI+] and/or the efficiency of translation termination by their effect on the synthetic lethality of the prion conformer Sup35 and mutant alleles of SUP45. This test system is suitable to search for proteins that affect the translation termination efficiency and/or prion maintenance in yeast cells. Gene library screening using this test system allowed us to identify the CUR1 gene, whose influence on another prion, [URE3], was shown earlier but the effect on translation termination factors was not known.
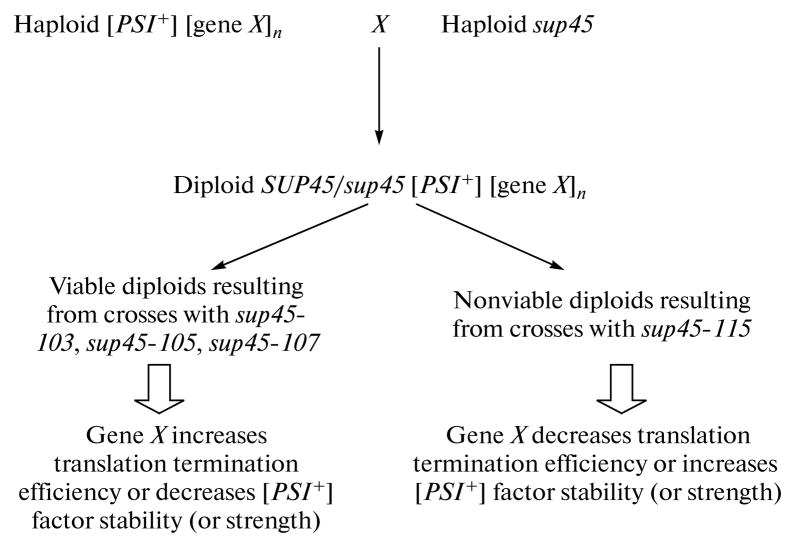
This test system is based on qualitative assessment of the viability of a diploid strain containing simultaneously (1) prion [PSI+], (2) sup45 mutation in the heterozygote, and (3) test gene(s) in a multicopy plasmid (Fig. 1). Synthetic lethality is manifested as the absence of growth of diploids with genotype SUP45/sup45 [PSI+] (Fig. 2, mutations 101–107, 111, and 116 are incompatible with the “strong” [PSI+]S). If the efficiency of translation termination increases due to the presence of additional copies of the test gene (X), formation of viable hybrids with genotype +/sup45 [PSI+] [gene X]n is observed. Conversely, a decrease in the translation termination efficiency in case of overexpression of the X gene is manifested as the death of diploids containing a mutant allele of sup45, which itself is not incompatible with [PSI+] (for example, sup45-113 (115)). Thus, tests can be performed in both directions. In addition, this method allows identification of proteins that affect the stability or strength of prion [PSI+] (Fig. 1).
Fig. 1.
Test system for identification of factors that influence the translation termination efficiency (see text for details).
Fig. 2.
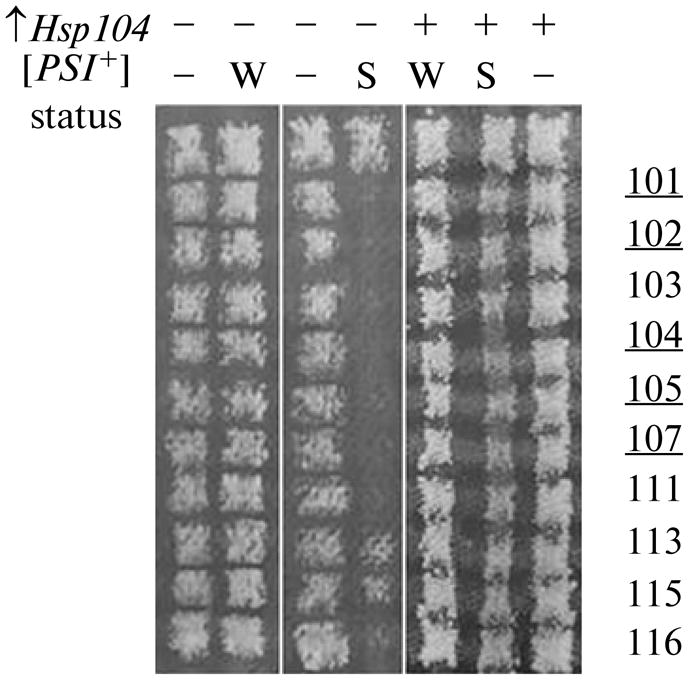
Increased expression of the HSP104 gene abolishes the incompatibility of factor [PSI+] and sup45 mutations. Strain 1A-D1628 [pRS315/SUP45] and its derivatives carrying pRS315/sup45 plasmid were crossed with [PSI+]- and [psi−] variants of strains OT55 [PSI+]W and OT56 [PSI+]S in the absence and presence of an extra copy of the HSP104 gene (designated “−” and “+”, respectively). Numbers of mutations are indicated on the right, the nonsense mutations are underlined. Designations: S, strong [PSI+]; W, weak [PSI+].
In the first stage, we tested whether the proposed test system allows identification of genes whose effect on prion [PSI+] was described in the literature. A key factor in the reproduction of prion [PSI+] is chaperone Hsp104. The maintenance of [PSI+] requires an intermediate level of Hsp104. Both overproduction and absence or inactivation of Hsp104 leads to the loss of the prion [6].
To test the effect of the dose of Hsp104 on the synthetic lethality of prion [PSI+] and sup45 mutations, strain OT56 (MATa ade1-14 his3Δ200 leu2-3,112 ura3-52 trp1-289 [PSI+]S) was transformed with a low-copy (centromeric) plasmid pYS104 carrying the HSP104 gene. The crosses of the transformants with derivatives of strain 1A-D1628 (MATa ade1-14 his3 lys2 trp1-289 ura3-52 leu2-3, 112 SUP45::HIS3), carrying the mutant allele of the SUP45 gene in the centromeric plasmid pRS315 [7] yielded viable hybrids in all combinations (Fig. 2). Thus, a moderate overproduction of Hsp104 restores the viability of sup45 mutants carrying [PSI+].
To search for proteins that affect the efficiency of translation termination, strain OT56 [PSI+]S was transformed with a library of yeast genes obtained using the multicopy plasmid YEp13 (American Type Culture Collection, ATCC no. 37323). The obtained transformants were crossed with derivatives of strain 1A-D1628 containing the pRS316 plasmid with one of the sup45 mutant alleles (nonsense alleles sup45-105 (UAA), sup45-107 (UGA) and missense alleles sup45-103, sup45-115). The use of nonsense alleles sup45 containing different stop codons made it possible to identify and distinguish the genes suppressing lethality due to increase in the amount of Sup45 protein as a result of suppression of one or both nonsense codons. The use of missense alleles sup45-103 made it possible to identify genes whose overexpression leads to a nonspecific (with respect to the sup45 mutation type) compensation of lethality. Mutation sup45-115, which does not cause synthetic lethality with the prion, was used to search for genes whose overexpression can increase lethality, leading to nonviability of the diploid strain. Strain 1A-D1628 carrying the wild-type SUP45 gene was used as a positive control.
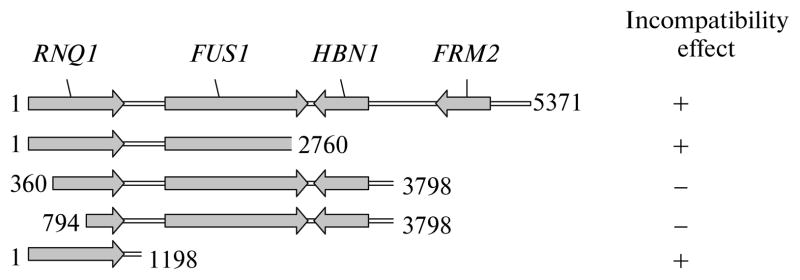
As a result of testing 16550 independent OT56 [PSI+]S transformants, we selected 135 candidates and divided them into six groups depending on the effect of the tested gene(s) on the formation of diploids in crosses with test strains (table). To evaluate the effectiveness of the test system, we performed a more detailed analysis of the six candidates from group I and five candidates from group V. Plasmids from group I led to nonspecific compensation of synthetic lethality. The analysis showed that two of them contained the SUP45 gene. The remaining four plasmids were divided into two pairs with identical restriction maps. For one plasmid from each pair, we sequenced the insert sites adjacent to the YEp13 plasmid sequence. The comparison with the published sequence of the yeast genome (http://seq.yeastgenome.org/cgi-bin/blast-sgd.pl) allowed us to determine the chromosomal localization of the insert. Deletion analysis (Fig. 3) showed that lethality compensation in one of the clones was determined by the 1198-bp fragment of chromosome III containing the 3′-terminal region of the RNQ1 gene.
Table.
Results of gene library screening
| Group | Viability of diploids* |
Number of selected clones | ||||
|---|---|---|---|---|---|---|
| SUP45 | sup45-105 | sup45-107 | sup45-103 | sup45-115 | ||
| I | + | + | + | + | + | 24 |
| II | + | + | – | – | + | 16 |
| III | + | – | + | – | + | 23 |
| IV | + | + | + | – | + | 8 |
| V | + | – | – | – | – | 64 |
| VI | + | – | – | – | + | 16515 |
| Total | 16650 | |||||
Viability of diploids was assessed on SC-Ura-Leu selective medium. Group VI combines the transformants that do not change the nature of incompatibility compared to the original strain.
Fig. 3.
Deletion analysis of fragment no. 275 from the S. cerevisiae gene library. The upper line shows fragment no. 275 corresponding to a region of S. cerevisiae chromosome III (nucleotides 70345–75715); the genes contained in this fragment are specified. Lines 2–5 correspond to a truncated variant of fragment no. 275. Numerals denote the nucleotides at the beginning and in the end of each fragment (enumeration was started with the first nucleotide of fragment no. 275). The compensation incompatibility of factor [PSI+] and sup45 mutations is marked with the “+” sign; its absence, with the “−” sign.
As was shown earlier by other methods, the expression of this fragment, indeed, leads to elimination of prion [PSI+] [8]. In the second clone, the insert leading to lethality compensation was located on chromosome XII and contained the HSP104 gene, the effect of which was already shown previously. A similar analysis of two plasmids from group V, which potentiate lethality in combination with sup45-115, showed that they both contain the CUR1 gene. When recloned into the multicopy plasmid pRS425, this gene was sufficient for the effect of synthetic lethality in combination with [PSI+]S and mutant alleles of sup45, which are usually compatible with the prion. Previously, it was found that overproduction of Cur1 leads to elimination of prion [URE3] [9]. Our data showed that it also affects other prions. The specific mechanisms and consequences of this effect remain to be elucidated.
Acknowledgments
We are grateful to O. Zemlyanko for her assistance in conducting some experiments.
This study was supported by the Russian Foundation for Basic Research (project no. 10-04-00237), NATO (project no. CBP.NR.981898), the program of the Presidium of the Russian Academy of Sciences “Biosphere Origin and Evolution,” the program of the President of the Russian Federation “Leading Scientific Schools” (project no. NSh-6455.2010.4), and the State Contract no. 02.740.11.0 698. The research was performed within the framework of the Federal Program Research and Scientific-Pedagogical Personnel of Innovative Russia in 2009–2013 (P799). The work of Y.O. Chernoff was supported by the National Institutes of Health, United States (grant no. R01 GM58763).
Footnotes
Presented by Academician S.G. Inge-Vechtomov October 7, 2010
References
- 1.Inge-Vechtomov S, Zhouravleva G, Philippe M. Biol Cell. 2003;95:195–209. doi: 10.1016/s0248-4900(03)00035-2. [DOI] [PubMed] [Google Scholar]
- 2.Haar T, Tuite MF. Trends Microbiol. 2007;15:78–86. doi: 10.1016/j.tim.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Tuite MF, Cox BS. Prion. 2007;1:101–109. doi: 10.4161/pri.1.2.4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cox BS, Tuite MF, McLaughlin CS. Yeast. 1988;4:159–178. doi: 10.1002/yea.320040302. [DOI] [PubMed] [Google Scholar]
- 5.Kiktev DA, Inge-Vechtomov SG, Zhouravleva GA. Prion. 2007;1/2:136–143. doi: 10.4161/pri.1.2.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, et al. Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 7.Moskalenko SE, Chabelskaya SV, Inge-Vechtomov SG, et al. BMC Mol Biol. 2003;4:2. doi: 10.1186/1471-2199-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurahashi H, Ishiwata M, Shibata S, Nakamura Y. Mol Cell Biol. 2008;28:3313–3323. doi: 10.1128/MCB.01900-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kryndushkin DS, Shewmaker F, Wickner RB. EMBO J. 2008;27:2725–2735. doi: 10.1038/emboj.2008.198. [DOI] [PMC free article] [PubMed] [Google Scholar]