Abstract
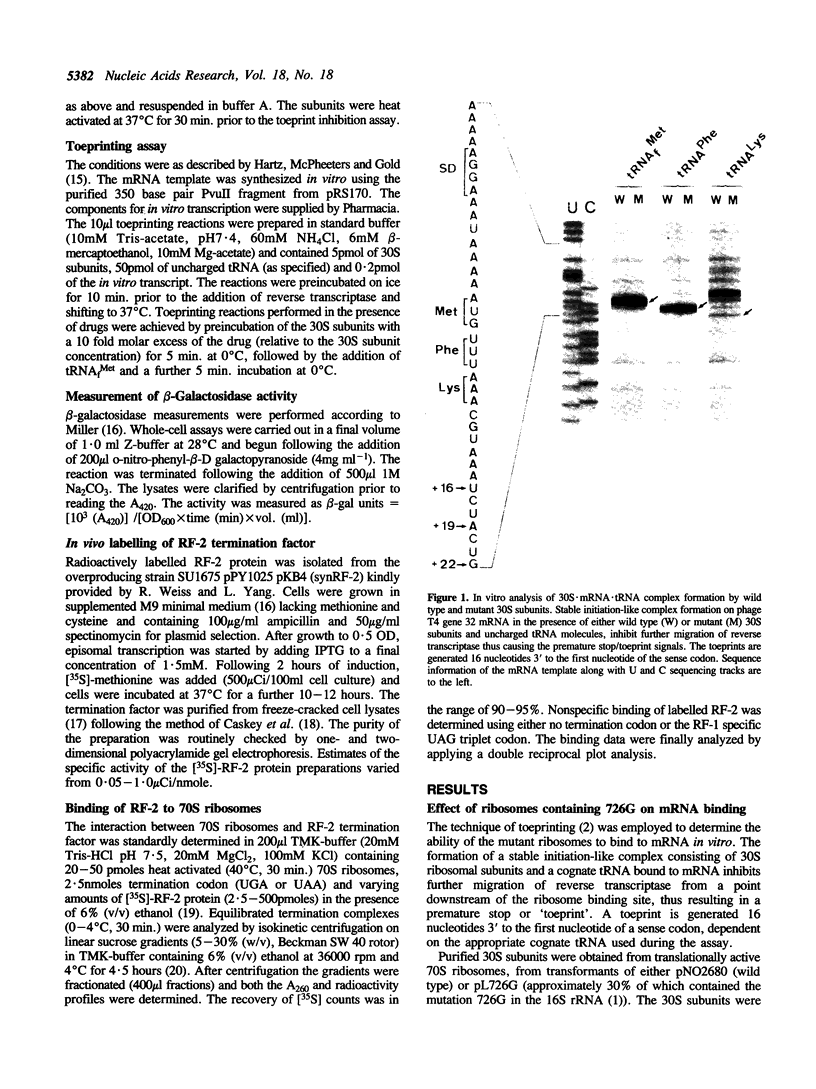
A single base change in 16S rRNA (C726 to G) has previously been shown to have a dramatic effect on protein synthesis in E. coli (1). This paper more specifically details the effects of the mutation on mRNA binding and translation-termination. The in vitro technique of toeprinting (2) was used to demonstrate that 30S subunits containing the mutation 726G had an altered binding affinity for mRNA by comparison to the wild type. In addition, expression of the mutant ribosomes in vivo resulted in exclusive suppression of the UGA nonsense codon. This effect was supported by in vitro studies that showed the mutant ribosomes to have an altered binding affinity for Release Factor-2.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Backendorf C., Ravensbergen C. J., Van der Plas J., van Boom J. H., Veeneman G., Van Duin J. Basepairing potential of the 3' terminus of 16S RNA: dependence on the functional state of the 30S subunit and the presence of protein S21. Nucleic Acids Res. 1981 Mar 25;9(6):1425–1444. doi: 10.1093/nar/9.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J., Dull T. J., Sleeter D. D., Noller H. F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981 May 15;148(2):107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- Dahlberg A. E. The functional role of ribosomal RNA in protein synthesis. Cell. 1989 May 19;57(4):525–529. doi: 10.1016/0092-8674(89)90122-0. [DOI] [PubMed] [Google Scholar]
- Draper D. E., Deckman I. C., Vartikar J. V. Physical studies of ribosomal protein-RNA interactions. Methods Enzymol. 1988;164:203–220. doi: 10.1016/s0076-6879(88)64044-4. [DOI] [PubMed] [Google Scholar]
- Fresno M., Carrasco L., Vazquez D. Initiation of the polypeptide chain by reticulocyte cell-free systems. Survey of different inhibitors of translation. Eur J Biochem. 1976 Sep 15;68(2):355–364. doi: 10.1111/j.1432-1033.1976.tb10822.x. [DOI] [PubMed] [Google Scholar]
- Ginzburg I., Miskin R., Zamir A. N-ethyl maleimide as a probe for the study of functional sites and conformations of 30 S ribosomal subunits. J Mol Biol. 1973 Sep 25;79(3):481–494. doi: 10.1016/0022-2836(73)90400-2. [DOI] [PubMed] [Google Scholar]
- Gourse R. L., Stark M. J., Dahlberg A. E. Regions of DNA involved in the stringent control of plasmid-encoded rRNA in vivo. Cell. 1983 Apr;32(4):1347–1354. doi: 10.1016/0092-8674(83)90315-x. [DOI] [PubMed] [Google Scholar]
- Hartz D., McPheeters D. S., Gold L. Selection of the initiator tRNA by Escherichia coli initiation factors. Genes Dev. 1989 Dec;3(12A):1899–1912. doi: 10.1101/gad.3.12a.1899. [DOI] [PubMed] [Google Scholar]
- Hartz D., McPheeters D. S., Traut R., Gold L. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 1988;164:419–425. doi: 10.1016/s0076-6879(88)64058-4. [DOI] [PubMed] [Google Scholar]
- Krisch H. M., Allet B. Nucleotide sequences involved in bacteriophage T4 gene 32 translational self-regulation. Proc Natl Acad Sci U S A. 1982 Aug;79(16):4937–4941. doi: 10.1073/pnas.79.16.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latif F. A., Schaup H. W. Escherichia coli 3'-terminal 16S rRNA sequence modulated fidelity during translation. Biochimie. 1988 Dec;70(12):1831–1839. doi: 10.1016/0300-9084(88)90045-4. [DOI] [PubMed] [Google Scholar]
- McPheeters D. S., Stormo G. D., Gold L. Autogenous regulatory site on the bacteriophage T4 gene 32 messenger RNA. J Mol Biol. 1988 Jun 5;201(3):517–535. doi: 10.1016/0022-2836(88)90634-1. [DOI] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature. 1987 Jun 4;327(6121):389–394. doi: 10.1038/327389a0. [DOI] [PubMed] [Google Scholar]
- Moazed D., Stern S., Noller H. F. Rapid chemical probing of conformation in 16 S ribosomal RNA and 30 S ribosomal subunits using primer extension. J Mol Biol. 1986 Feb 5;187(3):399–416. doi: 10.1016/0022-2836(86)90441-9. [DOI] [PubMed] [Google Scholar]
- Moazed D., Van Stolk B. J., Douthwaite S., Noller H. F. Interconversion of active and inactive 30 S ribosomal subunits is accompanied by a conformational change in the decoding region of 16 S rRNA. J Mol Biol. 1986 Oct 5;191(3):483–493. doi: 10.1016/0022-2836(86)90143-9. [DOI] [PubMed] [Google Scholar]
- Mougel M., Philippe C., Ebel J. P., Ehresmann B., Ehresmann C. The E. coli 16S rRNA binding site of ribosomal protein S15: higher-order structure in the absence and in the presence of the protein. Nucleic Acids Res. 1988 Apr 11;16(7):2825–2839. doi: 10.1093/nar/16.7.2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuyama A., Yoshikawa M., Tanaka N. Alteration of ribosomal protein S2 in kasugamycin-resistant mutant derived from Escherichia coli AB312. Biochem Biophys Res Commun. 1974 Oct 8;60(3):1163–1169. doi: 10.1016/0006-291x(74)90434-3. [DOI] [PubMed] [Google Scholar]
- Petrullo L. A., Gallagher P. J., Elseviers D. The role of 2-methylthio-N6-isopentenyladenosine in readthrough and suppression of nonsense codons in Escherichia coli. Mol Gen Genet. 1983;190(2):289–294. doi: 10.1007/BF00330653. [DOI] [PubMed] [Google Scholar]
- Prescott C. D., Dahlberg A. E. A single base change at 726 in 16S rRNA radically alters the pattern of proteins synthesized in vivo. EMBO J. 1990 Jan;9(1):289–294. doi: 10.1002/j.1460-2075.1990.tb08107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan V. R., Yabuki S., Sillers I. Y., Schindler D. G., Engelman D. M., Moore P. B. Positions of proteins S6, S11 and S15 in the 30 S ribosomal subunit of Escherichia coli. J Mol Biol. 1981 Dec 15;153(3):739–760. doi: 10.1016/0022-2836(81)90416-2. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigmund C. D., Ettayebi M., Morgan E. A. Antibiotic resistance mutations in 16S and 23S ribosomal RNA genes of Escherichia coli. Nucleic Acids Res. 1984 Jun 11;12(11):4653–4663. doi: 10.1093/nar/12.11.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark M. J., Gourse R. L., Dahlberg A. E. Site-directed mutagenesis of ribosomal RNA. Analysis of ribosomal RNA deletion mutants using maxicells. J Mol Biol. 1982 Aug 15;159(3):417–439. doi: 10.1016/0022-2836(82)90292-3. [DOI] [PubMed] [Google Scholar]
- Stöffler G., Tate W. P., Caskey C. T. Ribosomal proteins cross-linked to peptide chain termination release factor 2. J Biol Chem. 1982 Apr 25;257(8):4203–4206. [PubMed] [Google Scholar]
- Van Duin J., Wijnands R. The function of ribosomal protein S21 in protein synthesis. Eur J Biochem. 1981 Sep 1;118(3):615–619. doi: 10.1111/j.1432-1033.1981.tb05563.x. [DOI] [PubMed] [Google Scholar]