Abstract
ASF/SF2, a member of the serine-arginine (SR) protein family, has two RRM domains (RRM1 and RRM2) and a C-terminus domain rich in RS dipeptides. The SR protein kinase 1 (SRPK1) phosphorylates approximately 12 of these serines using a semi-processive mechanism. The x-ray structure of the ASF/SF2:SRPK1 complex revealed several features of the complex that raised intriguing questions of how the substrate is phosphorylated by the kinase: The part of the RS domain destined to be phosphorylated at later stages of the reaction docks to a kinase groove distal to the active site while the neighboring RRM2 binds near the active site (1). In this study we investigated the interplay between the RS domain and RRM2 for stable association and phosphorylation of ASF/SF2. Despite several contacts in the enzyme-substrate complex, free RRM2 does not bind efficiently to SRPK1 unless the docking groove is occupied by the RS domain. This domain cross-talk enhances the processive phosphorylation of the RS domain. The RRM:SRPK1 contact residues control the folding of a critical beta strand in RRM2. Unfolding of this structural element may force the N-terminal serines of the RS domain into the active site for sequential phosphorylation. Thus, ASF/SF2 represents a new class of substrates that use unique primary sequence to induce allosteric binding, processive phosphorylation, and product release.
The SR protein family plays essential roles in pre-mRNA splicing. These proteins are thought to be involved in every step during the assembly of the spliceosome, the catalyst for splicing reactions (2, 3). SR proteins are modular and contain one or two RNA recognition motifs (RRMs) at the N-terminus and an arginine-serine dipeptide rich domain at the C-terminus. One of the most well studied members of the SR protein family, ASF/SF2, contains two N-terminal RRMs followed by a 50-residue C-terminus RS domain. The RS domain can be divided into two modules, RS1 and RS2, whose sequence and structural properties allow them to be distinctly recognized and regulated by functionally unique kinases. Of the eighteen serines in the RS domain, twelve are present as every other residue, primarily as RS dipeptides within a 24 residue-stretch, which is referred to as the RS1 motif (residues 204 to 227). The other six serines are present within the C-terminus RS2 motif (residues 228–248).
Members of two kinase families, SR protein kinase (SRPK) and Cdc2-like-kinase/Serine-Threonine-Tyrosine (CLK/STY), phosphorylate the RS domain following the distribution rules of the serines: SRPK1 phosphorylates the RS1 serines in the cytoplasm generating hypo-phosphorylated ASF/SF2 (p-ASF/SF2). CLK/STY kinases are nuclear and they further phosphorylate the RS2 serines generating hyper-phosphorylated ASF/SF2 (pp-ASF/SF2) (4). Hypophosphorylation of ASF/SF2, p-ASF/SF2, is essential for the nuclear import of this SR protein (5, 6). Splicing experiments in vitro have demonstrated that ASF/SF2 undergoes cycles of phosphorylation and dephosphorylation during the assembly of the spliceosome (7, 8, 9). Several recent reports support this by showing that both SRPKs and protein phosphatases are present within the spliceosome (10, 11, 12). It is however unclear if phosphorylation and dephosphorylation of ASF/SF2 are subject to the entire RS domain (RS1 and RS2) or either one of the two modules (RS1 or RS2). The basic principles of SR protein phosphorylation by SRPK1 are emerging from recent kinetic and structural studies. The phosphorylation mechanism in the ASF/SF2:SRPK1 system defines a new paradigm for protein phosphorylation: SRPK1 has been shown to bind ASF/SF2 with high affinity (Kd = 50 nM) which promotes processive-phosphorylation of about 8 of the 12 serines in the RS1 motif. This extended sequential reaction requires sliding of the substrate along a highly charged groove in the kinase (5, 13).
The x-ray crystal structure of ASF/SF2: SRPK1: AMP-PNP complex provided clues for the basis of high affinity binding between SRPK1 and ASF/SF2 (1). The complex used for crystallization used the core ASF/SF2 fragment spanning residues 105–219 while SRPK1 in the complex is devoid of a long insert, known as the spacer domain, that bifurcates the two kinase domains and the N-terminus non-homologous regions (SRPK1ΔNS3) (Figure 1A). The structure of the core ASF/SF2: SRPK1 complex showed tripartite interaction between the kinase and substrate. The most surprising of which is the N-terminus half of the RS1 motif that is destined to be phosphorylated remains bound to a groove distal to active site (Figure 1B–1C). This interaction is dominated by electrostatics where a negatively charged surface interacts with the positive charged RS domain. A second interface is between a phospho-serine and a positively charged pocket of the kinase. We showed in a previous study that the pocket surrounding this phospho-serine plays an important role in maintaining high efficient phosphorylation of the RS domain (14).
Figure 1. Structural model of the ASF/SF2: SRPK1 complex.
(A) Schematic representation of the fragments of the kinase SRPK1ΔNS3 (deleted N-terminus region and spacer domain) and substrate ASF/SF2 (105–219) used for crystallization. (B) Ribbon diagram of the X-ray crystal structure of the complex between the core fragments of ASF/SF2 (ASF/SF2 (105–219)) and SRPK1 (SRPKΔNS3). (C) Cartoon depicting the structural model of the complex. The same cartoon is used throughout other figures.
The last interface is formed between the RRM2 of ASF/SF2 and the kinase where RRM2 makes a 3-point contact with both the small and large lobes at the front of the kinase (Figure 1B–1C). In particular, W134 and Q135 of ASF/SF2 are stacked against H90 and W88 of SRPK1, while R154 protrudes into a pocket formed by helices αD and αF, and is stabilized by interaction with E534 and Y549. Additionally, Q135 also contacts the backbone of the glycine-rich loop of SRPK1. In all, nearly 1100Å of surface area is buried upon the complex formation. The β4 motif of RRM2, which was previously thought to be involved in binding to the kinase remained in a folded conformation in the structure (13). Based on the structural information and correlated cross-linking and secondary structure melting analysis, we showed that the N-terminal portion of the RS domain (N′-RS1) translocates from the docking groove to the active site in a sequential manner during catalysis, which ultimately requires the melting of β4 strand in RRM2 (1). It was proposed that β4 docks into the kinase groove during the phopshorylation of the last few serines. However, β4 may not interact strongly in the kinase docking groove and may facilitate dissociation of the phosphorylated ASF/SF2. The β4 motif thus, may act as a dissociative docking motif.
Kinetic studies have shown that the RS domain linked to a fragment of RRM2 has an intrinsic capacity to initiate processive phosphorylation but that it is very limited in extending this reaction beyond 3 steps (14). These observations suggest a potential role for the RRM in the mechanism of ASF/SF2 phosphorylation. In this work we have investigated the role of RRM2 in kinase binding and phosphorylation. We find that the isolated RRM binds weakly to the kinase, and, alteration of all three contact residues in RRM2 enhance the kinase binding to a small extent suggesting that the binding interface is flexible. Surprisingly, binding is significantly enhanced in the presence of the RS peptide. Moreover, this RS dependent enhancement of RRM binding is observed mainly with the full length but not SRPK1ΔNS3. Finally, we show that in the presence of the RRM, SRPK1 phosphorylates the isolated RS domain in enhanced efficiency. We propose a model of substrate recognition, phosphorylation, and product release.
Materials & Methods
Cloning and Expression of Recombinant Proteins
Expression and purification of His-SRPK1 constructs with Ni-resin affinity chromatography have been previously described (Aubol et al., 2003). GST-ASF/SF2 constructs were generated in pGEX4T2 vector as described previously (13). E. coli BL21(DE3) cells harboring different plasmids were grown to optical density 0.4 (A600) followed by overnight induction with 0.1mM IPTG. Cells were then pelleted, resuspended in 150mL of lysis buffer (20mM Tris-HCl (pH 7.5), 300mM NaCl, 10% glycerol, 1mM DTT, and protease inhibitor cocktail), and lysed. The lysed cells were loaded on a glutathione sepharose column (GE Healthcare) for 3 hours at 4°C and bound protein was eluted with lysis buffer containing 20mM reduced glutathione. The eluted protein was dialyzed against lysis buffer for 4 hours at 4°C and concentrated. His-ASF/SF2(RS) construct was refolded and purified using previously published protocol (Velazquez-Dones et al., 2005).
GST Pulldown Assays
GST-fusion ASF/SF2 (10μM) was reacted with His-SRPK1 (20μM) for 30 minutes in binding buffer (20mM Tris-HCl (pH 7.5), 150mM NaCl, 0.2% Triton X-100, 1mM DTT). The mixture was incubated with 15μL of glutathione resin pre-equilibrated in same binding buffer for 25 minutes. The resin was washed 3 times with 700μL of binding buffer and the bound protein was eluted by boiling with 4μL of 4X SDS-PAGE loading dye for 2 minutes at 90°C. Samples were then loaded onto 12.5% SDS gel and visualized by Coomassie stain or Western blot.
Western Blotting
Protein samples were resolved on 12.5% SDS-PAGE, and transferred to an Immobilon polyvinylidene difluoride membrane (Millipore). Membrane was then probed with either α-His or α-GST antibodies (Santa Cruz Biotechnology).
Chemical Crosslinking
ASF/SF2 and SRPK1 were separately incubated with buffer containing (20mM Tris-HCl (pH 7.5), 300mM NaCl, 10% glycerol, and 10mM DTT) on ice for 1 hour, and exchanged with crosslinking buffer (20mM Tris-HCl (pH 7.5), 300mM NaCl, and 10% glycerol) using desalting spin columns (Pierce). 10μM ASF/SF2 was then mixed with 20μM SRPK1. 1mM BMB was added to the mixture and the crosslinking reaction was carried out at room temperature. Equal-volume aliquots were removed at each time point and quenched with 4X non-reducing SDS-PAGE loading dye. The reactions were resolved by Western blot with α-GST antibody.
In Vitro Kinase Assay
The phosphorylation of wild-type ASF/SF2 and RS domain by SRPK1 was carried out according to previously published procedures in the presence of 50 mM Mops (pH 7.4), 10 mM free Mg2+, 5 mg/mL BSA, and [γ-32P]ATP (600–1000 cpm pmol−1) at 23 °C (5). Reactions were typically initiated with the addition of 32P-ATP (100 mM) in a total reaction volume of 10 μL and then were quenched with 10 μL SDS-PAGE loading buffer. Each quenched reaction was loaded onto a 12% SDS-PAGE gel and the dried gels were exposed with Kodak imaging film (Biomax MR). The protein bands corresponding to phosphorylated ASF/SF2 were excised and counted on the 32P channel in liquid scintillant. All samples in competition and start-trap assay were processed through this SDS-PAGE gel method.
Steady-state Assay
Steady-state kinase assay was performed with 1 nM SRPK and 50 nM ASF construct at room temperature and pH 7.5, and stopped with SDS-quench buffer at different time points. We assessed possible competitive inhibition between GST-ASFΔRS and RS domain by using up to 1.6 μM GST-ASFΔRS in two-minute phosphorylation reaction. We fitted the data to a partial competitive inhibition equation. We collected and fitted kinetic data on RS domain phosphorylation to Michaelis-Menton equation to obtain Km,RS values at different GST-ASFΔRS concentrations. The Km,RS values were fitted to the partial competitive inhibition model.
Single-turnover Start-Trap Assay
The start-trap experiment was carried out according to previously published procedures (5). For start-trap, we first incubated 1 mM SRPK and 700 nM RS domain, initiated the reaction with ATP, and then challenged the complex by adding excess kinase-dead SRPK trap. GST-ASFΔβ4ΔRS (1 μM) was added for one set of start-trap experiment to assess its impact on processive phosphorylation. For positive control curve, we incubated full-length ASF/SF2 with SRPK1, initiated the reaction with ATP in the presence of no trap. Experiments were performed at room temperature and at pH 7.5. For trap-start control curve, we pre-mixed SRPK trap with SRPK1 and full-length ASF/SF2, and then initiated the reaction with ATP to assess for the trap inhibitory potential. The data was split into a fast and a slow phase for detailed kinetic analysis of processive phosphorylation. Processive phosphorylation would shift start-trap curve to the positive control curve, while distributive phosphorylation would shift start-trap curve to the trap-start control curve.
Results
Isolated RRM2 of ASF/SF2 Binds SRPK1 with Low Affinity
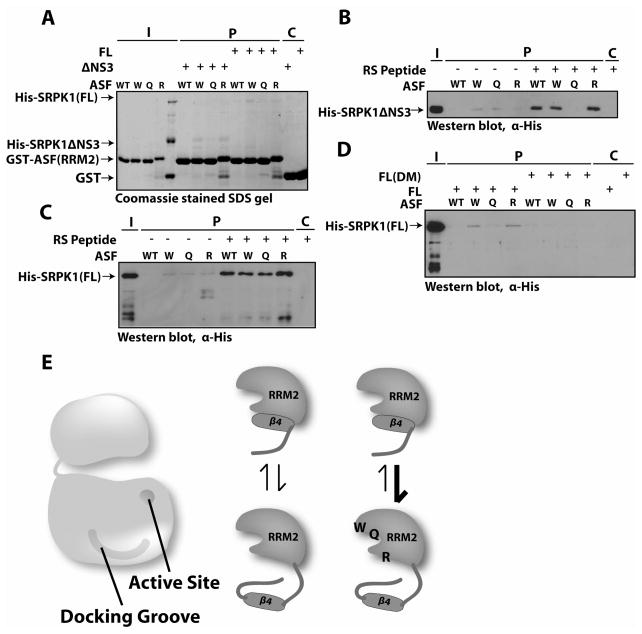
The x-ray structure reveals that the protein-protein interface between RRM2 and SRPK1, while extensive in surface area, has few direct residue contacts and lacks any recognizable hot-spot. Indeed, one of the four direct contacts is mediated by a glutamine with an energetically unfavorable conformation. These findings raise the question of how well RRM2 interacts with SRPK1 in the absence of other substrate binding components such as the RS domain. To investigate the role of the RRM2 residues in the formation of the binding interface with SRPK1, we expressed and purified ASF/SF2(RRM2) (residues 105–196) as a GST-fusion protein and performed binding experiments with both the full length SRPK1 and the truncated form used for co-crystallization which lacks the entire spacer region and first 56 residues at the N-terminus (SRPK1ΔNS3) (Figure 1A & 2A). Despite the extensive interface between RRM2 and the kinase (Figure 1B), RRM2 does not bind with sufficient affinity for SRPK1 and SRPK1dNS3 to be pulled down with it (Figure 2B–C).
Figure 2. RRM2 alone does not interact with SRPK1.
(A) Schematic representation of ASF/SF2 constructs used in the binding experiments with SRPK1. (B–C) GST co-precipitation of His-SRPK1(FL) and His-SRPKΔNS3 with GST-ASF(RRM2), visualized by Coomassie stain (B) and Western blot with α-His antibody (C). In all cases, I, P, C refer to input, precipitated (pulldown) fraction (bound), and GST only control, respectively. (D–E) GST co-precipitation of His-SRPK1(FL) and His-SRPKΔNS3 with GST-ASF(RRM2) in the presence of AMP-PNP, visualized by Coomassie stain (D) and Western blot with α-His antibody (E). (F–G) GST co-precipitation of His-SRPK1(FL) and His-SRPKΔNS3 with GST-ASF(RS), visualized by Coomassie stain (F) and Western blot with α-His antibody (G).
Since the current x-ray structure of the complex was solved with an ATP analog AMP-PNP, nucleotide binding may be essential for generating the interface between RRM2 and SRPK1. In the crystal structure of the ASF/SF2 (105–219):SRPK1ΔNS3 complex, AMP-PNP lies in the nucleotide pocket in the small lobe of the kinase and contacts residues in the glycine-rich loop, a critical segment for catalysis that also contains residues that interact with RRM2. To test the possibility that AMP-PNP could support association of RRM2 and SRPK1, we added the nucleotide in a 10:1 molar excess to the kinase and then performed pulldown assays. Our result clearly shows that AMP-PNP does not enhance binding interactions between ASF/SF2 and the full length or truncated kinases (Figure 2D–E). Overall, these findings indicate that RRM2 has poor intrinsic affinity for SRPK1, that ATP in the active site does not induce a conformation that promotes RRM2 binding, and that the insert does not play a role in stable RRM2 association.
Since a portion of the RS domain (N′-RS1) docks within the acidic groove of the large lobe of the kinase and buries approximately 900Å angstrom of free surface area in the x-ray structure, we wished to examine how well this domain interacts with SRPK1 as compared to the free RRM2. Pulldown assays using GST-ASF(RS) (residues 183–248) with the full length and truncated SRPK1 were carried out (Figure 2A). We observed strong interactions between the RS domain and both forms of the kinase (Figure 2F–G). These results suggest that the RS domain of ASF/SF2 binds with high affinity whether the SRPK1 insert is present or not. Overall, the pulldown assays suggest that while both the RS domain and RRM2 contacts bury significant and equivalent surface areas on SRPK1, the free RS domain and not RRM2 binds with apparent high affinity.
Docking of RS Peptide Induces RRM2 Binding
Since RRM2 does not appear to bind with high affinity to SRPK1, we wished to test the possibility that the occupancy of the docking groove by part of the RS domain is a prerequisite for stable association of RRM2 with the kinase. To test this model, we performed the same pulldown experiments in the presence of a 32-mer peptide with 16 Arg-Ser dipeptide repeats, (RS)16. The peptide was added in a 2:1 molar excess to SRPK1. Consistent with our hypothesis, we observed enhanced interactions between RRM2 and the kinase in the presence of the RS peptide (Figure 3A–B). In addition, the effect of enhancement seems to be stronger for the full length kinase than the truncated one. This observation led us to believe that the N-terminus and/or the spacer domain of the enzyme play an important role in binding. SR proteins are highly basic and bind non-specifically to cellular RNA. These RNAs remain bound to protein if care is not taken to separate them. We have tested the quality of protein by analyzing protein preparations by agarose gel and staining with ethidium bromide. We found that the protein used here does not contain RNA (Figure S1A). To further confirm that RNA contamination did not affect our result, we have carried out the pulldown in the presence of RNase. We found no difference in the binding efficiency (Figure S1B).
Figure 3. RRM2 interacts strongly with wild-type SRPK1 in the presence of RS peptide.
(A–B) GST co-precipitation of His-SRPK1(FL) and His-SRPKΔNS3 with GST-ASF(RRM2) in the presence of RS peptide, visualized by Coomassie stain (A) and Western blot with α-His antibody (B). (C) GST co-precipitation of His-SRPK1ΔN, His-SRPK1ΔS, and His-SRPK1ΔNS3 with GST-ASF(RRM2) in the presence of RS peptide, visualized by Coomassie stain. (D) Docking of RS peptide in the groove of the SRPK1 induces conformational change within the kinase, and allowing the N-terminus and spacer domain to stabilize the RRM2:SRPK1 association. (E) GST co-precipitation of full-length His-SRPK1containing the docking groove mutations (DM) with GST-ASF(RRM2) in the presence of RS peptide, visualized by Western blot with α-His antibody.
To further test which of the deleted segments in the kinase are involved in binding enhancement, we compared the interaction of RRM2 with three different SRPK1 constructs: SRPK1ΔNS3, SRPK1ΔS (spacer deleted), and SRPK1ΔN (N-terminal 69 residues deleted) (Figure 1A), all in the presence of the RS peptide. We saw strong interaction between RRM2 and ΔS, and only minor enhancement with ΔNS3 and ΔN (Figure 3C). We conclude that RRM2 alone has very weak binding affinity and can only associate efficiently after the RS domain has docked to SRPK1, inducing an allosteric change within the complex and allowing the N-terminus of the kinase to stabilize the association (Figure 3D). The spacer domain may still play a small role in stabilizing the complex. There may also be some communication between RRM2 and the RS domain after it has docked that helps RRM2 adopt a favorable folding state for binding.
To address whether this phenomenon is the result of interaction with the docking groove rather than other regions of the enzymes, we investigated the RS peptide-induced binding of RRM2 using a docking-defective form of SRPK1. We mutated six residues to alanine, (D548,E557,E558,D564,E571,K615) in the full length kinase that contact the RS peptide, generating the docking mutant, FL(DM). We then carried out the pulldown experiment with the RS peptide and FL(DM), and observed no enhancement in binding (Figure 3E). This confirmed that the RS peptide is docking to the kinase groove and assisting RRM2 binding in an allosteric manner.
Mutations at the RRM2:SRPK1 Interface Promote Binding
Having established that occupancy of the docking groove by the RS domain promotes stable RRM2 binding, we next wanted to examine the role of RRM2 residues that contact the enzyme. The interface formed between RRM2 of ASF/SF2 and the front of the kinase lobe is unusual in that although 1100Å of surface area is buried upon the complex formation, only four residues (W134, Q135, R154 and E184) make direct contact with the kinase and the primary attachment between the two molecules occurs at a single small area consisting of W134 and Q135 from RRM2. We mutated independently three of the four contact residues to alanine to generate RRM2-W134A, RRM2-Q135A and RRM2-R154A. We did not mutate E184 because the hydrogen bond mediated by this residue appears to be least favorable based on distance. To our surprise, pulldown experiments showed that the mutants bound with slightly improved affinity to both SRPK1ΔNS3 and full length SRPK1 in the absence of the RS peptide (Figure 4A). In the presence of the RS peptide, binding of the RRM2 mutants to the kinase was similar to that for RRM2 wild type and the kinase (Figure 4B–C). These findings suggest that another mode of binding besides these contact residues may also contribute to stabilizing the kinase-substrate complex.
Figure 4. RRM2 contact mutants bind SRPK1 better than wild type RRM2.
(A) GST co-precipitation of His-SRPK1(FL) and His-SRPKΔNS3 with GST-ASF(RRM2), GST-ASF(RRM2)-W134A, GST-ASF(RRM2)-Q135A, and GST-ASF(RRM2)-R15A, visualized by Coomassie staining. (B) GST co-precipitation of His-SRPKΔNS3 with GST-ASF(RRM2), GST-ASF(RRM2)-W134A, GST-ASF(RRM2)-Q135A, and GST-ASF(RRM2)-R15A in the absence and presence of RS peptide, visualized by Western blot with α-His antibody. (C) GST co-precipitation of His-SRPK(FL) with GST-ASF(RRM2), GST-ASF(RRM2)-W134A, GST-ASF(RRM2)-Q135A, and GST-ASF(RRM2)-R15A in the absence and presence of RS peptide, visualized by Western blot with α-His antibody.
We have shown previously that β4 (residues 191–196) in RRM2 unfolds and occupies the docking groove as the RS domain is phosphorylated in a C-to-N-terminus direction (1). These findings led us to speculate that β4 of the free RRM2 may not be a stable secondary structural element and may exist in equilibrium between folded and unfolded states (Figure 4E). In this model, when β4 is unfolded, it can bind to the docking groove of the kinase preventing interactions at sites seen in the structure. The fact that the RRM2 mutants bind with similar or slightly higher affinity than wild-type RRM2 in the absence of the RS peptide could be due to the fact that these mutants may have induced the unfolding of β4 and provides an additional docking motif for the domain. In other words, the mutations at the interface may change the folding-unfolding equilibrium of β4 such that this secondary structure unfolds and can be captured by the docking groove. To test this hypothesis, we performed binding experiments using the RRM2 mutants and the FL(DM) kinase version. If the interaction between RRM2 mutants and SRPK1 occurs through docking of the β4 motif into the groove of the kinase, then the docking-defective kinase should not be able to capture the ASF/SF2 mutants. Consistent with our hypothesis, we did not observe binding of the RRM2 mutants with FL(DM), but still observed binding of these mutants with wild type SRPK1 (Figure 4D). These findings suggest that the inability to destabilize RRM2-SRPK1 interactions may be due to an alternative binding mode for RRM2 that involves the docking groove of SRPK1.
To further test whether the alternative binding mode of RRM2 involves the unfolding of β4 and subsequent interactions with the docking groove, we carried out cross-linking experiments. In the context of RRM2, we mutated a lysine residue in β4 and an arginine in the kinase docking groove to cysteine and then probed for Cys-induced cross-linking (Figure S2C). We previously showed that these two residues come in close proximity during the last phase of RS1 phosphorylation (1). As shown in figure S2A, we found that SRPK1 cross-links with RRM2, suggesting that β4 interacts with the docking groove. Also, a cross-linked complex is not observed for RRM2 lacking K193C suggesting that the engineered cysteines are responsible for the intermolecular reaction between the proteins. We then tested whether cross-linking could occur with the W134A mutant. We found that RRM2-W134A cross-links more efficiently than wild type RRM2 (Figure S2B). It is important to note here that the efficiency of cross-linking is poor as the free RRM2 binds extremely weak to kinase. Nevertheless, while our results show that RRM2 does not interact favorably with the kinase, the interfacial residues discussed above are involved in modulating the stability of β4 and its interactions with the docking groove.
RRM2 Modulates RS Domain Phosphorylation
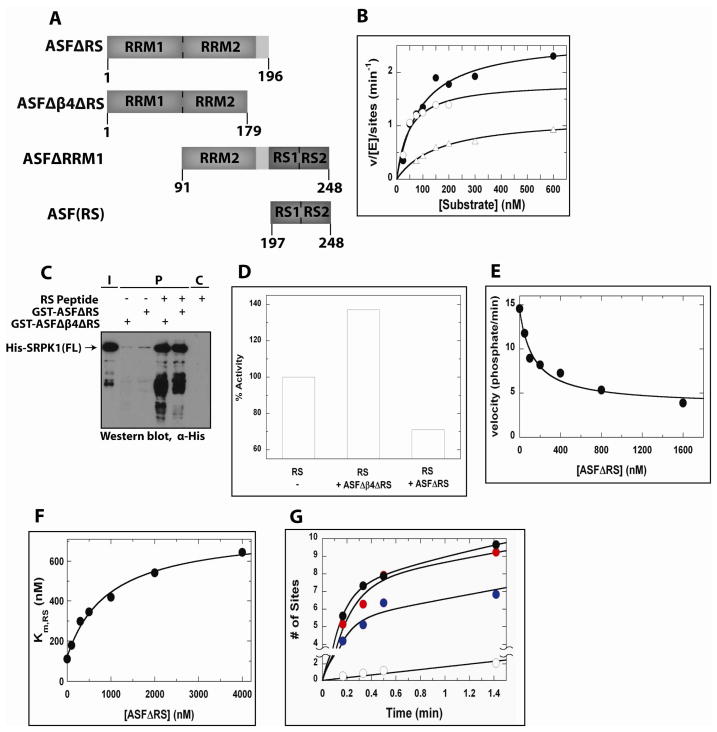
The above observations suggest that interactions between the RS domain and SRPK1 stabilize the RRM2: kinase interface. Given the RS domain-induced binding of RRM2 near the kinase catalytic site, we wished to investigate how this allosteric phenomenon alters the rate of phosphorylation. To verify that the RRMs play a catalytic role, the steady-state kinetics for the phosphorylation of wild-type ASF/SF2 and two substrates lacking RRM1 [ASFΔRRM1] and both RRM1 and RRM1 [ASF(RS)] were measured (Figure 5A). Overall, N-terminal truncation of ASF/SF2 led to decrease in Vmax consistent with a positive role for the RRMs in generating the phosphorylated product (Figure 5B). To further investigate how the RRMs control catalysis, we next studied a construct containing both RRM1 and RRM2 but lacking the RS domain [GST-ASFΔRS (residues 1–196)] and a second construct that also lacked β4 [GST-ASFΔβ4ΔRS (residues 1–179)] (Figure 5A). Consistent with our previous binding results, both truncated proteins exhibited very poor binding to full-length kinase, and the RS peptide can significantly enhance binding in both cases (Figure 5C). We next tested if the isolated RRM domain constructs could affect the phosphorylation of the isolated RS domain. We found that while GST-ASFΔβ4ΔRS enhanced the catalytic activity of SRPK1 toward the RS domain, GST-RRMΔRS actually lowered the activity in a steady-state kinetic assay containing stoichiometric amounts of GST-RRMΔRS and RS domain (Figure 5D). To more carefully evaluate the latter phenomenon we measured the reaction velocity as a function of GST-ASFΔRS and found that this RRM version inhibited phosphorylation by up to 3-fold (Figure 5E). Interestingly, the inability of GST-ASFΔRS to completely inhibit the reaction at high concentrations suggests that it possesses some non-overlapping binding determinants with the RS domain. To understand the nature of this inhibition, the Km for the RS domain was measured as a function of GST-ASFΔRS. We found that GST-ASFΔRS increased the Km in a hyperbolic fashion consistent with partial competitive inhibition (Figure 5F). These findings indicate that GST-ASFΔRS and the RS domain bind simultaneously to SRPK1 and negatively influence each other’s affinity. Since this inhibition occurs with GST-ASFΔRS and not with GST-ASFΔβ4ΔRS (Figure 5D), we presume that the RS domain and β4 of RRM2 may compete for the docking groove in SRPK1. Overall, these data suggest that GST-RRMΔRS and the RS domain possess both non-overlapping and overlapping regions.
Figure 5. The RRM domains regulate phosphorylation of RS domain.
(A) Schematic representation of ASF/SF2 constructs used for binding and phosphorylation experiments. (B) Steady-state assay showed deletion of RRM domain(s) decreased phosphorylation rate by SRPK1 ((●) ASF(WT), (○) ASF(ΔRRM1) and (Δ) ASF(RS)). (C) GST co-precipitation of His-SRPK1(FL) with GST-ASFΔβ4ΔRS and GST-ASFΔRS in the absence and presence of (RS)32-mer peptide, visualized by Western blot with α-His antibody. (D) Steady-state phosphorylation assay performed on RS domain showed kinetic enhancement in the presence of GST-ASFΔβ4ΔRS and inhibition in the presence of GST-ASFΔRS. Data at half-minute time point was taken from RS domain phosphorylation progress curve and compared. (E) Steady-state competition assay showed partial inhibition of RS domain phosphorylation by GST-ASFΔRS. (F) Saturating Km,RS values confirm partial competitive inhibition by GST-ASFΔRS. (G) Single-turnover start-trap assay showed enhanced processive phosphorylation of RS domain in the presence of GST-ASFΔβ4ΔRS. (●) Positive control curve: Phosphorylation of full-length ASF/SF2 by SRPK1 in presence of no trap. (○) Trap-start control curve: trap added to mixture of full-length ASF/SF2 and SRPK1 prior to initiation with ATP. ( ) Start-trap curve: pre-equilibrated mixture of RS domain and SRPK1 was initiated with ATP before excess trap was added. (
) Start-trap curve: pre-equilibrated mixture of RS domain and SRPK1 was initiated with ATP before excess trap was added. ( ) Start-trap curve with GST-ASFΔβ4ΔRS: presence of GST-ASFΔβ4ΔRS shifted start-trap curve towards the positive control curve with no SRPK trap.
) Start-trap curve with GST-ASFΔβ4ΔRS: presence of GST-ASFΔβ4ΔRS shifted start-trap curve towards the positive control curve with no SRPK trap.
RRM2 Enhances Processive Phosphorylation of the RS Domain
We showed that RRM2 can have varying effects on RS domain phosphorylation in a steady-state kinetic assay depending on the presence of a beta strand (β4) in RRM2 (Figure 5D). In the absence of β4, RRM2 enhances phosphorylation rates whereas in the presence of β4, the domain perturbs the binding of the RS domain. To study how RRM2 impacts the phosphorylation mechanism we asked whether this domain affects the processive character of the reaction. In prior studies we showed that ASF/SF2 is phosphorylated up to 8 sites in a processive manner and that removal of the RRMs lowers the extent of this processive phase (5). To investigate whether the RRMs can affect processivity, we used a start-trap assay described previously in which free forms of the substrate, as expected in a distributive reaction, may be trapped by an inactive form of SRPK1 (kdSRPK1). This experiment is performed in single turnover mode by first pre-equilibrating SRPK1 and the RS domain in the absence and presence of GST-ASFΔβ4ΔRS and then starting the reaction with the addition of ATP and excess kdSRPK1. We find that wild-type ASF/SF2 is processively phosphorylated at about 7 serines before the substrate dissociates and binds kdSRPK1. In trap-start experiments in which kdSRPK1 is added prior to ATP, the reaction is effectively inhibited indicating that the inactive kinase is an effective trap and can bind well to free phosphoforms of the substrate. In comparison, the free RS domain is processively phosphorylated at only 5 serines before SRPK1 dissociates. Interestingly, the addition of the GST-ASFΔβ4ΔRS permits a higher level of processive phosphorylation from 5 to 7 serines (Figure 5G). These finding suggest that the RRMs play an important role in governing both the binding mode and the phosphorylation mechanism of the RS domain.
Discussion
Protein kinases modify their substrates by recognizing sequences that are both local and distal to the site of phosphorylation. While the local residues provide a rudimentary platform for phosphorylation, the distal residues can offer greater specificity and overall binding affinity (15, 16). These distal contacts have been best characterized using crystallographic methods and there are now several x-ray structures where such docking sites have been identified (17). We recently solved a structure for SRPK1 with a portion of its protein substrate bound. This structure highlights the potential role of a docking site in SRPK1 for the RS domain and several contact points for one of the RRMs (RRM2). The structure suggests that the RS domain and RRM2 are uniquely positioned to initiate a highly directional (C-to-N-terminus) phosphorylation reaction where the RS domain slides through an extended electronegative channel separating the docking groove and the active site. RRM2 binds toward the periphery of the active site and guides the directional phosphorylation mechanism. That a dynamic interplay between substrate and kinase exists is evident in the observed unfolding and translocation of the final beta strand in RRM2 during the phosphorylation reaction (1). These studies highlight a unique characteristic of ASF/SF2 not seen in other protein kinase substrates thus far. Rather than displaying a simple rigid set of binding determinants for kinase recognition, the structure of ASF/SF2 accommodates an elaborate poly-phosphorylation reaction by changing its conformation during the multiple catalytic cycles. In this manuscript we now address the cooperativity between the RS domain and RRM2 and show how a flexible set of inter- and intramolecular contacts guide the phosphorylation reaction.
Allosteric Regulation of Complex Formation Through the Docking Groove and Kinase N-terminus
In a prior study we showed that an RRM1-RRM2 construct effectively displaced the full-length ASF/SF2 from SRPK1 indicating that the RRMs possess sufficient contacts for high affinity binding (14). These observations are made in catalytic assays where the docking groove is initially bound by the RS domain of ASF/SF2. In this new study we performed pulldown assays which do not require catalytic turnover and occupancy of the docking groove by the RS domain. Using this noncatalytic assay, we found that RRM2 or both RRMs (1 plus 2) actually bind SRPK1 poorly. Instead, high affinity binding of the RRMs is only attained in the presence of a peptide mimicking the RS domain. These findings suggest that occupancy of the docking groove by the RS domain induces a conformation in SRPK1 that promotes this interaction. Since this regulation is seen to greater extent with SRPK1 version still containing the N-terminus (full-length and ΔS), it is likely that the N-terminus plays a significant role in this phenomenon. At present, it is unclear how the N-terminus induces high affinity binding of the RRMs at a molecular level. One possibility is that structural changes in the kinase induced by the docking interaction allow the N-terminus to directly contact the RRMs. However, an indirect role of the N-terminus in enhancing the RRM-kinase core interaction cannot be ruled out. In such an allosteric model, the N-terminus would induce long-range changes in the kinase core that extend across the molecule impacting binding determinants for RRM2.
Domain Cross-Talk Promotes Processive Phosphorylation of ASF/SF2
Our results show that additional contacts between RRM2 and the full-length SRPK1 influence the rate and mechanism of substrate phosphorylation. We show that the catalytic activity of full-length SRPK1 may be enhanced through binding to both domains of ASF/SF2. Results here have provided insights into the function of the RRM domain in the regulation of ASF/SF2 phosphorylation. We showed that while the RS domain has an intrinsic capacity for processive phosphorylation, the presence of the RRMs enhances the length of the processive phase from 5 to 7 sites. These results suggest that the docking groove is sufficient to support a basal level of processivity and additional contacts from the RRMs further support processivity. These findings imply that communication between the docking groove and the RRM contacts is bidirectional. While the docking groove improves contacts with the RRMs through an allosteric mechanism requiring the N-terminus and possibly the insert domain, the RRMs can also impact catalytic events in the active site through enhancement of processive phosphorylation. Since processivity reflects the relationship between substrate affinity and forward catalysis, we propose that the enhanced effect in the presence of the RRMs is due to higher affinity binding of phospho-intermediates later in the reaction progress curve. While SRPK1 begins to release phospho-forms of the RS domain after approximately 5 serines are modified, the RRMs are likely to enhance the affinity of these forms and extend the processive phase for an additional two sites. Further experiments are required to dissect the contributions of rate and binding energetics in claming strategy.
Interfacial Residues Control Folding of the β4 Motif in RRM2
CD experiments showed structural transition of the SRPK1:ASF/SF2 (105–219) complex from beta to random coil only in the presence of ATP but not AMP-PNP (1). No such transition was observed when the SRPK1: ASF/SF2(RS) (residues 188–248) complex was used. We reasoned that β4 of RRM2 unfolds during RS domain phosphorylation to accommodate N-terminal sequences in the active site of SRPK1 (1). This structural model explains how SRPK1 can initiate phosphorylation at the C-terminus end and then sequentially modify RS1 in the N-terminus direction. In kinetic studies we now show that β4 possesses some flexibility as an RRM1-RRM2 construct containing this strand can inhibit RS domain phosphorylation in a partially competitive mode whereas a construct lacking this strand activates RS domain phosphorylation (Figure 5E–5F). How phosphorylation could induce strand unfolding and translocation is not well understood. We observe that mutation of several residues at the RRM2-SRPK1 binding interface slightly improve affinity and increases the flexibility of β4 in cross linking experiments (Figure S2B). Such findings suggest that domain-domain contacts may be coupled to the strand unfolding mechanism.
A Model of Allosteric Regulation in Processive Phosphorylation
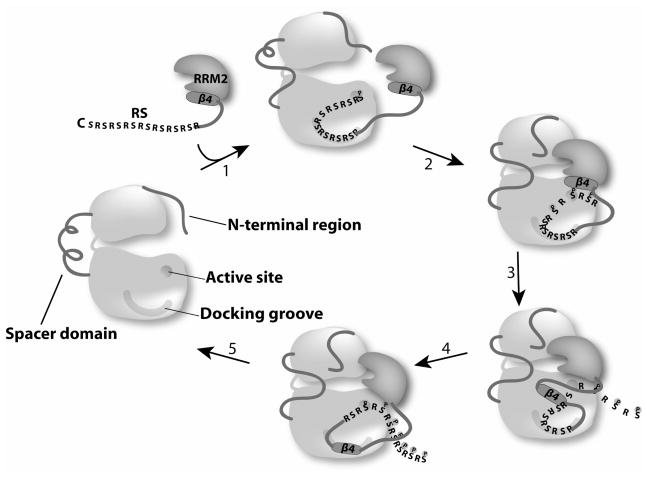
Previously we established binding mode between the kinase and ASF/SF2 that explained directionality of substrate binding and product dissociation. The current work clearly shows that both the β4 and RS domain can compete for the same binding site and therefore, the RS domain must bind first at the docking groove. This work further supports cooperation between RRM and RS domains and the positive role of the N-terminal domain in processive phosphorylation. Current observations led us to extend the model of processive phosphorylation described previously (1). In the current model, we propose that the N-terminus of the RS1 domain occupies the kinase docking groove first allowing the RRM domain to interact with the kinase. The RS and RRM2 domains cooperate and position the RS domain for phosphorylation initiation at the C-terminus end of RS1. We propose that as phosphates are added, force is applied to the RS domain and its contacts occur in ways that induce the modifications of RRM2 and the unfolding of the β4 strand. This phosphorylation-induced mechanism allows the strand to bind in the docking groove and N-terminus serines in RS1 to be phosphorylated. The N-terminus of SRPK1, and possibly also the insert domain, are likely to play an allosteric role in this model by adjusting the binding interactions with RRM2 and possibly promoting strand unfolding. Finally, given the strong dependence of RRM2 binding on docking groove occupancy, it is likely that the association of the multi-domain ASF/SF2 occurs in an orderly process where RS domain binding precedes RRM attachment.
Supplementary Material
Figure 6. A model of the processive phosphorylation cycles.
RS domain binds first. This induces certain structural changes in SRPK1 and/or RRM2 (communicate through the N-terminus and possibly also the spacer domain of SRPK1), allowing RRM2 to bind the kinase and initiate phosphorylation. The cycles continue for several phosphorylation steps in a processive manner (steps 1 to 8) until the last few phosphorylation steps (~steps 9 to12). During that time a mechanical stress induces the unfolding of the β4 motif, which then docks at the docking groove. This also signals RRM2 to begin to dissociate, which facilitate substrate dissociation after phosphorylation is completed.
Acknowledgments
We thank De-Bin Huang for figure 1. The work has been supported by NIH grants to GG (GM084277) and JA (GM067969).
Abbreviations
- SRPK1
SR protein kinase
- Clk/Sty
Cdc2-like-kinase/Serine-Threonine-Tyrosine
- SR protein
splicing factor containing arginine-serine repeats
- ASF/SF2
human alternative splicing factor
- RS domain
domain rich in arginine-serine repeats
- RRM
RNA recognition motif
- p-ASF/SF2
hypophosphorylated ASF/SF2
- pp-ASF/SF2
hyperphosphorylated ASF/SF2
- wt
wild-type
- fl
full-length
- fl(dm)
full-length docking mutants
Footnotes
This work is supported by NIH grants (GM067969 to JAA and GM084277 to GG).
Supporting Information Available: Data showing non-specific RNA contaminants has no role in kinase-substrate interaction. Cross-linking data shows the interaction between β4 of ASF/SF2 and the docking groove of the kinase. This material is available free of charge via the Internet at http://pub.acs.org.
References
- 1.Ngo JC, Giang K, Chakrabarti S, Ma CT, Huynh N, Hagopian JC, Dorrestein PC, Fu XD, Adams JA, Ghosh G. A roving docking interaction is essential for sequential and processive phosphorylation of an SR protein by SR protein kinase 1. Mol Cell. 2008;29:563–576. doi: 10.1016/j.molcel.2007.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tacke R, Manley JL. Determinants of SR protein specificity. Curr Opin Cell Biol. 1999;11:563–576. doi: 10.1016/S0955-0674(99)80050-7. [DOI] [PubMed] [Google Scholar]
- 3.Valcarcel J, Green MR. The SR protein family: pleiotropic functions in pre-mRNA splicing. Trends Biochem Sci. 1996;21:296–301. [PubMed] [Google Scholar]
- 4.Velazquez-Dones A, Hagopian JC, Ma CT, Zhong XY, Zhou H, Ghosh G, Fu XD, Adams JA. Mass spectrometric and kinetic analysis of ASF/SF2 phosphorylation by SRPK1 and Clk/Sty. J Biol Chem. 2005;280:41761–41768. doi: 10.1074/jbc.M504156200. [DOI] [PubMed] [Google Scholar]
- 5.Aubol BE, Chakrabarti S, Ngo JC, Shaffer J, Nolen B, Fu XD, Ghosh G, Adams JA. Processive phosphorylation of alternative splicing factor/splicing factor 2. Proc Natl Acad Sci. 2003;100:12601–12606. doi: 10.1073/pnas.1635129100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cazalla D, Zhu J, Manche L, Huber E, Krainer AR, Caceres JF. Nuclear export and retention signals in the RS domain of SR proteins. Mol Cell Biol. 2002;22:6871–6882. doi: 10.1128/MCB.22.19.6871-6882.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao W, Jamison SF, Garcia-Blanco MA. Both phosphorylation and dephosphorylation of ASF/SF2 are required for pre-mRNA splicing in vitro. RNA. 1997;3:1456–1467. [PMC free article] [PubMed] [Google Scholar]
- 8.Prasad J, Colwill K, Pawson T, Manley JL. The protein kinase Clk/Sty directly modulates SR protein activity: both hyper- and hypophosphorylation inhibit splicing. Mol Cell Biol. 1999;19:6991–7000. doi: 10.1128/mcb.19.10.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao SH, Manley JL. Phosphorylation-dephosphorylation differentially affects activities of splicing factor ASF/SF2. The EMBO J. 1998;17:6359–6367. doi: 10.1093/emboj/17.21.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allemand E, Hastins ML, Murray MV, Myers MP, Krainer AR. Alternative spliing regulation by interaction of phosphatase PP2Cgamma with nucleic acid-binding protein YB-1. Nat Struct Mol Biol. 2005;14:630–638. doi: 10.1038/nsmb1257. [DOI] [PubMed] [Google Scholar]
- 11.Mathew R, Hartmuh K, Mohlmann S, Urlaub H, Ficner R, Luhrmann R. Phosphorylation of human PRP28 by SRPK2 is required for integration of the U4/U6-U5 tri-snRNP into the spliceosome. Nat Struct Mol Biol. 2008;15:426–428. doi: 10.1038/nsmb.1415. [DOI] [PubMed] [Google Scholar]
- 12.Shi Y, Reddy B, ad Manley JL. PP1/PP2A phosphatases are required for the second step of pre-mRNA splicing and target specific snRNP proteins. Mol Cell. 2006;23:819–829. doi: 10.1016/j.molcel.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 13.Ngo JC, Chakrabarti S, Ding JH, Velazquez-Dones A, Nolen B, Aubol BE, Adams JA, Fu XD, Ghosh G. Interplay between SRPK and Clk/Sty kinases in phosphorylation of the splicing factor ASF/SF2 is regulated by a docking motif in ASF/SF2. Mol Cell. 2005;20:77–89. doi: 10.1016/j.molcel.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 14.Hagopian JC, Ma CT, Meade BR, Albuquerque CP, Ngo JC, Ghosh G, Jennings PA, Fu XD, Adams JA. Adaptable molecular interactions guide phosphorylation of the SR protein ASF/SF2 by SRPK1. J Mol Biol. 2008;382:894–909. doi: 10.1016/j.jmb.2008.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aubol BE, Nolen B, Shaffer J, Ghosh G, Adams JA. Novel destabilization of nucleotide binding by the gamma phosphate of ATP in the yeast SR protein kinase Sky1p. Biochemistry. 2003;42:12813–12820. doi: 10.1021/bi035200c. [DOI] [PubMed] [Google Scholar]
- 16.Lieser SA, Aubol BE, Wong L, Jennings PA, Adams JA. Coupling phosphoryl transfer and substrate interactions in protein kinases. Biochim Biophys Acta. 2005;1754:191–199. doi: 10.1016/j.bbapap.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 17.Goldsmith EJ, Akella R, Min X, Zhou T, Humphreys JM. Substrate and docking interactions in serine/threonine protein kinases. Chem Rev. 2007;107:5065–5081. doi: 10.1021/cr068221w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.