Abstract
Deleted in liver cancer (DLC1), a tumor suppressor gene in multiple cancers, is recurrently down regulated or inactivated by epigenetic mechanisms in primary prostate carcinomas (PCAs). In this study the methylation and acetylation profile of the DLC1 promoter region was examined in three PCA cell lines with low or undetectable DLC1 expression: LNCaP, its derivative C4-2B-2, and 22Rv1. Two histone deacetylase inhibitors (HDAC), suberoylanilide hydroxamic acid (SAHA) and trichostatin A (TSA) induced histone acetylation of the DLC1 promoter in all three lines. DLC1 promoter methylation and deacetylation were detected in LNCaP and C4-2B-2 cells while in 22Rv1cells DLC1 is silenced by deacetylation. Treatment with SAHA or TSA efficiently increased DLC1 expression in all lines, particularly in 22Rv1 cells, and activated the DLC1 promoter through the same Sp1 sites. The 22Rv1 cell line was selected to evaluate the efficacy of combined DLC1 transduction and SAHA treatment on tumor growth in athymic mice. Individually, DLC1 transduction and SAHA exposure reduced the tumor size by 75–80% compared to controls and in combination almost completely inhibited tumor growth. The antitumor effect was associated with the induction of apoptosis and inhibition of RhoA activity. SAHA alone significantly reduced RhoA activity, showing that this RhoGTPase is a target for SAHA. These results, obtained with a reliable preclinical in vivo test, predict that combined therapeutic agents targeting the pathways governing DLC1 function and HDAC inhibitors may be beneficial in management of prostate cancer.
Keywords: DLC1, tumor suppressor gene, SAHA, apoptosis, RhoA activity, inhibition prostate tumor growth in vivo
1. Introduction
Human cancer is regarded as a genetic disorder originating from a single precursor cell by cumulative acquisition of multiple genetic or epigenetic alterations. Over the past several years the trend in cancer therapy has been increasingly focused on therapeutics based on epigenetic changes in cancer cells. Epigenetic changes commonly down regulate or inactivate tumor suppressor genes (TSGs) by promoter hypermethylation or histone deacetylation [1]. Agents that reverse the inhibitory effect of epigenetic processes such as Vidaza, Decitabine, Zebularine or suberoylanilide hydroxamic acid (SAHA), have been developed and shown promise in clinical trials used alone or in combination.
DLC-1 (Deleted in Liver Cancer) that encodes a Rho GTPase-activating protein is an established TSG and functions as a metastasis suppressor in several common cancers [2–5]. Down regulation of the DLC-1 expression is mediated by genetic and epigenetic changes in many forms of cancer and in certain cancers is more frequently deleted than the well-known TSGs [4,5]. However, the epigenetic mechanisms are the major contributing factors in deregulation of DLC-1 in various cancers [4].
Among solid tumors prostate carcinoma (PCA) provides a good example for the role of epigenetic mechanisms in negative regulation of DLC1 expression. DLC1 has been found to be down regulated or silenced by epigenetic modifications in a high number of primary PCAs [6].
Because DLC1 transduction and histone deacetylase (HDAC) inhibitors exert antineoplastic functions, their combined action could be exploited for a more effective cancer therapy. We opted to test the antioncogenic action of SAHA, the most advanced HDAC inhibitor proven to have therapeutic efficacy in certain types of cancer [7,8]. Based on these considerations, we undertook present study to evaluate the combinatorial effect of SAHA and DLC-1 in vivo, as a conclusive preclinical test. The major finding in this study is the robust inhibitory effect of combined SAHA and DLC1 transduction on tumor growth in nude mice of highly tumorigenic PCA 22Rw1 cells. Importantly, inhibition of tumor growth was associated with reduction of RhoA activity and induction of apoptosis.
2. Materials and methods
2.1 Cell lines and culture conditions
The human PCA cell lines 22Rv1 and LNCaP were obtained from the American Type Culture Collection (ATCC, Manassas, VA). LNCaP and 22Rv1 cells were cultured in RPMI 1640 and DMEM/F12 medium, respectively (Invitrogen, Carlsbad, CA). C4-2B cell, a metastatic androgen-independent PCA cell line derived from LNCaP, was purchased from ViroMed (Minneapolis, MN) and was cultured in T medium (Invitrogen). All cell culture medium were supplemented with fetal bovine serum (10%), penicillin/streptomycin (100U/ml) and the cell cultures were maintained in a humidified atmosphere containing 5% CO2 at 37°C.
2.2 Chemicals and Administration
SAHA (Selleck Chemicals LLC, Houston, TX) and trichostatin A (TSA) (Sigma, St. Louis, MO) were prepared as 10 mM or 1mM stock solution in dimethylsulphoxide (DMSO), respectively. For in vitro experiments, cells were treated with 5 μM of SAHA or 0.5 μM TSA of for 24 or 48 hrs after seeding. For in vivo experiments, mice received 50 mg/kg SAHA daily by i.p injection for 21 days. The injection volume was kept constant at 1 μl/g body weight.
2.3 Methylation-specific PCR (MSP)
MSP for detection of DLC1 promoter methylation was carried out with minor modifications as previously described [6].
2.4 Chromatin Immunoprecipitation assay (ChIP)
Histone acetylation detection was carried out using an acetyl-histone H3 immunoprecipitation (ChIP) assay Kit (Millipore, Billerica, MA) according to the manufacturer’s instructions. Two sets of primers spanning different DLC1 promoter regions were used, the primer sets are Chip-F1: 5-GCT AGA GGG CGG CCT GAG GC-3, Chip-R1: 5-CAG TCG GAG CGA ACT GTC TC-3; Chip-F2: 5-AGAGGAGAGGCGGGGCCT-3, Chip-R2: 5-CTTAGCGACGGGCTGTTCTCC-3, which yield products of 124bp and 215 bp in length, respectively.
2.5 Real-time RT-PCR
PCA cell lines, 22Rv1, C4-2-B2, and LNCaP were treated with 5 μM SAHA or 0.5 μM TSA or DMSO for 24 hrs and harvested. Total RNAs were isolated using RNeasy kit (Qiagen, Valencia, CA) and first-strand cDNAs were synthesized from 1μg of total RNA by a SuperScript III system (Invitrogen) according to the manufacture’s instructions. Real-time PCR reactions were performed using TaqMan® Universal PCR Master Mix (Applied Biosystems, Carlsbad, CA). The 2−ΔΔCt method was used to calculate the relative fold difference of DLC1 mRNA.
2.6 Construction of fragments from DLC-1 promoter region
DLC1 promoter construct −577/+117-pGL3b was kindly provided by Drs. Yung-Jue Bang and Tai Young Kim. Five truncated fragments from DLC1 promoter region (−374/−203, −374/186, − 293/+26, −203/+26, −189/+26, numbered relative to the transcription start site+1) were generated by PCR using the following primers: F (−374): 5- CTT GTG ACC TTT GCC TTT GC-3; F (−293): 5-GGG AAA CAT TCC AGC CTT C-3; F (−203): 5-CTG GGC GGG GCG GGG CTA G-3; F (−189): 5-GCT AGA GGG CGG CCT GAG GC-3; R (203): 5-CCC CGC CTC TCC TCT GTC CCG -3; R (−186): 5-TAG CCC CGC CCC GCC CAG-3; R (+26): 5-CAG TCG GAG CGA ACT GTC TC-3, respectively. The resulting PCR products were inserted into the Topo TA pCR2.1-vector (Invitrogen) and subcloned into KpnI/XhoI-cut pGL3-basic vector (Promega, Madison, WI). The sequences and orientations of the inserts were verified directly by DNA sequencing.
2.7 Transient transfection and luciferase assay
The effect of SAHA and TSA on DLC-1 promoter activity was determined by transient transfection and luciferase assay. The truncated DLC-1 promoter-luciferase constructs were transfected into 22Rv1 cells using Lipofectamine 2000 (Invitrogen). After 24 hrs post-transfection, cells were treated with 5 μM SAHA or 0.5 μM of TSA, for additional 24 hrs. Cell lysates were collected for luciferase assays with britelite™ plus Reporter Gene Assay System (Waltham, MA 02451) according to the manufacturer’s instructions. Luciferase activities were measured using a Vector2, 1420 Multulabel counter (Perkin Elmer, San Jose, CA) and normalized for total protein concentration determined using BCA assay (Pierce, Rockford, IL).
2.8 In vivo tumorigenicity assay
All procedures involving animals were reviewed and approved by NCI Animal Care and Use Committee. The stably transfected 22Rv1 cells expressing a DLC1-luciferasion protein were generated by inserting the full-length wtDLC1 into pPK-CMV-F4 vector (PromoKine, Heidelberg, Germany) followed by selection with 750 μg/ml of neomycin. To establish subcutaneous tumors, 6-week-old BALB/c AnNCr-nu/nu male mice (NCI-Frederick, Frederick, MD) were injected with 3 × 106 exponentially growing 22Rv1 cells. When tumors in each treatment group reached a similar size (5 × 5 mm, around 6 to 8 days), 50 mg/kg (50 ug/g) SAHA or DMSO were administrated daily by intraperitoneal injections for 21 days. The injection volume was kept constant at 1 ul/g body weight. Tumor size was measured using a digital caliper and average tumor volume was calculated using the standard formula: tumor volume = (W2 × L) × 0.5. To quantify the tumor size, luciferase bioluminescent images were measured using Xenogen IVIS system according to the manufacturer’s instruction (Caliper Life Sciences, Hopkinton, MA). Data presented are representative of three independent experiments. The significant differences between means was determined by Student’s t test. A P value of <0.05 was considered statistically significant.
2.9 Rho Activation, apoptosis and cell cycle
An adenoviral vector carrying human DLC1 cDNA was prepared and purified as previously described [9]. The effects of DLC1 and SAHA alone or in combination on cell cycle, apoptosis and RhoA activation were tested as previously described [10]. The DNA histograms have been gated to include only single cells and the data were analyzed with CellQuest software (BD Bioscience, San Jose, CA). Active RhoA level in SAHA and solvent treated cells were determined by an ELISA-based RhoA activation assay (G-LISA, Cytoskeleton, Inc. Denver, CO) according to the manufacturer’s instructions.
3. Results
3.1 Methylation and acetylation profile of PCA cells
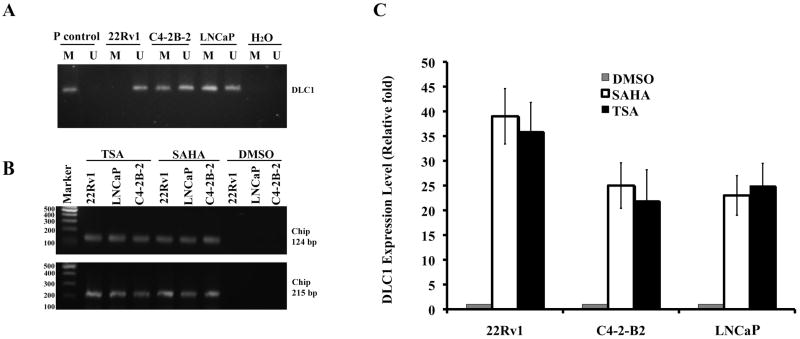
Three PCA cell lines, LNCaP, its derivative metastatic C4-2B-2 and highly tumorigenic 22Rv1 lines were selected for this study. The cells from all three lines express either a negligible level of or lacking endogenous DLC1 [6, 9]. LNCaP and 22Rv1 but not C4-2B-2 cells that are metastatic to bone have been previously examined for the epigenetic modifications responsible for DLC1 down regulation. Thus, we decided to reexamine all three lines under identical conditions. DLC1 promoter methylation was detected in LNCaP and C4-2B-2 cells and not in 22Rv1(Figure 1A). In our previous study using 22Rv1 cells, direct evidence showing that TSA induced DLC-1 mRNA re-expression is mediated through acetylation of the DLC1 promoter region has been presented [6]. To determine whether promoter methylation was exclusively responsible for DLC1 deregulation in LNCaP and C4-2B-2 cells, we compared the effect of SAHA and TSA on histone acetylation using ChIP assay for two sets of primers from DLC1 promoter region. In all three cell lines, DLC1 specific amplification products of fragments 124bp and 201bp were detected in cells treated with either SAHA or TSA, indicating that DLC1 promoter is both methylated and acetylated in LNCaP and C4-2B-2 cells (Figure 1B).
Fig. 1. SAHA and TSA restore DLC1 expression in prostate cancer cells.
(A): Methylation of DLC1 promoter in 22Rv1, LNCaP and C4-2-B2 cells. P control, universal methylated human DNA; M, methylated; U, unmethylated. (B): Chromatin immunoprecipitation analysis of Histone H3 acetylation on the DLC1 promoter. 22Rv1, LNCaP, and C4-2-B2 were treated with TSA or SAHA. Two sets of primers spanning different DLC1 promoter regions were used, which yield products of 124bp and 215 bp in length, respectively. M, DNA marker. (C): Restoration of DLC1 expression lines 22Rv1, C4-2-B2 and LNCaP cells by SAHA and TSA. DLC1 expression level was measured by real-time PCR.
3.2 Restoration of DLC1 expression by HDAC inhibitors
Based on these results, we then tested the effectiveness of SAHA and TSA in inducing DLC1 mRNA expression in these lines. The level of DLC1 transcriptional re-expression after exposure to SAHA or TSA was 22 to 35 fold higher over the control samples and as expected, DLC1 expression was 13 fold higher in 22Rv1 cells than in LNCaP and C4-2B-2 cells (Figure 1C).
3.3 Activation of DLC1 promoter by HDAC inhibitors
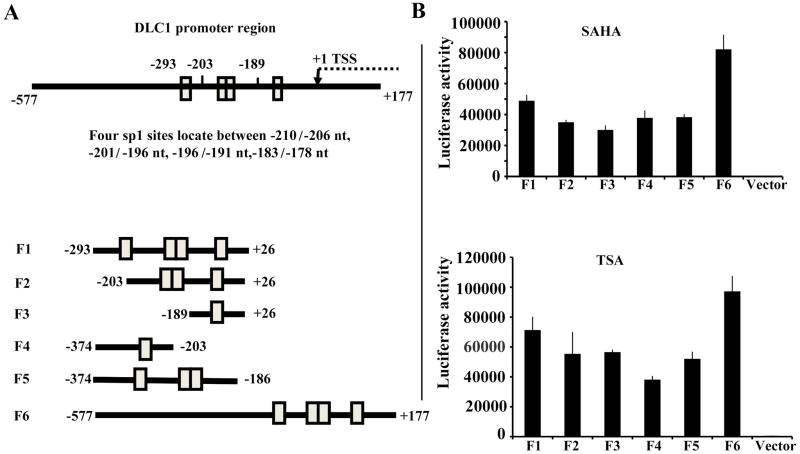
In gastric cancer cells, others have demonstrated that TSA activates DLC1 promoter activity through Sp1 sites located at −219 and −174 relative to the transcription start site, thus providing possible clues for the mechanism responsible for the induction of DLC1 expression through TSA- mediated chromatin modifications [11]. To see whether SAHA and TSA activate the DLC1 promoter through the same sites we constructed several DLC1 promoter fragments covering a series of Sp1 binding sites and linked them to a luciferase reporter gene (Figure 2A). The promoter activity driven by these fragments was measured after exposure to SAHA or TSA. The DLC1 promoter fragments most responsive to both HDAC inhibitors were fragment F1 (−293/+26) and F6 (−577/+177) (Figure 2B). Seemingly, these Sp1 sites are essential for both SAHA and TSA -mediated DLC1 promoter activation. Fragment F2 (−203/+26) containing three Sp1 sites, and fragment F3 (−189/+26) containing the fourth Sp1 site, also elicit a significant capacity to activate the DLC1 promoter (Figure 2B). Fragment F4 (−374/−203) containing the first Sp1 site and F5 (−374/−186) carrying three sp1 sites had a similar capacity. Overall, these observations suggest that not all Sp sites are equally responsive on HDAC inhibitors -mediated DLC-1 promoter activation.
Fig. 2. SAHA and TSA activate DLC1 promoter with the same SP1 sites.
A: Schematic representation of the DLC1 promoter region, which spans from −577 to + 177 and contains four SP1sites locate between −210/−206, −201/−196, −196/−191, −183/−178. Fragments with different combinations of SP1 sites are shown in the bottom. The numbers refer to the transcription start site (left panel). B: Activation of the truncated constructs of DLC-1 promoter in 22Rv1 cells. The constructs were transiently transfected into 22Rv1 cells followed by treatment with TSA or SAHA. Luciferase activity was measured and normalized to protein concentration. Data presented are representative of three independent experiments (right panel).
3.4 SAHA and/or DLC1 inhibition of tumor growth in vivo
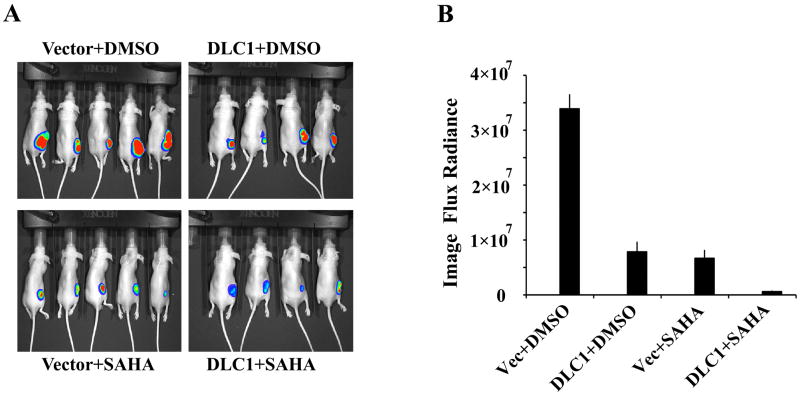
Once relevant information on interaction of the two HDAC inhibitors were generated, we proceeded with the central aim of present study, to assess the therapeutic efficacy of combined DLC1 transduction and SAHA treatment on 22Rv1 tumor growth in nude mice. 22Rv1 cells are highly tumorigenic, as only 6 to 10 days after injection of all animals developed small palpable tumors. At this time SAHA or solvent was administrated i.p daily for 21 days. The kinetics of tumor growth was monitored regularly using digital caliper and quantitative measurements of bioluminescent images. Either DLC1 transduction or exposure to SAHA had an inhibitory effect on tumor growth as early as 13 days, and after 21 days at the end point of the experiment, the size of the tumors was reduced by 70 to 88 % (Figure 3A and B). The size of tumors developed in mice inoculated with DLC1 transfected cells and treated with SAHA was reduced as compared to either DLC1 or SAHA treatment alone (Figure 3A and B). Quantitative data analysis showed that combined DLC1 transduction and exposure to SAHA had a robust inhibitory effect on tumor growth, and bioluminescent signals of the tumors were reduced to less than 2% (Figure 3B).
Fig. 3. SAHA and/or DLC1 effect of in vivo tumor growth of 22Rv1 cells.
Exponentially growing 22Rv1 cells were injected subcutaneously in BALB/c AnNCr-nu/nu male mice, 50 mg/kg (50 ug/g) SAHA or DMSO were administrated daily by intraperitoneal injections for 21 days beginning after all animals developed palpable tumors. The bioluminescent signal was acquired and illustrated as a pseudocolor image (A) and quantitative data from the region of interest are shown as the photon counts (B).
To evaluate the toxicity of SAHA, the weight of all animals was checked twice a week and their general condition monitored daily. Among 40 mice from both control and DLC1 vector injected with SAHA for 16 and 19 days, 4 did not survive, most likely due to SAHA toxicity. In contrast, DLC1 transduction did not affect their survival, but contributed to slight weight loss.
3.5 Reduction of RhoA activity, induction of apoptosis and. cell cycle progression
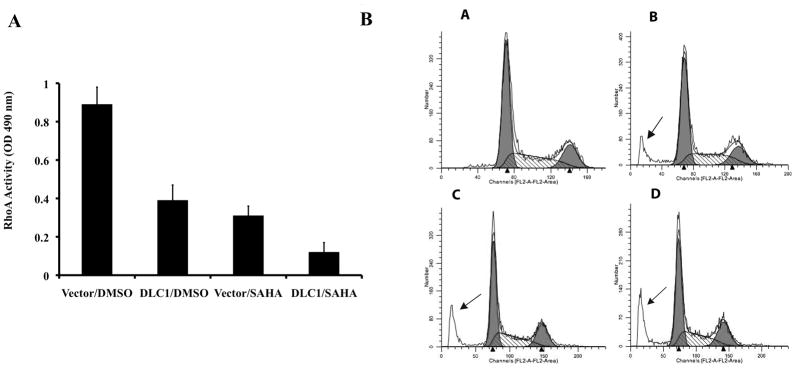
To gain insights into mechanisms involved in the process of SAHA and DLC1 inhibitory effects of tumor growth, we next examined the effects on RhoA activity, induction of apoptosis and cell cycle progression in 22Rv1 cells (Figure 4). An ELISA-based RhoA activation assay was employed to determine whether re-expression of DLC1 and exposure to SAHA affect RhoA activity in 22Rv1 cells. The level of active GTP-bound RhoA was 57 % lower in cells transduced with DLC1, 66% lower in SAHA treated cells and up to 87% in combination as compared with controls vector and solvent treated cells (P=0.035, Figure 4A).
Fig. 4. Effects of SAHA on RhoA activity, apoptosis and cell cycle progression.
A: Effect of SAHA and DLC1 on RhoA activity. DLC1 or vector only transduced 22Rv1 cells were treated with SAHA or DMSO. Twenty-four hrs after treatment, cells were lysed and equal amounts of cell lysates were subjected to G-Lisa RhoA assay. B: Effects of SAHA and DLC1 on apoptosis and cell cycle. DLC1 or vector only transduced 22Rv1 cells were treated with SAHA or DMSO. The percentage of cells in the sub-G1 phase (arrow) was significantly increased upon DLC1 or/and SAHA treatment for 24 hrs. Vector and DMSO (a), DLC1 and DMSO (b), Vector and SAHA (c), DLC1 plus SAHA (d).
For apoptosis and cell cycle analysis, 22Rv1 cells were transfected with the DLC1 expression vector, the vector only or exposed to SAHA for 24 hrs. We detected a significant accumulation of subG1 cells in cells treated with DLC1, SAHA, and DLC1 plus SAHA (arrows, Figure 4B, C, D), indicative of apoptosis, compared to the control (Figure 4A). The populations in G1 and G2 phase were not significantly affected (Figure 4).
4. Discussions
Recurrent loss of DNA copy number and loss of heterozygosity on the short arm of chromosome 8, non randomly affecting the region 8p21–22 harboring the DLC1 gene, have been detected in several earlier studies with in PCAs. Subsequently, it has been firmly demonstrated that promoter methylation and histone deacetylation are major mechanisms for the down regulation and silencing of DLC1 expression in primary PCAs [6]. In a recent genome-wide array analysis of copy number alterations of PCAs, the highest frequencies of loss DNA copy number was found to be confined on chromosomes 8p and 6q and affected the DLC1 and WWOX gene loci, respectively. Both genes were found to be involved in the apoptosis pathway and associated with increasing disease grade. Based on these results and our previous data with epigenetic deregulation of DLC1, this study concluded that DLC1 is involved in pathogenesis of prostate cancer [12].
Here we examined three PCA cell lines with low level, or lacking DLC1 expression to identify the mechanism responsible for DLC1 deregulation. Both promoter methylation and histone deacetylation were detected in LNCaP and C4-2B-2 cells but only deacetylation in 22Rv1cells. Treatment of cells from all three lines with SAHA or TSA efficiently restored DLC1 expression and activated the DLC1 promoter through the same Sp1 sites. Also, we showed that not all the Sp1 sites confer the same DLC1 promoter responsiveness to both HDAC inhibitors, and this difference might due to the uneven distribution of these Sp1 sites.
22Rv1 cell line was used for in vivo tumorigenicity assay because these cells do not express endogenous DLC1 due to histone deacetylation. Restoration of DLC1 expression combined with SAHA treatment resulted in significant inhibition of tumor development in nude mice. This effect on tumor growth in vivo is consistent with the synergistic inhibitory effect of DLC1 and SAHA on cell migration and anchorage independent growth of 22Rv1 cells in vitro [13]. 22Rv1 cell line was established from PCA xenograft, CWR22 [14]. SAHA treatment of xenotransplanted CWR22 tumors in nude mice caused a 97 % reduction of tumor size compared to controls [15]. Current results and those with CWR22 transplanted tumors clearly show that these tumor cells lacking DLC1 expression are highly susceptible to SAHA.
The antitumor effect of DLC1 and SAHA in 22Rv1 cells was associated with the induction of apoptosis and inhibition of RhoA activity, both known contributing factors in the inhibition of tumor cell growth. Accumulating evidence indicates that inactivation of DLC1 may represent the most frequent mechanism for aberrant activation of RhoGTPases in human oncogenesis [16]. The suppressive effect on tumor cell growth and tumorigenicity induced by DLC1 requires RhoGAP activity, which can negatively regulate Rho GTPases, most commonly RhoA [5,17–19]. As expected, DLC1 transduction of 22Rv1 cells reduced RhoA activity and the addition of SAHA enhanced this effect. However, the most important and unanticipated finding with multiple implications, was the significant inhibitory effect of SAHA alone on RhoA activity. This finding certainly requires further confirmation with other types of cancer cells, yet raises the possibility that RhoA and perhaps other RhoGTPases are targets for SAHA or different HDAC inhibitors. It also adds a new antitumor action of SAHA [7,8, 20–24].
In clinical trials SAHA has been considerably more effective in hematological malignancies than in solid tumors [7]. As previously discussed in detail, DLC1 is most frequently inactivated by epigenetic mechanisms in over 80% of patients with acute lymphoblastic leukemia and non-Hodgkin’s lymphoma and multiple myeloma [10]. Because inactivation or down regulation of DLC1 expression is commonly associated with an increase RhoA activity, SAHA might be more effective in hematological malignancies than in solid tumors.
Despite the antitumor effects of HDAC inhibitors in multiple cancers, it is increasingly documented that combinatorial treatment with DNA demethylating agents may enhance the therapeutic efficacy. Both agents efficiently restore DLC1 expression in cancer cells and their combined use has been highlighted as an attractive therapeutic approach [25]. Both promoter methylation and acetylation are responsible for the reduction of DLC1 expression in androgen-independent C4-2B-2 cells that metastasize to bone. Thus, these cells are ideal for testing the effect of combined of DNA demethylating agents and HDAC inhibitors in DLC1-negative and DLC1 transduced cells. Such experiments are in progress.
Given the high incidence of primary PCA with low or lacking DLC1 expression, we propose that DLC1 deficiency may serve as an independent diagnostic factor in prostate cancer. In a recent article, it has been suggested that DLC1 is a therapeutic target for future cancer treatment in conjunction with other factors [26]. Current results with in vivo inhibition tumor growth show that DLC1-negative PCA cells are susceptible to antitumor action of SAHA. The development of novel therapies of prostate cancer is a clinical priority. Thus, taking into consideration the large number of patients with disabled DLC1 expression, SAHA alone or in combination with other agents may be therapeutically beneficial in prostate cancer.
Highlights.
Prostate tumor cells lacking DLC1 expression are sensitive to SAHA.
Re-expression of DLC1 and SAHA inhibit prostate tumor cells growth in vivo.
Antitumor effect correlated with apoptosis and reduction of RhoA activity.
RhoA appears to be an important target for SAHA.
Treatment with SAHA might be therapeutically beneficial for prostate cancer.
Acknowledgments
This work was supported by the Intramural Research Program of the National Cancer Institute, NIH.
Abbreviations
- DLC1
deleted in liver cancer
- PCAs
prostate carcinomas
- HDAC
histone deacetylase inhibitors
- SAHA
suberoylanilide hydroxamic acid
- TSA
trichostatin A
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yuan BZ, Miller MJ, Keck CL, Zimonjic DB, Thorgeirsson SS, Popescu NC. Cloning, characterization and chromosomal localization of a gene frequently deleted in human liver cancer (DLC-1) homogous to rat RhoGAP. Cancer Res. 1998;58:2196–2199. [PubMed] [Google Scholar]
- 3.Goodison S, Yuan J, Sloan D, Kim R, Li C, Popescu NC, Urquidi V. The RhoGAP protein DLC-1 functions as a metastasis suppressor in breast cancer cells. Cancer Res. 2005;65:6042–6053. doi: 10.1158/0008-5472.CAN-04-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Durkin ME, Yuan BZ, Zhou X, Zimonjic DB, Lowy DR, Thorgeirsson SS, Popescu NC. DLC-1: a Rho GTPase-activating protein and tumour suppressor. J Cell Mol Med. 2007;11:1185–1207. doi: 10.1111/j.1582-4934.2007.00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xue W, Krasnitz A, Lucito R, Sordella R, Vanaelst L, Cordon-Cardo C, Singer S, Kuehnel F, Wigler M, Powers S, Zender L, Lowe SW. DLC1 is a chromosome 8p tumor suppressor whose loss promotes hepatocellular carcinoma. Genes Dev. 2008;22:1439–1444. doi: 10.1101/gad.1672608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guan M, Zhou X, Soulitzis N, Spandidos DA, Popescu NC. Aberrant methylation and deacetylation of deleted in liver cancer-1 gene in prostate cancer: potential clinical applications. Clin Cancer Res. 2006;12:1412–1419. doi: 10.1158/1078-0432.CCR-05-1906. [DOI] [PubMed] [Google Scholar]
- 7.McGuire C, Lee J. Brief review of Vorinostat. Clin Med Insights:Therapeutics. 2010;2:83–87. [Google Scholar]
- 8.Kim HJ, Bae SC. Histone deacetylase inhibitors: molecular mechanisms of action and clinical trials as anti-cancer drugs. Am J Transl Res. 2011;3:166–179. [PMC free article] [PubMed] [Google Scholar]
- 9.Guan M, Tripathi V, Zhou X, Popescu NC. Adenovirus-mediated restoration of expression of the tumor suppressor gene DLC1 inhibits the proliferation and tumorigenicity of aggressive, androgen-independent human prostate cancer cell lines:prospects for gene therapy. Cancer Gene Ther. 2008;15:371–381. doi: 10.1038/cgt.2008.13. [DOI] [PubMed] [Google Scholar]
- 10.Ullmannova-Benson V, Guan M, Zhou X, Tripathi V, Yang XY, Zimonjic DB, Popescu NC. DLC1 tumor suppressor gene inhibits migration and invasion of multiple myeloma cells through RhoA GTPase pathway. Leukemia. 2009;23:383–390. doi: 10.1038/leu.2008.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim TY, Kim IS, Jong HS, Lee JW, Kim TY, Jung M, Bang YJ. Transcriptional induction of DLC-1 gene through Sp1 sites by histone deacetylase inhibitors in gastric cancer cells. Exp Mol Med. 2008;40:639–646. doi: 10.3858/emm.2008.40.6.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng, Levin AM, Tai YC, Plummer S, Chen GK, Neslund-Dudas C, Casey G, Rybicki BA, Witte JS. Copy number alterations in prostate tumors and disease aggressiveness. Genes Chromosomes Cancer. 2012;51:66–76. doi: 10.1002/gcc.20932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou X, Yang XY, Popescu NC. Synergistic antineoplastic effect of DLC1 tumor suppressor protein and histone deacetylase inhibitor, suberoylanilide hydroxamicacid (SAHA), on prostate and liver cancer cells: perspectives for therapeutics. Int J Oncol. 2010;36:999–1005. doi: 10.3892/ijo_00000580. [DOI] [PubMed] [Google Scholar]
- 14.Sramkoski RM, Pretlow TG, 2nd, Giaconia JM, Pretlow TP, Schwartz S, Sy MS, Marengo SR, Rhim JS, Zhang D, Jacobberger JW. A new human prostate carcinoma cell line, 22Rv1. In Vitro Cell Dev Biol Anim. 1999;35:403–409. doi: 10.1007/s11626-999-0115-4. [DOI] [PubMed] [Google Scholar]
- 15.Butler LM, Agus DB, Scher HI, Higgins B, Rose A, Cordon-Cardo C, Thaler HT, Rifkind RA, Marks PA, Richon VM. Suberoylanilide hydroxamic acid, an inhibitor of histone deacetylase, suppresses the growth of prostate cancer cells in vitro and in vivo. Cancer Res. 2000;60:5165–5170. [PubMed] [Google Scholar]
- 16.Yang XY, Guan M, Vigil D, Der CJ, Lowy DR, Popescu NC. p120Ras-GAP binds the DLC1 Rho-GAP tumor suppressor protein and inhibits its RhoA GTPase and growth suppressing activities. Oncogene. 2009;28:1401–1409. doi: 10.1038/onc.2008.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sahai E, Marshall CJ. RHO-GTPases and cancer. Nat Rev Cancer. 2002;2:133–142. doi: 10.1038/nrc725. [DOI] [PubMed] [Google Scholar]
- 18.Jaffe AB, Hall A. Rho GTPases in transformation and metastasis. Adv Canc Res. 2002;84:57–80. doi: 10.1016/s0065-230x(02)84003-9. [DOI] [PubMed] [Google Scholar]
- 19.Vigil D, Cherfils J, Rossman KL, Der CJ. Ras superfamily GEFs andGAPs: validated and tractable targets for cancer therapy? Nat Rev Cancer. 2010;10:842–857. doi: 10.1038/nrc2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnstone RW. Histone-deacetylase inhibitors: novel drugs for the treatment ofcancer. Nat Rev Drug Discov. 2002;1:287–299. doi: 10.1038/nrd772. [DOI] [PubMed] [Google Scholar]
- 21.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006;6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 22.Lane AA, Chabner BA. Histone deacetylase inhibitors in cancer therapy. J Clin Oncol. 2009;27:5459–5468. doi: 10.1200/JCO.2009.22.1291. [DOI] [PubMed] [Google Scholar]
- 23.Batty N, Malouf GG, Issa JP. Histone deacetylase inhibitors as anti-neoplastic agents. Cancer Lett. 2009;280:192–200. doi: 10.1016/j.canlet.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 24.Richon VM, Garcia-Vargas J, Hardwick JS. Development of vorinostat: current applications and future perspectives for cancer therapy. Cancer Lett. 2009;280:201–210. doi: 10.1016/j.canlet.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 25.Lahoz, Hall A. DLC1: a significant GAP in the cancer genome. Genes Dev. 2008;22:1724–1730. doi: 10.1101/gad.1691408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao X, Voss C, Zhao B, Kaneko T, Li SSC. Differential regulation of the activity of deleted in liver cancer 1(DLC1) by tensin control cell migration and transformation. Proc Natl Acad Sci USA. 2012;109:1455–146. doi: 10.1073/pnas.1114368109. [DOI] [PMC free article] [PubMed] [Google Scholar]