Abstract
Background
Hyperhomocysteinemia is regarded as a risk factor for cardiovascular diseases, diabetes and obesity. Manifestation of these chronic metabolic disorders starts in early life marked by increase in body mass index (BMI). We hypothesized that perturbations in homocysteine metabolism in early life could be a link between childhood obesity and adult metabolic disorders. Thus here we investigated association of common variants from homocysteine metabolism pathway genes with obesity in 3,168 urban Indian children.
Methodology/Principal Findings
We genotyped 90 common variants from 18 genes in 1,325 children comprising of 862 normal-weight (NW) and 463 over-weight/obese (OW/OB) children in stage 1. The top signal obtained was replicated in an independent sample set of 1843 children (1,399 NW and 444 OW/OB) in stage 2. Stage 1 association analysis revealed association between seven variants and childhood obesity at P<0.05, but association of only rs2796749 in AMD1 [OR = 1.41, P = 1.5×10-4] remained significant after multiple testing correction. Association of rs2796749 with childhood obesity was validated in stage 2 [OR = 1.28, P = 4.2×10-3] and meta-analysis [OR = 1.35, P = 1.9×10-6]. AMD1 variant rs2796749 was also associated with quantitative measures of adiposity and plasma leptin levels that was also replicated and corroborated in combined analysis.
Conclusions/Significance
Our study provides first evidence for the association of AMD1 variant with obesity and plasma leptin levels in children. Further studies to confirm this association, its functional significance and mechanism of action need to be undertaken.
Introduction
Childhood obesity is a growing public health issue worldwide [1]. Prevalence of overweight/obesity has increased from 16% in 2002 to 24% in 2006 in urban school children in Delhi, India [2]. Both genetic and environmental factors play an important role in the development of obesity. Studies investigating the genetic component of obesity have primarily focused on adult obesity and around 32 obesity susceptibility genes have been identified through large scale genome wide association studies (GWAS) [3]. The search for genetic risk factors for childhood obesity and related phenotypes are mainly limited to the replication of variants identified through genome wide association studies (GWAS) in adults [3].
Childhood obesity is one of the major determinants of many chronic diseases in adulthood such as type 2 diabetes, hypertension and cardio-vascular diseases [4]. Manifestation of these chronic metabolic disorders, which have become pandemic in India [5]–[7], starts in early life [8]. Indian subjects with impaired glucose tolerance or diabetes are shown to have typically low BMI up to the age of two years, followed by increasing BMI at the age of 12 years [8]. This suggests that molecular events leading to obesity in childhood predispose individuals to chronic metabolic disorders in later life. This makes identification of factors linking childhood obesity and adult chronic disorders very imperative.
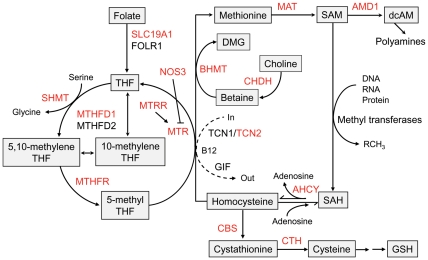
Elevated level of homocysteine, termed as hyperhomocysteinemia, is regarded as a potential risk factor for cardiovascular diseases, diabetes, hypertension and number of other pathologies [9]–[12]. Homocysteine is a thiol containing amino acid which plays an important role in cell metabolism. The metabolic traffic of homocysteine occurs either via remethylation to methionine or through irreversible trans-sulfuration to cysteine. Homocysteine metabolism involves a series of enzymatic reactions that produce variety of metabolic intermediates which are important for cellular processes such as transmethylation, transulfuration and polyamine biosynthesis (Figure 1) [13]. Perturbations in the activities of enzymes involved in these processes such as methylene tetrahydrofolate reductase (MTHFR), methylene tetrahydrofolate dehydrogenase (MTHFD), methionine synthase reductase (MTRR), cystathionine bsynthase (CBS) may results in altered levels of homocysteine and thus metabolic disorders.
Figure 1. Homocysteine metabolism pathway.
The genes selected for the study are marked in red fonts. Refer to Table 2 for gene names. Figure is adapted from Souto JC et al. (2005) Am J Hum Genet 76: 925–933 and Finkelstein JD (1998) Eur J Pediatr 157: S40–44.
Contribution of variants in homocysteine pathway genes to susceptibility of diabetes, obesity and vascular diseases has been suggested by previous studies [14]–[16]. Genetic variants of MTHFR, MTR and MTRR have been shown to be associated with obesity [17]. The C677T polymorphism of MTHFR is reported to be associated with hyperhomocysteinemia, type 2 diabetes and related complications [18], [19]. However there is no report of association of homocysteine pathway genes with childhood obesity. Here we hypothesized that genetic variant in homocysteine metabolism pathway genes leading to perturbation in homocysteine metabolism might be a link between childhood obesity and adult metabolic disorders. To test this, we designed the present two stage case-control study to investigate association of 90 common variants from homocysteine metabolism pathway genes with obesity in 3,168 urban Indian children.
Materials and Methods
Ethics Statement
The study was carried out in accordance with the principles of Helsinki Declaration and was approved by Human Ethics Committee of Institute of Genomics and Integrative Biology (CSIR) and All India Institute of Medical Sciences Research Ethics Committee. Informed written consent was obtained from all the participants. Prior informed written consent was obtained from parents/guardians of the children while verbal consent from children themselves was taken.
Study Population
A total of 3,168 children (aged 11–17 years) were recruited from school health survey from four different zones of Delhi (India) after obtaining prior informed consent from school authorities, parents/guardians and verbal consent from children themselves. For stage 1, we selected 1,325 children [862 NW and 463 OW/OB] to identify SNPs showing association with obesity. This was followed by further selection of 1843 children (1,399 NW and 444 OW/ OB) in stage 2 for replication analysis. Subjects were classified as normal-weight (NW) and over-weight/obese (OW/OB) based on age and sex specific BMI cutoff provided by Cole et al [20], that gives BMI cut offs for overweight and obesity by sex for children between 2 and 18 years corresponding to the cut off points of 25 and 30 kg/m2 for adults.
Anthropometric and Clinical Measurements
Anthropometric measurements including height, weight, waist circumference (WC) and hip circumference (HC) were taken using standard methods. BMI and waist-to-hip ratio (WHR) were calculated. Plasma levels of glucose, insulin and C-peptide were measured as described earlier [21] and HOMA-IR was calculated as described by Matthews et al. [22]. Plasma levels of leptin, resistin and adiponectin were measured using commercial ELISA kits (R&D Systems, Minneapolis, MN). Anthropometric and clinical characteristics of subjects are provided in Table 1.
Table 1. Clinical characteristics of subjects.
| Character | Stage 1 | Stage 2 | P † | P ‡ | ||
| NW children | OW/OB children | NW children | OW/OB children | |||
| N (M/F) | 830 (370/464) | 453 (173/279) | 1399 (420/979) | 444 (132/312) | ||
| Age (years) | 14.00 (12.50–15.00) | 13.00 (12.00–15.00) | 13.00 (12.00–14.50) | 13.2 (12.0–15.0) | 3.0×10-17 | 0.43 |
| Height (m) | 1.54 (1.48–1.61) | 1.56 (1.50–1.62) | 1.52 (1.46–1.58) | 1.56 (1.51–1.62) | 4.8×10-8 | 0.24 |
| Z-Height* | -0.11±0.99 | 0.27±0.89 | -0.11±1.00 | 0.35±0.93 | 0.93 | 0.18 |
| Weight (kg) | 42.30 (37.00–48.85) | 64.00 (55.45–73.90) | 42.00 (36.00–48.40) | 63.5 (56.0–71.0) | 0.18 | 0.73 |
| Z-Weight* | -0.51±0.75 | 1.20±0.54 | -0.40±0.70 | 1.17±0.55 | 4.4×10-4 | 0.35 |
| BMI (kg/m2) | 17.58 (15.86–19.44) | 25.85 (23.97–28.97) | 17.91 (16.26–19.78) | 25.6 (24.0–28.0) | 2.9×10-3 | 0.14 |
| Z-BMI* | -0.56±0.76 | 1.27±0.49 | -0.40±0.65 | 1.18±0.50 | 7.3×10-7 | 0.01 |
| WC (m) | 0.66 (0.61–0.72) | 0.84 (0.77–0.90) | 0.65 (0.60–0.71) | 0.85 (0.78–0.90) | 4.2×10-5 | 1.2×10-3 |
| Z-WC* | -0.42±0.77 | 1.11±0.59 | -0.40±0.79 | 1.11±0.63 | 1.49×10-4 | 2.7×10-13 |
| HC (m) | 0.81 (0.76–0.86) | 0.97 (0.92–1.04) | 0.80 (0.75–0.86) | 0.97 (0.91–1.02) | 8.3×10-3 | 2.1×10-8 |
| Z-HC* | -0.42±0.76 | 1.23±0.63 | -0.41±0.71 | 1.11±0.62 | 0.11 | 1.4×10-14 |
| WHR | 0.82 (0.78–0.87) | 0.86 (0.81–0.90) | 0.82 (0.78–0.86) | 0.87 (0.83–0.92) | 0.05 | 0.21 |
| Z-WHR* | -0.22±0.89 | 0.41±0.93 | -0.20±0.98 | 0.62±0.96 | 0.14 | 1.3×10-9 |
| Glucose (mmol/L) | 5.04 (4.72–5.33) | 5.00 (4.72–5.33) | 4.77 (4.44–5.11) | 4.60 (4.22–4.90) | 3.3×10-23 | 6.3×10-27 |
| Insulin (pmol/L) | 37.80 (23.40–54.60) | 76.80 (52.20–108.00) | 42.00 (28.80–58.80) | 71.40 (47.40–111.60) | 4.4×10-5 | 1.8×10-3 |
| C-peptide (nmol/L) | 0.46 (0.33–0.59) | 0.61 (0.39–0.80) | 0.48 (0.37–0.62) | 0.70 (0.53–0.89) | 0.86 | 0.19 |
| HOMA-IR | 1.41 (0.83–2.09) | 2.83 (1.86–4.13) | 1.44 (0.99–2.09) | 2.41 (1.52–3.89) | 8.1×10-3 | 1.4×10-4 |
| hsCRP (mg/L) | 0.25 (0.1–0.76) | 1.26 (0.48–3.06) | 0.34 (0.13–0.84) | 1.11 (0.53–2.38) | 3.0×10-3 | 0.94 |
| Adiponectin (µg/mL) | 8.45 (5.95–12.50) | 4.62 (2.81–7.18) | 8.61 (4.80–13.59) | 7.79 (4.76–10.83) | 0.07 | 0.18 |
| Leptin (ng/mL) | 6.68 (4.30–11.51) | 17.37 (10.72–26.81) | 8.14 (5.10–12.75) | 19.53 (13.43–30.34) | 5.4×10-5 | 3.0×10-3 |
| Resistin (ng/mL) | 5.59 (4.39–7.82) | 5.76 (4.33–7.25) | 5.30 (4.25–6.80) | 5.67 (4.60–7.25) | 9.7×10-3 | 7.6×10-8 |
Data are presented as median with interquartile ranges.
Data presented are mean values with standard deviation for Z-scores of the parameters.
Genes and SNPs Selection
We selected 16 genes which have central role in the homocysteine metabolism pathway (Figure 1) [13], [23] and 2 genes (ACE and NOS3) reported to be associated with plasma homocysteine levels [24], [25]. Common variants (MAF >0.05) of the selected genes were prioritized based on previous reports of association with metabolic disorders, tagging and linkage disequilibrium (LD) pattern information, and spacing between the SNPs to cover the gene. Finally a set of 90 SNPs were selected for genotyping in stage 1 (Table S1).
Genotyping
Genotyping in stage 1 was performed using Illumina GoldenGate assay (Illumina Inc., San Diego, CA, USA). Stringent quality control (QC) were applied to the genotyping data that included genotype confidence score >0.25, SNP call rate >0.9, GenTrans score >0.6, cluster separation score >0.4, Hardy Weinberg Equilibrium (HWE) [P>0.01 in NW, OW/OB and all subjects], minor allele frequency (MAF) >0.05. After excluding 35 SNPs based on QC criteria (Table S1), the successfully genotyped SNPs had an average call rate of 98% and concordance rate >99% based on 5% duplication.
In stage 2, variant showing strongest association with childhood obesity and standing multiple testing correction was validated by genotyping in an independent sample set of 1843 children (1,399 NW and 444 OW/OB) using iPLEX (Sequenom, San Deigo, CA, USA). Genotyping success rate was 94% with >99.9% consistency rate in genotype calls in 5% duplicates.
Statistical Analysis
Hardy Weinberg Equilibrium (HWE) was tested using χ2. Pairwise LD between the SNPs and haplotype blocks were determined using Haploview 4.0. Continuous variables were inverse normal transformed and were converted to age and sex specific internal Z-scores by dividing differences of mean value of study population and individual values by standard deviation. Logistic and linear regression analyses were performed to assess associations of variants with obesity and quantitative traits respectively under additive model adjusting for age, sex and Z- BMI as appropriate. A P value of <7.7×10-4 [0.05/(65] was considered significant after Bonferroni correction. Meta-analysis was performed by combining summary estimates while association by combining raw genotyped data of two phases was also tested. β values are presented as Z-score units. For continuous traits, P value of <5.5×10-5 was considered statistically significant [0.05/(65*14)]. Haplotype association analysis adjusted for age and sex was carried out at 10,000 permutations. Statistical analyses were performed using PLINK v. 1.07 (http://pngu.mgh.harvard.edu/purcell/plink) and SPSS v. 17.0 (SPSS, Chicago, IL, USA).
Statistical power of the study was determined using Quanto software (http://hydra.usc.edu/gxe/) assuming log additive model of inheritance and 24% prevalence of overweight/obesity (1) at α = 0.05. Sample size in stage 1 and combined analysis provided 44–76% and 76–97% power respectively to detect association of variant with allele frequency of 0.20 and effect size of 1.20–1.30.
Results
Association analysis in stage 1 suggested association of seven variants-rs2796749 (AMD1), rs7768897 (AMD1), rs1985908 (MAT1A), rs9397028 (MTHFD1L), rs1021737 (CTH), rs6693082 (CTH) and rs1801133 (MTHFR) with childhood obesity at P<0.05 (Tables 2 and 3). The association of rs2796749 in AMD1 [OR = 1.41, P = 1.5×10-4] remained significant even after multiple testing correction. Linear regression analysis to assess the association of variants with BMI as continuous trait also revealed association of rs2796749 with BMI [β = 0.16 Z-score unit, P = 4.2×10-4] (Table 3). AMD1 variant rs2796749 was also associated with other quantitative measures of adiposity (Table 4). With increase in per copy of risk allele G of rs2796749, significant increase in height and weight by 0.10 Z-score units (P = 0.02) and 0.17 (P = 1.3×10-4) respectively was observed (Table 4). Similar trend in increase in WC and HC was found with the increase in risk allele of rs2796749 [β = 0.18, P = 3.2×10-5 for WC and β = 0.18, P = 4.4×10-5 for HC]. The rs2796749 was also associated with plasma leptin level [β = 0.14, P = 4.2×10-4].
Table 2. Homocysteine metabolism pathway genes and association analysis of their variants with obesity in Indian children.
| Gene ID | Gene name | SNP ID | Position in gene | Base change | AA change | MAF NW | MAF OW/OB | OR (95%CI) | P |
| MTHFR | Methylenetetrahydrofolate reductase | rs3737965 | Intron 1 | G/A | 0.09 | 0.08 | 0.86 (0.63–1.18) | 0.36 | |
| rs9651118 | Intron 2 | T/C | 0.28 | 0.25 | 0.84 (0.69–1.01) | 0.07 | |||
| rs1801133 | Exon 4 | C/T | A222V | 0.17 | 0.20 | 1.24 (1.01–1.52) | 0.04 | ||
| rs1801131 | Exon 7 | A/C | E429A | 0.38 | 0.41 | 1.12 (0.95–1.32) | 0.19 | ||
| CTH | Cystathionase | rs663465 | 5' flank | C/T | 0.43 | 0.45 | 1.1 (0.93–1.30) | 0.27 | |
| rs663649 | Intron 7 | C/A | 0.18 | 0.19 | 1.04 (0.84–1.29) | 0.70 | |||
| rs1021737 | Exon 12 | G/T | S403I | 0.25 | 0.29 | 1.26 (1.05–1.52) | 0.01 | ||
| rs6693082 | 3' flank | T/G | 0.25 | 0.30 | 1.26 (1.05–1.51) | 0.01 | |||
| MTR | 5-methyltetrahydrofolate-homocysteine | rs946403 | Intron 13 | A/G | 0.37 | 0.41 | 1.16 (0.98–1.38) | 0.09 | |
| methyltransferase | rs1770449 | Intron 24 | A/G | 0.30 | 0.28 | 0.91 (0.76–1.08) | 0.27 | ||
| rs1805087 | Exon 26 | T/C | D919G | 0.31 | 0.30 | 0.95 (0.79–1.13) | 0.55 | ||
| rs16834521 | Exon 28 | A/G | A1048A | 0.31 | 0.34 | 1.15 (0.97–1.37) | 0.12 | ||
| rs2229276 | Exon 29 | A/G | A1048A | 0.31 | 0.34 | 1.16 (0.97–1.38) | 0.10 | ||
| rs1050993 | 3' UTR | C/T | 0.29 | 0.28 | 0.91 (0.76–1.09) | 0.31 | |||
| CHDH | Choline dehydrogenase | rs6445607 | Intron 1 | T/G | 0.25 | 0.27 | 1.14 (0.94–1.39) | 0.17 | |
| rs2241808 | Exon 4 | A/G | A240A | 0.41 | 0.43 | 1.09 (0.92–1.28) | 0.34 | ||
| MTRR | Methionine synthase reductase | rs1801394 | Exon 2 | G/A | I22M | 0.45 | 0.50 | 1.18 (1.00–1.40) | 0.05 |
| rs162036 | Exon 7 | T/C | K350R | 0.14 | 0.16 | 1.14 (0.90–1.43) | 0.27 | ||
| rs10380 | Exon 14 | G/A | H595Y | 0.14 | 0.15 | 1.10 (0.87–1.39) | 0.42 | ||
| BHMT | Betaine-homocysteine S-methyltransferase | rs492842 | Intron 1 | T/C | 0.43 | 0.40 | 0.87 (0.74–1.03) | 0.10 | |
| rs3733890 | Exon 6 | C/T | R239Q | 0.28 | 0.27 | 0.95 (0.78–1.14) | 0.56 | ||
| rs585800 | 3' UTR | A/T | 0.14 | 0.13 | 0.87 (0.68–1.12) | 0.27 | |||
| AMD1 | Adenosylmethionine decarboxylase 1 | rs2796749 | 5' flank | C/G | 0.34 | 0.27 | 0.71 (0.59–0.85) | 1.5×10-4 | |
| rs1007274 | Intron 1 | G/A | 0.24 | 0.27 | 1.10 (0.91–1.33) | 0.31 | |||
| rs7768897 | Intron 4 | C/T | 0.25 | 0.20 | 0.77 (0.63–0.93) | 7.5×10-3 | |||
| NOS3 | Nitric oxide synthase 3 | rs1799983 | Exon 7 | G/T | D298E | 0.17 | 0.19 | 1.14 (0.92–1.41) | 0.22 |
| rs2566514 | Exon 18 | G/C | A666A | 0.35 | 0.34 | 0.94 (0.80–1.12) | 0.50 | ||
| MAT1A | Methionine adenosyltransferase I | rs17677908 | 5' flank | T/C | 0.26 | 0.29 | 1.16 (0.96–1.39) | 0.12 | |
| rs2282367 | Intron 4 | G/A | 0.15 | 0.14 | 0.94 (0.75–1.19) | 0.63 | |||
| rs10788546 | Exon 7 | C/T | V290V | 0.16 | 0.15 | 0.97 (0.77–1.22) | 0.79 | ||
| rs2993763 | Exon 9 | A/G | Y377Y | 0.38 | 0.35 | 0.88 (0.74–1.04) | 0.14 | ||
| rs1985908 | 3' UTR | T/C | 0.48 | 0.42 | 0.78 (0.66–0.92) | 4.0×10-3 | |||
| FOLH1 | Folate hydrolase 1 | rs202719 | Intron 11 | T/C | 0.11 | 0.13 | 1.24 (0.97–1.59) | 0.086 | |
| rs6485965 | 3' flank | G/A | 0.38 | 0.37 | 0.96 (0.80–1.15) | 0.67 | |||
| MTHFD1 | Methylenetetrahydrofolate dehydrogenase 1 | rs8006686 | Intron 2 | T/C | 0.17 | 0.17 | 0.99 (0.79–1.23) | 0.90 | |
| rs1950902 | Exon 6 | G/A) | K134R | 0.08 | 0.07 | 1.00 (0.72–1.38) | 0.99 | ||
| rs8016556 | Intron 16 | A/G | 0.26 | 0.24 | 0.90 (0.74–1.09) | 0.27 | |||
| rs2236225 | Exon 20 | T/C | R653Q | 0.50 | 0.49 | 0.93 (0.78–1.11) | 0.41 | ||
| rs2281603 | Intron 26 | T/C | 0.21 | 0.20 | 0.95 (0.78–1.17) | 0.64 | |||
| MTHFD1L | Methylenetetrahydrofolate dehydrogenase 1-like | rs9397028 | 5' flank | G/A | 0.42 | 0.37 | 0.80 (0.67–0.95) | 0.01 | |
| rs2073063 | Intron 4 | A/G | 0.43 | 0.39 | 0.84 (0.71–1.00) | 0.05 | |||
| rs1555179 | Intron 15 | C/T | 0.24 | 0.23 | 0.95 (0.79–1.15) | 0.61 | |||
| rs509474 | Exon 23 | G/C | S832S | 0.33 | 0.32 | 0.98 (0.81–1.17) | 0.78 | ||
| rs1047662 | 3' UTR | C/G | 0.28 | 0.27 | 0.97 (0.80–1.16) | 0.71 | |||
| SHMT1 | Serine hydroxymethyltransferase 1 | rs2273028 | Intron 7 | C/T | 0.16 | 0.17 | 1.01 (0.81–1.25) | 0.94 | |
| rs1979277 | Exon 12 | G/A | L474F | 0.16 | 0.15 | 0.99 (0.80–1.24) | 0.96 | ||
| rs12949119 | 3' UTR | A/T | 0.36 | 0.35 | 0.97 (0.81–1.16) | 0.71 | |||
| ACE | Angiotensin I converting enzyme | rs4291 | 5' flank | A/T | 0.39 | 0.39 | 1.03 (0.87–1.23) | 0.71 | |
| rs4331 | Exon 16 | C/T | A731A | 0.41 | 0.43 | 1.13 (0.96–1.33) | 0.16 | ||
| rs4362 | Exon 24 | C/T | F1129F | 0.35 | 0.38 | 1.16 (0.97–1.39) | 0.10 | ||
| AHCY | Adenosylhomocysteinase | rs819146 | 5' UTR | A/C | 0.22 | 0.21 | 0.94 (0.77–1.15) | 0.56 | |
| rs819147 | Intron 2 | T/C | 0.23 | 0.21 | 0.89 (0.73–1.09) | 0.25 | |||
| rs864702 | Intron 10 | C/T | 0.23 | 0.21 | 0.88 (0.72–1.08) | 0.22 | |||
| rs819173 | 3' flank | T/C | 0.24 | 0.21 | 0.88 (0.72–1.08) | 0.21 | |||
| CBS | Cystathionine-beta-synthase | rs706208 | 3' UTR | T/C | 0.38 | 0.37 | 0.94 (0.80–1.12) | 0.51 | |
| rs6586282 | Intron 15 | G/A | 0.08 | 0.07 | 0.89 (0.65–1.20) | 0.43 | |||
| rs2014564 | Intron 12 | G/A | 0.47 | 0.46 | 0.98 (0.83–1.16) | 0.83 | |||
| rs234706 | Exon 9 | C/T | Y233Y | 0.23 | 0.24 | 1.05 (0.87–1.28) | 0.60 | ||
| SLC19A1 | Solute carrier family 19 member 1 | rs1051296 | 3' UTR | A/C | 0.47 | 0.45 | 0.91 (0.77–1.07) | 0.24 | |
| rs12659 | Exon 3 | G/A | P114P | 0.34 | 0.34 | 1.03 (0.87–1.22) | 0.76 | ||
| rs1051266 | Exon 2 | G/A | H27R | 0.38 | 0.38 | 0.99 (0.84–1.17) | 0.89 | ||
| TCN2 | Transcobalamin II | rs5749131 | 5' flank | A/G | 0.37 | 0.37 | 0.98 (0.83–1.16) | 0.81 | |
| rs9606756 | Exon 2 | T/C | I23V | 0.09 | 0.11 | 1.11 (0.84–1.46) | 0.47 | ||
| rs1801198 | Exon 6 | G/C | R259P | 0.41 | 0.43 | 1.08 (0.92–1.28) | 0.35 | ||
| rs9621049 | Exon 7 | G/A | S348F | 0.08 | 0.09 | 1.18 (0.88–1.58) | 0.26 | ||
| rs10418 | 3' UTR | G/A | 0.21 | 0.19 | 0.88 (0.72–1.09) | 0.24 |
AA: Amino acid; MAF: minor allele frequency; OR: odds ratio; CI: confidence interval. OR and P values presented were calculated using logistic regression analysis under additive model with age and sex as covariates.
Table 3. Association of homocysteine metabolism pathway genes with obesity in urban Indian children.
| SNP | Gene | RAF NWchildren | RAF OW/OBchildren | Obesity | BMI | |||
| OR (95%CI) | P | β | SE | P | ||||
| Stage 1 | ||||||||
| rs2796749 (C/G) | AMD1 | 0.657 | 0.733 | 1.41 (1.18–1.69) | 1.5×10-4 | 0.16 | 0.05 | 4.2×10-4 |
| rs7768897 (C/T) | AMD1 | 0.745 | 0.795 | 1.30 (1.07–1.59) | 7.5×10-3 | 0.11 | 0.05 | 0.03 |
| rs1985908 (T/C) | MAT1A | 0.524 | 0.581 | 1.28 (1.09–1.51) | 4.0×10-3 | 0.14 | 0.04 | 1.5×10-3 |
| rs9397028 (G/A) | MTHFD1L | 0.577 | 0.627 | 1.25 (1.05–1.49) | 0.01 | 0.15 | 0.05 | 1.3×10-3 |
| rs1021737 (G/T) | CTH | 0.250 | 0.292 | 1.26 (1.05–1.52) | 0.01 | 0.16 | 0.05 | 1.6×10-3 |
| rs6693082 (T/G) | CTH | 0.250 | 0.300 | 1.26 (1.05–1.51) | 0.01 | 0.16 | 0.05 | 9.1×10-4 |
| rs1801133 (C/T) | MTHFR | 0.169 | 0.203 | 1.23 (1.01–1.52) | 0.04 | 0.12 | 0.06 | 0.02 |
| Stage 2 | ||||||||
| rs2796749 (C/G) | AMD1 | 0.668 | 0.723 | 1.28 (1.09–1.54) | 4.2×10-3 | 0.10 | 0.03 | 2.6×10-3 |
| Combined | ||||||||
| rs2796749 (C/G) | AMD1 | 0.664 | 0.728 | 1.35 (1.19–1.52) | 1.9×10-6 | 0.13 | 0.03 | 2.5×10-6 |
| Meta-analysis* | ||||||||
| rs2796749 (C/G) | AMD1 | 1.35 | 2.7×10-6 | 0.12 | 6.5×10-6 | |||
RAF: risk allele frequency; OR: odds ratio. Odds ratio are presented with respect to risk allele that are highlighted as underlined text. β: change in Z score of BMI per increase in risk allele; SE: standard error. Combined analysis was performed by combining raw genotype data from stage 1 and stage 2. Meta-analysis was performed by combining the summary estimates (OR/β, 95%CI and S.E). No heterogeneity in two study population was observed (P value for Cochrane's Q statistic = 0.48 and Î2 heterogeneity index = 0.0). *OR/β and P value for fixed effect meta-analysis.
Table 4. Association of AMD1 variant rs2796749 with clinical traits in urban Indian children.
| Stage 1 | Stage 2 | Combined | Mean | ||||||
| Trait | β | P | β | P | β | P | CC | CG | GG |
| Height† | 0.10 | 0.02 | 0.09 | 0.01 | 0.09 | 6.7×10–4 | −0.20 | 0.01 | 0.04 |
| Weight† | 0.17 | 1.3×10–4 | 0.12 | 2.8×10–4 | 0.14 | 9.1×10–8 | −0.17 | −0.05 | 0.10 |
| BMI† | 0.16 | 4.2×10–4 | 0.10 | 2.6×10–3 | 0.13 | 2.5×10–6 | −0.11 | −0.06 | 0.10 |
| WC† | 0.18 | 3.2×10–5 | 0.11 | 2.5×10–3 | 0.14 | 3.2×10–7 | −0.16 | −0.06 | 0.10 |
| HC† | 0.18 | 4.4×10–5 | 0.12 | 3.3×10–4 | 0.16 | 3.7×10–8 | −0.17 | −0.05 | 0.12 |
| WHR† | 0.07 | 0.07 | 0.04 | 0.27 | 0.06 | 0.05 | −0.07 | −0.04 | 0.03 |
| Adiponectin‡ | −0.03 | 0.49 | −0.01 | 0.71 | −0.02 | 0.41 | −0.08 | 0.04 | 0.00 |
| Leptin‡ | 0.14 | 4.2×10–4 | 0.09 | 0.02 | 0.12 | 2.2×10–5 | −0.01 | −0.01 | 0.06 |
| Resistin‡ | 0.01 | 0.82 | 0.02 | 0.60 | 0.01 | 0.74 | −0.01 | 0.01 | 0.01 |
| hsCRP‡ | 0.08 | 0.08 | 0.06 | 0.11 | 0.07 | 0.01 | 0.03 | −0.03 | 0.02 |
| Total Cholesterol‡ | −0.009 | 0.84 | 0.07 | 0.03 | 0.04 | 0.12 | 0.01 | −0.05 | −0.003 |
| HDL‡ | −0.07 | 0.05 | 0.009 | 0.81 | −0.03 | 0.35 | −0.003 | −0.001 | −0.01 |
| LDL‡ | 0.02 | 0.65 | 0.08 | 0.02 | 0.06 | 0.04 | −0.01 | −0.02 | 0.03 |
| TG‡ | 0.06 | 0.10 | −0.03 | 0.45 | 0.01 | 0.73 | 0.07 | 0.05 | 0.05 |
| FPG‡ | −0.03 | 0.47 | 0.002 | 0.94 | 0.07 | 0.01 | −0.02 | 0.05 | 0.001 |
| FPI‡ | 0.06 | 0.15 | 0.07 | 0.06 | −0.01 | 0.67 | −0.08 | 0.03 | −0.005 |
| C-peptide‡ | 0.08 | 0.05 | 0.03 | 0.36 | 0.06 | 0.04 | −0.09 | 0.06 | −0.01 |
| HOMA-IR‡ | 0.05 | 0.27 | 0.07 | 0.05 | 0.07 | 0.02 | −0.09 | 0.04 | −0.01 |
β represents change in Z score per increase in risk allele. The mean values presented were obtained by ANCOVA analysis adjusted for appropriate covariates.
Analysis performed after adjusting for age and sex.
Analysis performed after adjusting for age, sex and BMI.
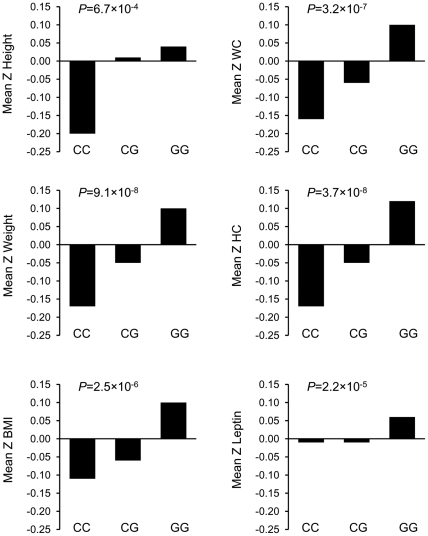
Because of strong association of AMD1 variant rs2796749 with childhood obesity and measures of adiposity, we attempted to replicate the signal obtained at rs2796749 in additional sample set of 1843 children in stage 2. Association of rs2796749 with childhood obesity was validated in stage 2 [OR = 1.28, P = 4.2×10–3] and was confirmed by meta-analysis [OR = 1.35, P = 2.7×10–6] (Table 3). Similar results were obtained in combined analysis by pooling genotype data from two stages [OR = 1.35, P = 1.9×10–6]. Consistently, association of rs2796749 with BMI was also replicated [β = 0.10, P = 2.6×10–3]. The meta-analysis and combined analysis corroborated the association of rs2796749 [β = 0.12, P = 6.5×10-6 in meta-analysis and β = 0.13, P = 2.5× 10–6 in combined analysis]. Association of rs2796749 with measures of adiposity and plasma leptin levels was also replicated in stage 2 and was corroborated in combined analysis (Figure 2). No association of rs2796749 with other biochemical traits investigated could be detected (Table 4).
Figure 2. Association of rs2796749 with clinical traits in urban Indian children.
Mean Z-scores adjusted for age and sex are plotted on the y-axis for the corresponding genotypes on x-axis for all the traits except leptin for which the mean Z-scores are adjusted for age, sex and Z-BMI.P values presented were determined by linear regression analysis for Z-score change per risk allele of the respective trait.
Stage 1 analysis also suggested association of MAT1A variant rs1985908 with childhood obesity [OR = 1.28, P = 4.0×10–3], BMI [β = 0.14, P = 1.5×10–3] and leptin levels [β = –0.12, P = 2.7×10–3]. CTH variant rs6693082 which showed nominal association with obesity, was also found to be associated with BMI [P = 9.1×10–4], WC [P = 4.0×10–4], HC [P = 6.3×10-4] and adiponectin levels [P = 4.5×10–3] (Table S2). However the associations of rs1985908 and rs6693082 with obesity and other traits did not remain significant after correcting for multiple comparisons.
Further, we investigated the association of all the SNPs genotyped in stage 1 with quantitative traits related to obesity that included measures of glucose homeostasis, lipid metabolism and adipokines. We observed suggestive association of some variants with quantitative traits (Table S2). CTH variant rs1021737 showed association with total cholesterol (P = 6.8×10-3]while MAT1A variant rs17677908 and MTR variant rs16834521 were found to be associated with TG [P = 6.8×10-3 and 3.56×10-3 respectively]. Besides AMD1 and MAT1A, levels of leptin were also associated with MTRR variant rs1801394 [P = 6.8×10-3]. However, none of the associations remained significant after accounting for multiple tests. We also performed haplotype association analysis that suggested association of haplotypes of variants SNPs in AMD1, CTH, MTHFR and MTRR (Table S3). None of the haplotype associations remained significant after 10,000 permutations.
Discussion
Several GWAS and large-scale meta-analysis for adult BMI have identified susceptibility loci for increased BMI, explaining only a small proportion of variation in BMI [3]. Investigation of genetic risk factors for childhood obesity has been restricted mainly to the replication of GWAS findings [3]. Though modern genotyping arrays capture most of the common variations, there remain substantial additional contributions to phenotypic variation from the variants not well captured by these arrays [26]. Investigation of variants prioritized based on their functional and positional significance provides an opportunity to identify those not detected by GWAS. Employing functional candidate analysis, we recently identified association of common variants of IL6, LEPR, and PBEF1 with increased susceptibility to obesity and measures of adiposity in Indian children [27]. Homocysteine metabolism pathway seems to be a promising candidate pathway by virtue of its involvement in important cellular processes. Thus, here we comprehensively examined association of common variants of homocysteine metabolism pathway genes with obesity in children.
The study reveals association of AMD1 variant rs2796749 with obesity, measures of adiposity and plasma leptin levels in urban Indian children. To the best of our knowledge, this is the first study demonstrating the association of AMD1 variant with obesity. In our previous study of association of homocysteine metabolism pathway genes with type 2 diabetes and adult obesity [28], rs2796749 did not show association with adult obesity or BMI but was directionally consistent. Further, we investigated effect of this variant on adult BMI using the association data from GIANT consortium GWA meta-analysis which included 249,796 individuals of European ancestry [29]. In this recent meta-analysis also, rs2796749 showed consistency in direction of association with BMI, though could not reach statistical significance (P = 0.19). This suggests that AMD1 variant rs2796749 might have influence on adult BMI also, but with smaller effect size.
AMD1 codes for an important regulatory enzyme adenosyl-methionine decarboxylase 1 (AdoMetDC) which catalyzes the decarboxylation of S-adenosyl methionine (SAM), a methyl-group donor in transmethylation reactions. The imbalances in decarboxylation of SAM might result in disruption of DNA methylation process leading to abnormal gene expressions [30], which is implicated in various metabolic disorders including obesity and diabetes [31]. In addition to its role in transmethylation reaction, AdoMetDC is also a regulatory enzyme in polyamine biosynthesis. SAM is committed to polyamine biosynthesis after decarboxylation and serves as an aminopropyl donar in spermidine and spermine synthesis [32]. The polyamines are ubiquitous and are implicated in cellular growth and differentiation, regulation of adipocyte formation and glucose metabolism [33].
A recent study reported increased levels of polyamines in childhood obesity and their correlation with biomarkers of oxidative stress, inflammation and leptin [34]. The study supported the role of polyamines on growth and development on adipose tissue. On similar lines, our study demonstrated association of rs2796749 in 5? flank region of AMD1 with obesity in Indian children. The same variant also showed association with quantitative measures of adiposity including weight, BMI, hip and waist circumferences and plasma levels of leptin. Moreover, AMD1 mRNA is shown to increase in response to insulin via insulin response element in 5' flank region of AMD1 [35]. The variant rs2796749 might also have role in influencing the transcription of AMD1 in response to insulin, which needs to be investigated.
MAT1A codes for methionine adenosyltransferase (MAT), which catalyzes the conversion of methionine to S- adenosylmethionine (SAM). MAT1A variants have been shown to be associated with hypertension and stroke [36]. Our study also suggested association of MAT1A variant rs1985908 with childhood obesity. MAT1A variants also showed association with the plasma levels of leptin, adiponectin and triglyerides. But these associations could not stand multiple testing corrections, hence further replication in larger sample set and in other ethnic groups are required to substantiate the observations in the present study.
Although the present study design limits the potential to identify the causal relationships, based on previous studies, we can speculate that AMD1 variant might influence the susceptibility to obesity by modulating either the polyamines metabolism or DNA methylation. Dietary habits can affect these processes as nutrients such as vitamin B12 and folate are required as important cofactors in these enzymatic reactions (Figure 1). The plasma levels of folate and vitamin B12 as well as the dietary intake of these are inversely correlated with the plasma homocysteine levels [37]. Moreover, individuals consuming only vegetarian diet tend to have low vitamin B12 and elevated homocysteine levels [38], [39]. Majority of Indian population adheres to a vegetarian diet that predisposes them to vitamin B12 deficiency. Thus diet which can influence the availability of vitamins may modulate the impact of genetic variants on obesity phenotypes. Previous studies including investigation in Indian populations suggested the possibility of interaction between diet and genetic variants of homocysteine pathway genes in influencing the homocysteine levels [40], however no study provided strong evidence for this interaction and its impact on impact on phenotypes. Unfortunately, lack of data on the levels of folate and vitamin B12 and the dietary habits of our study subjects limited further investigation of the interaction of these factors with the genetic variants and their combined effect on obesity phenotypes.
In case-control association studies, limited statistical power and population stratification can lead to spurious association results. Though the sample size of our study is considerable, there is a likelihood of false negative observations for variants with small effect sizes, as the present study is sufficiently powered to capture only large effects (OR>1.3) of the common variants with frequencies more than 0.20. Further, to minimize the effect of population stratification, we have collected samples from a small geographical region that forms a homogenous cluster as reported by the Indian Genome Variation Consortium [41]. Moreover, the multidimensional scaling (MDS) analysis using the genotype data for 595 unlinked markers (r2<0.20) for stage 1 samples shows that the study population belongs to one cluster (Figure S1).
In conclusion, we demonstrate here, for the first time, association of AMD1 variant with susceptibility to obesity and measures of adiposity in Indian children. Further studies to confirm the association of AMD1 variant with childhood, its functional significance and molecular mechanism need to be undertaken. Moreover, our study provides a lead for future investigations towards understanding the genetic predisposition of obesity in childhood and exploring therapeutic options for prevention of childhood obesity.
Supporting Information
Multidimensional scaling for the study population in stage 1. The 595 unlinked markers (r2<0.20) were used to obtain the positions on the first and second dimensions using PLINK.
(DOC)
SNPs selected for the study and their association with obesity in urban Indian children AA: Amino acid; HWE: Hardy Weinberg Equillibrium; MAF: minor allele frequency; QC: quality control; OR: odds ratio; CI: confidence interval. OR and P values presented were calculated with respect to minor allele using logistic regression analysis under additive model with age and sex as covariates.
(DOC)
Association of analyzed SNPs with biochemical parameters in urban Indian children β, L95 and U95 represent change in Z-score units of the parameters with per increase in risk allele with 95% confidence interval. β values are presented with respect to the minor alleles. Analysis for height, weight, BMI, WC and HC were adjusted for age and sex whereas analyses for all other parameters were adjusted for age, sex and Z-BMI.
(DOC)
Association of haplotypes with obesity in urban Indian children OR: odds ratio; NW: normal-weight; OW/OB: overweight and obese.
(DOC)
Acknowledgments
We are thankful to all the participating subjects, their parents and school authorities for their support and cooperation in carrying out the study. We thank Mr. Kuntal Bhadra from Institute of Nuclear Medicine and Allied Sciences for his help in sample collection. We thank Dr. Anurag Agrawal from CSIR-Institute of Genomics and Integrative Biology for his critical evaluation of the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was supported by 'Diabetes mellitus-New drug discovery R&D, molecular mechanisms and genetic & epidemiological factors' (NWP0032-OB2) funded by Council of Scientific and Industrial Research (CSIR), Government of India. RT received postdoctoral fellowship from the Fogarty International Center and the Eunice Kennedy Shriver National Institute of Child Health & Human Development at the National Institutes of Health (1 D43 HD065249). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript."
References
- 1.Lobstein, T, Baur, L, Uauy R. IASO International Obesity Task Force. Obesity in children and young people: a crisis in public health. Obesity Reviews. 2004;5:4–104. doi: 10.1111/j.1467-789X.2004.00133.x. [DOI] [PubMed] [Google Scholar]
- 2.Bhardwaj S, Misra A, Khurana L, Gulati S, Shah P, et al. Childhood obesity in Asian Indians: a burgeoning cause of insulin resistance, diabetes and sub-clinical inflammation. Asia Pac J Clin Nutr. 2008;17(Suppl 1):172–175. [PubMed] [Google Scholar]
- 3.Fernandez JR, Klimentidis YC, Dulin-Keita A, Casazza K. Int J Obes (Lond); 2011. Genetic influences in childhood obesity: recent progress and recommendations for experimental designs. doi: 10.1038/ijo.2011.236. [DOI] [PubMed] [Google Scholar]
- 4.Biro FM, Wien M. Childhood obesity and adult morbidities. Am J Clin Nutr. 2010;91:1499S–1505S. doi: 10.3945/ajcn.2010.28701B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wild S, Roglic G, Green A, Sicree R, King H. Global prevalence of diabetes: estimates for the year 2000 and projections for 2030. Diabetes Care. 2004;27:1047–1053. doi: 10.2337/diacare.27.5.1047. [DOI] [PubMed] [Google Scholar]
- 6.Gupta R, Deedwania PC, Gupta A, Rastogi S, Panwar RB, et al. Prevalence of metabolic syndrome in an Indian urban population. Int J Cardiol. 2004;97:257–261. doi: 10.1016/j.ijcard.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 7.Gupta R, Joshi P, Mohan V, Reddy KS, Yusuf S. Epidemiology and causation of coronary heart disease and stroke in India. Heart. 2008;94:16–26. doi: 10.1136/hrt.2007.132951. [DOI] [PubMed] [Google Scholar]
- 8.Bhargava SK, Sachdev HS, Fall CH, Osmond C, Lakshmy R, et al. Relation of serial changes in childhood body-mass index to impaired glucose tolerance in young adulthood. N Engl J Med. 2004;350:865–875. doi: 10.1056/NEJMoa035698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Humphrey LL, Fu R, Rogers K, Freeman M, Helfand M. Homocysteine level and coronary heart disease incidence: a systematic review and meta-analysis. Mayo Clinic Proceedings. 2008;83:1203–1212. doi: 10.4065/83.11.1203. [DOI] [PubMed] [Google Scholar]
- 10.Buysschaert M, Dramais AS, Wallemacq PE, Hermans MP. Hyperhomocysteinemia in type 2 diabetes: relationship to macroangiopathy, nephropathy, and insulin resistance. Diabetes Care. 2000;23:1816–1822. doi: 10.2337/diacare.23.12.1816. [DOI] [PubMed] [Google Scholar]
- 11.Ntaios G, Savopoulos C, Chatzopoulos S, Mikhailidis D, Hatzitolios A. Iatrogenic hyperhomocysteinemia in patients with metabolic syndrome: A systematic review and metaanalysis. Atherosclerosis. 2011;214:11–19. doi: 10.1016/j.atherosclerosis.2010.08.045. [DOI] [PubMed] [Google Scholar]
- 12.Meigs JB, Jacques PF, Selhub J, Singer DE, Nathan DM, et al. Fasting plasma homocysteine levels in the insulin resistance syndrome: the Framingham Offspring Study. Diabetes Care. 2001;24:1403–1410. doi: 10.2337/diacare.24.8.1403. [DOI] [PubMed] [Google Scholar]
- 13.Finkelstein JD. The metabolism of homocysteine: pathways and regulation. Eur J Pediatr. 1998;157:S40–44. doi: 10.1007/pl00014300. [DOI] [PubMed] [Google Scholar]
- 14.Mehri S, Koubaa N, Nakbi A, Hammami S, Chaaba R, et al. Relationship between genetic polymorphisms of angiotensin-converting enzyme and methylenetetrahydrofolate reductase as risk factors for type 2 diabetes in Tunisian patients. Clinical Biochemistry. 2010;43:259–266. doi: 10.1016/j.clinbiochem.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 15.Bazzaz JT, Shojapoor M, Nazem H, Amiri P, Fakhrzadeh H, et al. Methylenetetrahydrofolate reductase gene polymorphism in diabetes and obesity. Molecular Biology Reports. 2010;37:105–109. doi: 10.1007/s11033-009-9545-z. [DOI] [PubMed] [Google Scholar]
- 16.Frosst P, Blom HJ, Milos R, Goyette P, Sheppard CA, et al. A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nature Genetics. 1995;10:111–113. doi: 10.1038/ng0595-111. [DOI] [PubMed] [Google Scholar]
- 17.Terruzzi I, Senesi P, Fermo I, Lattuada G, Luzi L. Are genetic variants of the methyl group metabolism enzymes risk factors predisposing to obesity? J Endocrinol Invest. 2007;30:747–753. doi: 10.1007/BF03350812. [DOI] [PubMed] [Google Scholar]
- 18.Benes P, Kanková K, Muzík J, Groch L, Benedík J, et al. Methylenetetrahydrofolate reductase polymorphism, type II diabetes mellitus,coronary artery disease, and essential hypertension in the Czech population. Mol Genet Metab. 2001;73:188–195. doi: 10.1006/mgme.2001.3188. [DOI] [PubMed] [Google Scholar]
- 19.Rahimi M, Hasanvand A, Rahimi Z, Vaisi-Raygani A, Mozafari H, et al. Synergistic effects of the MTHFR C677T and A1298C polymorphisms on the increased risk of micro- and macro-albuminuria and progression of diabetic nephropathy among Iranians with type 2 diabetes mellitus. Clin Biochem. 2010;43:1333–1339. doi: 10.1016/j.clinbiochem.2010.08.019. [DOI] [PubMed] [Google Scholar]
- 20.Cole TJ, Bellizzi MC, Flegal KM, Dietz WH. Establishing a standard definition for child overweight and obesity worldwide: international survey. BMJ. 2000;320:1240–1243. doi: 10.1136/bmj.320.7244.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tabassum R, Chavali S, Dwivedi OP, Tandon N, Bharadwaj D. Genetic variants of FOXA2: risk of type 2 diabetes and effect on metabolic traits in North Indians. J Hum Genet. 2008;53:957–965. doi: 10.1007/s10038-008-0335-6. [DOI] [PubMed] [Google Scholar]
- 22.Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, et al. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- 23.Souto JC, Blanco-Vaca F, Soria JM, Buil A, Almasy L, et al. A genomewide exploration suggests a new candidate gene at chromosome 11q23 as the major determinant of plasma homocysteine levels: results from the GAIT project. Am J Hum Genet. 2005;76:925–933. doi: 10.1086/430409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mehri S, Koubaa N, Nakbi A, Hammami S, Chaaba R, et al. Relationship between genetic polymorphisms of angiotensin-converting enzyme and methylenetetrahydrofolate reductase as risk factors for type 2 diabetes in Tunisian patients. Clin Biochem. 2010;43:259–66. doi: 10.1016/j.clinbiochem.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 25.Brown KS, Kluijtmans LA, Young IS, Woodside J, Yarnell JW, et al. Genetic evidence that nitric oxide modulates homocysteine: the NOS3 894TT genotype is a risk factor for hyperhomocystenemia. Arterioscler Thromb Vasc Biol. 2003;23:1014–1020. doi: 10.1161/01.ATV.0000071348.70527.F4. [DOI] [PubMed] [Google Scholar]
- 26.1000 Genomes Project Consortium, Durbin RM, Abecasis GR, Altshuler DL, Auton A, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. [Google Scholar]
- 27.Tabassum R, Yuvaraj M, Dwivedi OP, Chauhan G, Ghosh S, et al. Diabetes 2012; DOI: 10.2337/db11-1501 (In Press); 2012. Common variants of IL6, LEPR and PBEF1 are associated with obesity in Indian children. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chauhan G, Kaur I, Tabassum R, Dwivedi OP, Ghosh S, et al. Exp Diabetes Res 2012: 960318; 2012. Common variants of homocysteine metabolism pathway genes and risk of type 2 diabetes and related traits in indians. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brooks WH. Autoimmune Diseases and Polyamines. Clin Rev Allergy Immunol. 2012;42:58–70. doi: 10.1007/s12016-011-8290-y. [DOI] [PubMed] [Google Scholar]
- 31.Park LK, Friso S, Choi SW. Nutritional influences on epigenetics and age-related disease. Proc Nutr Soc. 2011;4:1–9. doi: 10.1017/S0029665111003302. [DOI] [PubMed] [Google Scholar]
- 32.Vuohelainen S, Pirinen E, Cerrada-Gimenez M, Keinänen TA, Uimari A, et al. Spermidine is indispensable in differentiation of 3T3-L1 fibroblasts to adipocytes. J Cell Mol Med. 2010;14:1683–1692. doi: 10.1111/j.1582-4934.2009.00808.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pirinen E, Kuulasmaa T, Pietila M, Heikkinen S, Tusa M, et al. Enhanced polyamine catabolism alters homeostatic control of white adipose tissue mass, energy expenditure, and glucose metabolism. Mol Cell Biol. 2007;27:4953–4967. doi: 10.1128/MCB.02034-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Codoner-Franch P, Tavarez-Alonso S, Murria-Estal R, Herrera-Martin G, Alonso-Iglesias E. Polyamines Are Increased in Obese Children and Are Related to Markers of Oxidative/Nitrosative Stress and Angiogenesis. J Clin Endocrinol Metab. 2011;96:2821–2825. doi: 10.1210/jc.2011-0531. [DOI] [PubMed] [Google Scholar]
- 35.Soininen T, Liisanantti MK, Pajunen AE. S-adenosylmethionine decarboxylase gene expression in rat hepatoma cells: regulation by insulin and by inhibition of protein synthesis. Biochem J 316 (Pt. 1996;1):273–277. doi: 10.1042/bj3160273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lai CQ, Parnell LD, Troen AM, Shen J, Caouette H, et al. MAT1A variants are associated with hypertension, stroke, and markers of DNA damage and are modulated by plasma vitamin B-6 and folate. Am J Clin Nutr. 2010;91:1377–1386. doi: 10.3945/ajcn.2009.28923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Selhub J, Jacques PF, Wilson PW, Rush D, Rosenberg IH. Vitamin status and intake as primary determinants of homocysteinemia in an elderly population. JAMA. 1993;270:2693–2698. doi: 10.1001/jama.1993.03510220049033. [DOI] [PubMed] [Google Scholar]
- 38.Kumar J, Das SK, Sharma P, Karthikeyan G, Ramakrishnan L, et al. Homocysteine levels are associated with MTHFR A1298C polymorphism in Indian population. J Hum Genet. 2005;50:655–663. doi: 10.1007/s10038-005-0313-1. [DOI] [PubMed] [Google Scholar]
- 39.Refsum H, Yajnik CS, Gadkari M, Schneede J, Vollset SE, et al. Hyperhomocysteinemia and elevated methylmalonic acid indicate a high prevalence of cobalamin deficiency in Asian Indians. Am J Clin Nutr. 2001;74:233–241. doi: 10.1093/ajcn/74.2.233. [DOI] [PubMed] [Google Scholar]
- 40.Kumar J, Garg G, Kumar A, Sundaramoorthy E, Sanapala KR, et al. Single nucleotide polymorphisms in homocysteine metabolism pathway genes: association of CHDH A119C and MTHFR C677T with hyperhomocysteinemia. Circ Cardiovasc Genet. 2009;2:599–606. doi: 10.1161/CIRCGENETICS.108.841411. [DOI] [PubMed] [Google Scholar]
- 41.Indian Genome Variation Consortium. Genetic landscape of the people of India: a canvas for disease gene exploration. J Genet. 2008;87:3–20. doi: 10.1007/s12041-008-0002-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multidimensional scaling for the study population in stage 1. The 595 unlinked markers (r2<0.20) were used to obtain the positions on the first and second dimensions using PLINK.
(DOC)
SNPs selected for the study and their association with obesity in urban Indian children AA: Amino acid; HWE: Hardy Weinberg Equillibrium; MAF: minor allele frequency; QC: quality control; OR: odds ratio; CI: confidence interval. OR and P values presented were calculated with respect to minor allele using logistic regression analysis under additive model with age and sex as covariates.
(DOC)
Association of analyzed SNPs with biochemical parameters in urban Indian children β, L95 and U95 represent change in Z-score units of the parameters with per increase in risk allele with 95% confidence interval. β values are presented with respect to the minor alleles. Analysis for height, weight, BMI, WC and HC were adjusted for age and sex whereas analyses for all other parameters were adjusted for age, sex and Z-BMI.
(DOC)
Association of haplotypes with obesity in urban Indian children OR: odds ratio; NW: normal-weight; OW/OB: overweight and obese.
(DOC)