Abstract
Kohlschütter-Tönz syndrome (KTS) is an autosomal-recessive disease characterized by the combination of epilepsy, psychomotor regression, and amelogenesis imperfecta. The molecular basis has not yet been elucidated. Here, we report that KTS is caused by mutations in ROGDI. Using a combination of autozygosity mapping and exome sequencing, we identified a homozygous frameshift deletion, c.229_230del (p.Leu77Alafs∗64), in ROGDI in two affected individuals from a consanguineous family. Molecular studies in two additional KTS-affected individuals from two unrelated Austrian and Swiss families revealed homozygosity for nonsense mutation c.286C>T (p.Gln96∗) and compound heterozygosity for the splice-site mutations c.531+5G>C and c.532-2A>T in ROGDI, respectively. The latter mutation was also found to be heterozygous in the mother of the Swiss affected individual in whom KTS was reported for the first time in 1974. ROGDI is highly expressed throughout the brain and other organs, but its function is largely unknown. Possible interactions with DISC1, a protein involved in diverse cytoskeletal functions, have been suggested. Our finding that ROGDI mutations cause KTS indicates that the protein product of this gene plays an important role in neuronal development as well as amelogenesis.
Main Text
Kohlschütter-Tönz syndrome (KTS, MIM 226750) is a rare genetic disorder characterized by the combination of epilepsy, psychomotor delay and regression, and amelogenesis imperfecta. So far, 24 individuals with the clinical diagnosis of KTS have been reported.1–9 Pedigrees suggest an autosomal-recessive mode of inheritance, but genetic heterogeneity cannot be excluded. The molecular basis of KTS has not yet been elucidated. The most striking feature is global enamel deficiency (amelogenesis imperfecta) of the hypoplastic or hypocalcified type; this deficiency affects primary as well as permanent teeth right from the moment of eruption. The enamel is very thin, rough, prone to disintegration, and stained in various shades of brown. Onset of epilepsy usually occurs in the first year of life; seizures are difficult to treat or might be refractory to therapy. Affected children show severe psychomotor delay or regression, which might be present after birth but more frequently develops after the onset of seizures. Both gross and fine motor skills are usually impaired, and intellectual disability might be severe. The natural course is variable; several affected individuals developed spastic tetraplegia, and some died in childhood. There are no consistent dysmorphic features or metabolic abnormalities, although nonspecific facial anomalies have been reported in some affected individuals. Cranial imaging frequently shows mild brain atrophy.
In order to identify the genetic basis of KTS, we investigated four affected children from three families as well as healthy members of the index family reported in 1974.1 Clinical features of the affected individuals are summarized in Table 1. Family A is a consanguineous Moroccan family with two affected children (A-IV:3 and A-IV:4; Figure 1);9 the parents are first cousins. Initial development of the affected boy (A-IV:3) appeared normal, but treatment-resistant epilepsy started when he was 4 months old and led to loss of fixation and global developmental delay. The affected younger sister (A-IV:4) showed psychomotor delay from birth onward. Epileptic seizures, which were difficult to treat, started when she was 12 months old. The first teeth in both children erupted when they were 13 and 14 months old, respectively; from the beginning, their teeth were lusterless and had a brownish discoloration. Family B has been reported previously;8 the parents of the affected boy (B-II:1) are not knowingly related but come from neighboring villages in East Tyrol (Austria). Epilepsy started when the boy was 5 months old but later improved; there were no seizures after 7 years of age, and medication was discontinued when he was 15 years old. Primary and permanent teeth were yellow, hypoplastic, and crowded. Family C has one affected girl (C-XI:2) who has not yet been reported. Left-sided hemiconvulsive seizures started when she was 6 months old and were initially difficult to treat, but when she was 6 years old, anticonvulsive treatment could be discontinued. Primary and secondary dentition showed enamel abnormalities typical of KTS (Figure 2). Genealogical studies revealed that this girl is distantly related to the mother of the affected individuals (C-IX:3) reported in 19741 via both the maternal line (six generations ago) and the paternal line (nine generations ago). The parents of individual C-XI:2 are also eighth cousins (see family C in Figure 1).
Table 1.
Clinical Features of KTS and ROGDI Genotypes in the Affected Individuals
| A-IV:3 | A-IV:4 | B-II:1 | C-XI:2 | |
|---|---|---|---|---|
| ROGDI genotype | homozygous for c.229_230del (p.Leu77Alafs∗64) | homozygous for c.229_230del (p.Leu77Alafs∗64) | homozygous for c.286C>T (p.Gln96∗) | compound heterozygous for c.531+5G>C and c.532-2A>T |
| Age at time of last evaluation | 12 years | 9 years | 18 years | 9 years |
| Growth parameters | mild microcephaly | normal | normal | normal |
| Initial development | normal until onset of seizures | developmental delay since birth | normal until onset of seizures | normal until onset of seizures |
| Language skills and social interaction at time of last evaluation | no expressive language | some words; deterioration of social interaction after onset of seizures | 35 single words and sentences with two words; social and friendly behavior | competent to talk in short and simple sentences |
| Age of walking without support | 4.5 years | 2.2 years | 2.5 years | 2 years |
| Age of seizure onset | 4 months | 12 months | 5 months | 6 months |
| EEG findings (generalized or partial traits) | multifocal epileptic activity and poorly developed background activity | focal epileptic activity and poorly developed background activity | multifocal epileptic activity (later generalized) and abnormal background activity | focal epileptic activity; normalization at 6 years of age |
| Seizure type and frequency | episodes of cyanosis and apnea; later generalized tonic-clonic seizures (1–5 per day); only seizures with fever since start of levetiracetam at 3.5 years of age | mostly myoclonic seizures (1–5 per day); only seizures with fever since start of levetiracetam at 1.8 years of age | focal and generalized seizures (1–5 per day); seizure free since 7 years of age and no medication since 15 years of age | left-sided hemiconvulsive seizures and various anticonvulsants; seizure free without treatment since 6 years of age |
| Hearing | normal | normal | normal | normal |
| Vision | loss of visual fixation after onset of seizures | normal | normal | normal |
| Dentition | eruption of first teeth at 13 months of age; discoloration from the beginning | eruption of first teeth at 14 months of age; lusterless and rapid discoloration | primary and permanent teeth with discoloration and enamel defects | primary and permanent teeth with discoloration and enamel defects |
The following abbreviation is used: EEG, electroencephalography.
Figure 1.
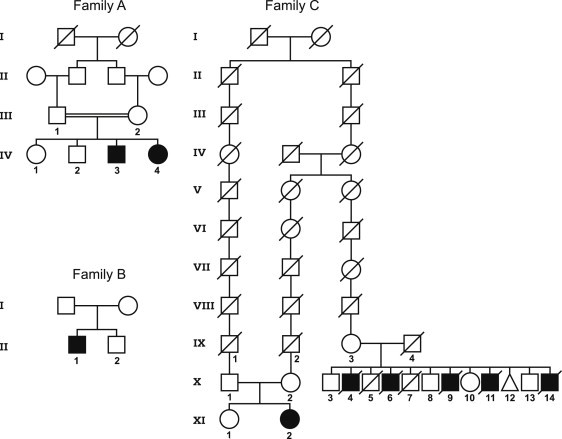
Pedigrees of Investigated Families
The pedigree for family A shows a consanguineous Moroccan family9 in which linkage analysis and exome sequencing were performed. The pedigree for family B shows a Tyrolean family affected by KTS,8 and the parents are not knowingly related. The pedigree for family C shows the newly diagnosed Swiss family (the parents are X:1 and X:2) and its relationship with the distantly related index family reported in 1974 (parents IX-3 und IX-4).1 Note that the parents in the newly identified family are distantly related to each other, but the affected child is compound heterozygous for two different mutations.
Figure 2.
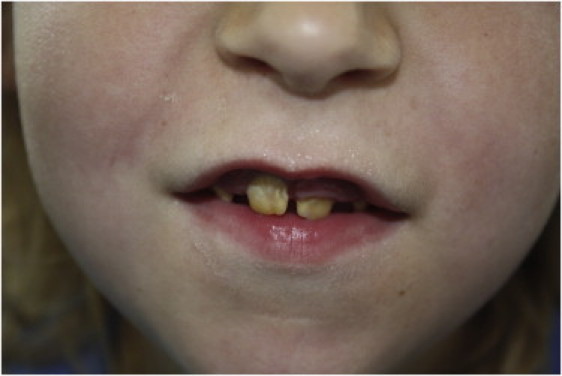
Dental Phenotype in the So Far Unreported Individual C-XI:2
Tooth discoloration due to global enamel defect (amelogenesis imperfecta).
In order to identify the candidate gene for KTS, we performed linkage analysis and autozygosity mapping in family A. Analyses of all families were carried out with informed consent and were approved by the institutional review board at Medical University Innsbruck. Affected individuals, siblings, parents, and grandparents were investigated. We analyzed 250 ng of genomic DNA from each individual on the SNP-based mapping-chip GeneChip Human Mapping 10K Array (Affymetrix, Santa Clara, CA, USA); we used the operating software (Affymetrix GCOS 1.4) and genotyping-analysis software (Affymetrix GTYP 4.0) according to the manufacturer's instructions. A multipoint LOD score was calculated with the software programs Allegro10 and ALOHOMORA.11 The haplotype analysis and the LOD-score estimation based on the model of autosomal-recessive inheritance showed four possible linkage regions in chromosomal regions 3q13.31–q13.32, 11q24.1–q24.2, 16p13.3, and 17q25.1–q25.3 (Figure 3A). LOD scores in these regions ranged between 1.05 and 2.06. The autozygous regions had a total size of 15.83 Mb and contained 326 known protein-coding genes (see Table S1).
Figure 3.
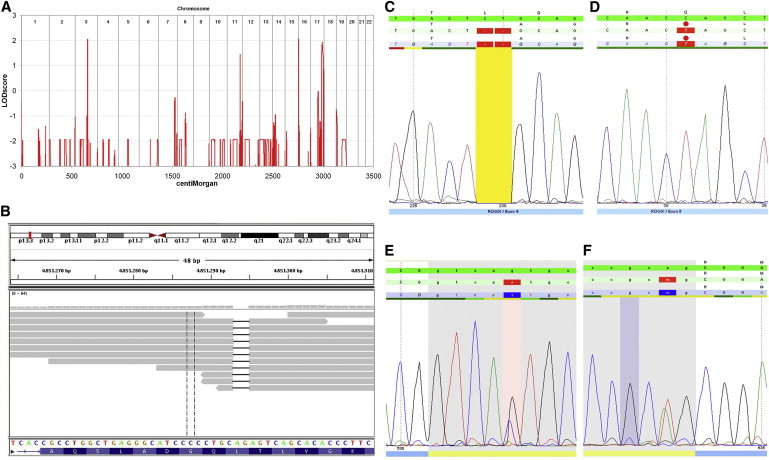
Linkage and Genomic Sequence Analyses
(A) Linkage analysis in family A revealed four autozygous regions in chromosomes 3, 11, 16, and 17.
(B) Exome sequencing in family A revealed a homozygous 2 bp deletion, c.229_230del, in exon 4 of ROGDI.
(C–F) Identification of mutations by Sanger sequencing. Homozygous deletion c.229_230del (C) is present in family A, homozygous nonsense mutation c.286C>T (D) is present in family B, and heterozygous splice-site mutations c.531+5G>C (E) and c.532-2A>T (F) are present in family C.
Considering the large number of genes in the autozygous regions, we decided to use whole-exome sequencing (carried out by ServiceXS, Leiden, The Netherlands) for the genetic analysis of one affected individual (A-IV:4) from family A. Exome capturing was performed with the Agilent SureSelect Human All Exon Kit (Agilent, Santa Clara, CA), and the sample was sequenced on an Illumina Genome Analyzer II platform (Illumina, San Diego, CA). Data analysis was carried out with the SIMPLEX pipeline, which uses the Burrows Wheeler Aligner12 to map the reads to the human reference-genome sequence (USCS hg19, February 2009, Genome Reference Consortium GRCh37). For SNP and DIP (deletion-insertion polymorphism) calling, as well as for realignment around indels, we applied the Genome Analysis Toolkit (GATK).13 Exon boundaries were specified by the Consensus Coding Sequence (CCDS).10 An exome coverage depth of 23× was achieved: 46% of exons showed high coverage (≥20×), and around 10% of exons showed low coverage (≤5×). Variant detection identified 20,454 SNPs as well as 1,208 DIPs. We annotated all variants with additional information by using GATK and ANNOVAR14 to facilitate the identification of disease-causing mutations. Subsequently, we applied the auto_annovar functionality to filter variants against dbSNP (build 132), the 1,000 Genomes Project (Nov 2010), and previously assigned conservation scores (for filtering details, see Table S2). After all filtering steps, only a single strong candidate gene, ROGDI (rogdi homolog [Drosophila], RefSeq accession number NM_024589.1) in chromosomal region 16p13.3, remained in the autozygous regions of interest. In exon 4 of this gene, we found a homozygous frameshift deletion, c.229_230del (p.Leu77Alafs∗64), which is predicted to disrupt the amino acid structure and cause a premature stop codon (Figure 3B). The filtering algorithm also called a missense variant, c.2273G>C (p.Cys758Ser), in EVPL (envoplakin [MIM 601590]) in the linkage region of 17q25.1; this variant (rs142251448) was included in build 134/135 of dbSNP and had a heterozygote frequency of 0.3% in the North American population.
After completing exome sequencing, we found a PhD thesis that reports the results of autozygosity mapping in five families affected by KTS.15 That study identified 30 candidate genes, including ROGDI, but did not find any linkage to chromosomal region 17q25.1. There is no other report on a possible link between KTS and ROGDI or EVPL. Also considering the expected severity of the frameshift deletion found in family A, we focused our subsequent studies on ROGDI (Ensembl accession number ENSG00000067836). This gene stretches over 5.98 kb in chromosomal region 16p13.3 and contains 11 exons, all of which are coding. Bioinformatics analysis showed that the transcript of ROGDI codes for 287 amino acids and results in a molecular weight of 32 kDa (RefSeq accession number NP_078865.1). There is only one known functional transcript. Dye-terminator sequencing of all exons and adjacent intron sequences of ROGDI (NM_024589.1) (ABI Prism 7000 sequence detection system, Applied Biosystems, Carlsbad, CA; primer sequences are available in Table S3) confirmed the homozygous presence of the mutation c.229_230del in both affected siblings of family A (Figure 3C). As expected, both parents were found to be heterozygous. Sequence analysis in family B revealed a homozygous nonsense mutation, c.286C>T, in exon 5 of ROGDI in affected individual B-II:1 (Figure 3D). This mutation is predicted to change a CAG triplet that codes for glutamine into a TAG stop codon, denoted p.Gln96∗. Both parents in the family were heterozygous for this mutation. In family C, two heterozygous splice-site mutations, c.531+5G>C and c.532-2A>T, in intron 7 of ROGDI were identified in affected individual C-XI:2 (Figures 3E and 3F). In silico analysis indicated that both mutations destroy the respective splice donor and acceptor sites of intron 7 (Alamut [Interactive Biosoftware, Rouen, France], data not shown). The mother (C-X:2) was found to be heterozygous for c.532-2A>T, and the father (C-X:1) was found to be heterozygous for c.531+5G>C, confirming compound heterozygosity in the affected child. The unaffected sister (C-XI:1) was found to be heterozygous for c.531+5G>C. Finally, we acquired archival DNA from the unaffected mother (C-IX:3) and four healthy siblings (C-X:3, C-X:8, C-X:10, and C-X:13) of the original family reported by Kohlschütter et al.1 (family C in Figure 1); none of these individuals have epilepsy and all have normal intelligence and normal teeth with intact enamel. The affected family members as well as the father of that family are deceased, and their DNA samples are not available. The mother and all investigated siblings are heterozygous for splice-site mutation c.532-2A>T, which is also found in the mother of affected individual C-XI:2. It can be assumed that the mothers from both family branches have a common ancestor who lived in the Swiss valley of Schächental in the 18th century and who was a carrier for this mutation (family C in Figure 1).
All mutations identified were frameshift, nonsense, or splice-site mutations that are expected to either cause premature mRNA degradation by nonsense-mediated decay or dramatically alter protein structure and consequently cause complete loss of protein function. They are not listed in publicly available genome-variant databases and are absent from the 1,000 Genomes Project. In order to assess the functional effects of the different mutations, we obtained fresh peripheral-blood samples from the affected individuals in families B and C. Peripheral-blood mononuclear cells (PBMC) were isolated from blood samples and cultivated in the presence of phytohemagglutinin (Quantum PBL by PAA Laboratories GmbH, Pasching, Austria) for three days. Thereafter, RNA was isolated, and cDNA synthesis was performed by standard methods. RT-PCR amplification spanning exons 6–9 of the transcripts in affected individual C-XI:2 (primer sequences are available as Table S4) showed that the wild-type amplicon (386 bp) was absent but that a strong shorter band (approximately 290 bp) and a weak band somewhat larger than the wild-type band were present (Figure 4A). The other family members showed the wild-type amplicon, but the father and sister (both heterozygous for c.531+5G>C) also showed the shorter band, and the mother (heterozygous for c.532-2A>T) also showed the weak larger band. Dye-terminator sequencing of these products revealed that the short band reflects an in-frame deletion of exon 7 caused by mutation c.531+5G>C (Figure 4C). The other amplicon associated with mutation c.532-2A>T was detectable as background sequencing trace in the mother and the affected child. The aberrant transcript is a result of the use of an intron 7 cryptic splice acceptor site that leads to the inclusion of an additional 83 nucleotides before exon 8 (data not shown). The predicted effect is the inclusion of two abnormal amino acids followed by a stop codon. cDNA sequence analysis was not performed in affected individual B-II:1, who is homozygous for the nonsense mutation c.286C>T, which is not expected to affect splicing.
Figure 4.
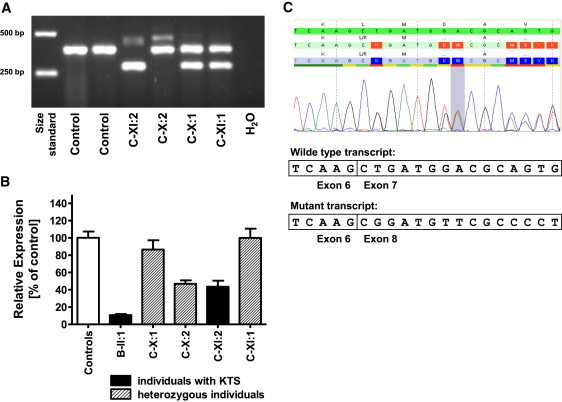
cDNA Analyses
(A) RT-PCR analysis of ROGDI in family C. Note the absence of the wild-type amplicon as well as the presence of two aberrant bands in affected individual C-XI:2. One of the aberrant bands is approximately 100 bp shorter than the wild-type band and is also found in the father (C-X:1) and sister (C-XI:1), who are both heterozygous for c.531+5G>C. The other aberrant band is weak, approximately 80 bp larger than the wild-type band, and is also observed in the mother (C-X:2), who is heterozygous for c.532-2A>T.
(B) RT-qPCR analysis of ROGDI in affected individuals, healthy family members, and controls shows markedly reduced mRNA transcript in affected individual B-II:1. Heterozygosity for c.532-2A>T in C-X:2 is associated with a cDNA reduction of approximately 50%, most likely reflecting nonsense-mediated decay of that allele. In contrast, heterozygosity for c.531+5G>C is not associated with the loss of cDNA in C-X:1 and C-XI:1. The fact that affected individual C-XI:2 has half normal cDNA reflects the combination of both alleles. The error bars represent means and standard deviations of three independence measurements of the probands and four controls.
(C) cDNA sequence analysis of the RT-PCR product of exons 6–9 in individual C-X:1, heterozygous for c.531+5G>C, shows skipping of in-frame ROGDI exon 7.
We quantified the expression of ROGDI with real time PCR by using specific primers spanning exons 3–4 (primer sequences are available in Table S4) and Maxima SYBR Green/ROX qPCR Master Mix (Fermentas) in an Applied Biosystems Prism 7000 sequence detection system. PCR reaction was carried out under standard conditions. The cycle threshold (Ct) values were calculated with sequence-detection system (SDS) software v1.2 (Applied Biosystems). We quantified relative gene expression with the comparative ΔΔCt method by using HPRT1 (RefSeq accession number NM_000194.2) as a reference gene. These analyses showed that the amount of ROGDI cDNA was markedly reduced to 10.6% (much lower than the mean of the four controls) in affected individual B-II:1 (Figure 4B). The amount of cDNA in affected individual C-XI:2 was 43.6%, similar to the value of 46.9% in her mother (C-X:2). The amount of ROGDI cDNA in the father and sister was in the normal range (86.6% and 100.2%, respectively).
In summary, the cDNA analyses confirm that the mutations in affected individuals B-II:1 and C-XI:2 severely disrupt the normal ROGDI transcript. Mutation c.531+5G>C causes skipping of in-frame exon 7 but does not lead to a translational frameshift and is not associated with nonsense-mediated decay. In contrast, mutation c.532-2A>T triggers the use of a cryptic intronic splice acceptor site, explaining both a larger size of the cDNA amplicon and nonsense-mediated decay. The latter effect was also observed for nonsense mutation c.286C>T. Thus, the mutations in all three KTS-affected families are expected to be severe (null) mutations that are likely to cause complete loss of ROGDI function.
The exact function of the protein encoded by ROGDI is unknown. Using ANNIE,16 sequence-structure analysis showed neither relevant features (e.g., transmembrane regions or signal peptides) nor relevant protein domains. Protein prediction methods17 indicate that ROGDI is a globular protein and that the secondary structure consists of 45% helix motifs, 37% loop structures, and 17% strands. The gene is highly conserved and has orthologs in many species, including Drosophila melanogaster. It shows particularly high expression levels in various human brain regions,18 in line with the CNS phenotype of KTS. A Drosophila mutant of this gene showed a possible deficiency in olfactory memory.19 Yeast two-hybrid screens20 suggested a possible interaction between ROGDI and DISC1 (MIM 605210), a protein implicated in the development of schizophrenia and involved in processes of cytoskeletal stability and organization, neuronal migration, intracellular transport, and cell division.21 There are no published studies that examined the role of ROGDI in tooth development and amelogenesis. Our own data provide robust information on the clinical effects of the loss of ROGDI function in humans and provide interesting perspectives for research into the molecular causes of epilepsy and other conditions.
In conclusion, we report that KTS is caused by putative loss-of-function mutations in ROGDI. All mutations identified are predicted to be severe (null) mutations that are likely to cause complete loss of protein function. Heterozygosity for ROGDI-null mutations does not appear to have any adverse effects. It is possible that individuals with homozygosity or compound heterozygosity for hypomorphic missense mutations in ROGDI could present with isolated epilepsy independently from minor enamel defects or vice versa. Assessing potential genotype-phenotype correlations will require molecular studies on additional affected individuals. Although we found ROGDI mutations in all KTS-affected individuals investigated so far, we cannot rule out genetic heterogeneity. Future work will hopefully elucidate the exact function of ROGDI in neuronal development and amelogenesis.
Acknowledgments
This work was supported by a grant from the Standortagentur Tirol. We wish to thank Josef Muheim (Greppen, Switzerland) for considerable help with the genealogical studies that allowed us to link the two Swiss nuclear families into a single pedigree. Long-term medical care to the affected individuals in the study was provided by Thomas Schmitt-Mechelke and Petra Kolditz, (both from Children's Hospital, Lucerne, Switzerland). Bart Janssen and Thomas Chin-A-Woeng (both from ServiceXS, Leiden, The Netherlands) assisted with the exome sequencing. We gratefully acknowledge expert technical assistance by Brunhild Schagen (Department of Oral, Dental, and Maxillofacial Diseases, Heidelberg University, Germany) as well as by Pia Traunfellner, Sandra Unterkirchner, and Ramona Berberich (all from the Division of Human Genetics, Medical University Innsbruck, Austria).
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
Ensembl, http://www.ensembl.org
GenBank, http://www.ncbi.nlm.nih.gov/genbank/
Online Mendelian Inheritance in Man (OMIM), http://www.omim.org
References
- 1.Kohlschütter A., Chappuis D., Meier C., Tönz O., Vassella F., Herschkowitz N. Familial epilepsy and yellow teeth—a disease of the CNS associated with enamel hypoplasia. Helv. Paediatr. Acta. 1974;29:283–294. [PubMed] [Google Scholar]
- 2.Christodoulou J., Hall R.K., Menahem S., Hopkins I.J., Rogers J.G. A syndrome of epilepsy, dementia, and amelogenesis imperfecta: Genetic and clinical features. J. Med. Genet. 1988;25:827–830. doi: 10.1136/jmg.25.12.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petermöller M., Kunze J., Gross-Selbeck G. Kohlschütter syndrome: Syndrome of epilepsy—dementia—amelogenesis imperfecta. Neuropediatrics. 1993;24:337–338. doi: 10.1055/s-2008-1071567. [DOI] [PubMed] [Google Scholar]
- 4.Zlotogora J., Fuks A., Borochowitz Z., Tal Y. Kohlschütter-Tönz syndrome: Epilepsy, dementia, and amelogenesis imperfecta. Am. J. Med. Genet. 1993;46:453–454. doi: 10.1002/ajmg.1320460422. [DOI] [PubMed] [Google Scholar]
- 5.Musumeci S.A., Elia M., Ferri R., Romano C., Scuderi C., Del Gracco S. A further family with epilepsy, dementia and yellow teeth: The Kohlschütter syndrome. Brain Dev. 1995;17:133–138. doi: 10.1016/0387-7604(95)00013-2. discussion 142–133. [DOI] [PubMed] [Google Scholar]
- 6.Wygold T., Kurlemann G., Schuierer G. Kohlschütter syndrome—an example of a rare progressive neuroectodermal disease. Case report and review of the literature. Klin. Padiatr. 1996;208:271–275. doi: 10.1055/s-2008-1046481. [DOI] [PubMed] [Google Scholar]
- 7.Donnai D., Tomlin P.I., Winter R.M. Kohlschutter syndrome in siblings. Clin. Dysmorphol. 2005;14:123–126. [PubMed] [Google Scholar]
- 8.Haberlandt E., Svejda C., Felber S., Baumgartner S., Günther B., Utermann G., Kotzot D. Yellow teeth, seizures, and mental retardation: A less severe case of Kohlschütter-Tönz syndrome. Am. J. Med. Genet. A. 2006;140:281–283. doi: 10.1002/ajmg.a.31071. [DOI] [PubMed] [Google Scholar]
- 9.Schossig A., Wolf N., Grond-Ginsbach C., Schagen B., Koch M., Rating D., Zschocke J. Epileptische Enzephalopathie und Zahnschmelzdefekt (Kohlschütter-Tönz-Syndrom): Drei Fallberichte und Literaturübersicht. Med. Genetik. 2007;19:422–426. [Google Scholar]
- 10.Pruitt K.D., Tatusova T., Maglott D.R. NCBI reference sequences (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35(Database issue):D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rüschendorf F., Nürnberg P. ALOHOMORA: A tool for linkage analysis using 10K SNP array data. Bioinformatics. 2005;21:2123–2125. doi: 10.1093/bioinformatics/bti264. [DOI] [PubMed] [Google Scholar]
- 12.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang K., Li M., Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lo, C. (2009). Genetics in Epilepsy. PhD thesis, University College London, London, UK.
- 16.Ooi H.S., Kwo C.Y., Wildpaner M., Sirota F.L., Eisenhaber B., Maurer-Stroh S., Wong W.C., Schleiffer A., Eisenhaber F., Schneider G. ANNIE: Integrated de novo protein sequence annotation. Nucleic Acids Res. 2009;37(Web Server issue) doi: 10.1093/nar/gkp254. W435–W440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rost B., Yachdav G., Liu J. The PredictProtein server. Nucleic Acids Res. 2004;32(Web Server issue) doi: 10.1093/nar/gkh377. W321–W326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu C., Orozco C., Boyer J., Leglise M., Goodale J., Batalov S., Hodge C.L., Haase J., Janes J., Huss J.W., 3rd, Su A.I. BioGPS: An extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dubnau J., Chiang A.S., Grady L., Barditch J., Gossweiler S., McNeil J., Smith P., Buldoc F., Scott R., Certa U. The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr. Biol. 2003;13:286–296. doi: 10.1016/s0960-9822(03)00064-2. [DOI] [PubMed] [Google Scholar]
- 20.Camargo L.M., Collura V., Rain J.C., Mizuguchi K., Hermjakob H., Kerrien S., Bonnert T.P., Whiting P.J., Brandon N.J. Disrupted in Schizophrenia 1 Interactome: Evidence for the close connectivity of risk genes and a potential synaptic basis for schizophrenia. Mol. Psychiatry. 2007;12:74–86. doi: 10.1038/sj.mp.4001880. [DOI] [PubMed] [Google Scholar]
- 21.Brandon N.J., Sawa A. Linking neurodevelopmental and synaptic theories of mental illness through DISC1. Nat. Rev. Neurosci. 2011;12:707–722. doi: 10.1038/nrn3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


