Abstract
Hydrophilic poly(ethylene glycol) diacrylate (PEGDA) hydrogel surfaces resist protein adsorption and are generally thought to be unsuitable for anchorage dependent cells to adhere. Intriguingly, our previous findings revealed that PEGDA superporous hydrogel scaffolds (SPHs) allow anchorage of bone marrow derived human mesenchymal stem cells (hMSCs) and support their long term survival. Therefore, we hypothesized that the physicochemical characteristics of the scaffold impart properties that could foster cellular responses. We examined if hMSCs alter their microenvironment to allow cell attachment by synthesizing their own extracellular matrix (ECM) proteins. Immunofluorescence staining revealed extensive expression of collagen type I, collagen type IV, laminin and fibronectin within hMSC-seeded SPHs by the end of the third week. Whether cultured in serum-free or serum-supplemented medium, hMSC ECM protein gene expression patterns exhibited no substantial changes. The presence of serum proteins is required for initial anchorage of hMSCs within the SPHs but not for the hMSC survival after 24 hours. In contrast to 2D expansion on tissue culture plastic (TCP), hMSCs cultured within SPHs proliferate similarly in the presence or absence of serum. To test whether hMSCs retain their undifferentiated state within the SPHs, cell-seeded constructs were cultured for 3 weeks in stem cell maintenance medium and the expression of hMSC-specific cell surface markers were evaluated by flow cytometry. CD105, CD90, CD73 and CD44 were present to a similar extent in the SPH and in 2D monolayer culture. We further demonstrated multi lineage potential of hMSCs grown in the PEGDA SPHs whereby differentiation into osteoblasts, chondrocytes and adipocytes could be induced. The present study demonstrates the potential of hMSCs to alter the “blank” PEGDA environment to a milieu conducive to cell growth and multi-lineage differentiation by secreting adhesive ECM proteins within the porous network of the SPH scaffolds.
Keywords: Porous, hydrogels, scaffolds, poly(ethylene glycol), mesenchymal stem cells, extracellular matrix, cell proliferation, cell differentiation
Introduction
Human mesenchymal stem cells (hMSCs) derived from adult tissues have shown increasing potential in regenerative medicine due to their multi-lineage differentiation capability and their immunomodulatory effects.1-3 In the human body MSCs ensure homeostasis by supporting the repair and rejuvenation of degenerated tissues and organs. To induce repair, hMSCs undergo differentiation into a variety of mature cell types including osteoblasts, chondrocytes and adipocytes and secrete bioactive factors as “trophic mediators” that alter the milieu of dysfunctional tissues.4, 5 Current tissue engineering approaches typically involve the development of stem cell-carrying extracellular matrix (ECM)-mimicking scaffolds that mature in vitro before implantation into the patient. To control for lineage specific differentiation, ideal scaffold materials should be able to maintain stem cells in their undifferentiated phenotype and promote differentiation only after induction.
Our group has demonstrated that poly(ethylene glycol) diacrylate (PEGDA) superporous hydrogels (SPHs) promote long-term survival and mineralization of hMSCs in vitro.6 Host cell infiltration and neovascularization of hMSC-seeded SPHs was also observed in vivo.7 SPHs were fabricated utilizing a gas foaming technique, wherein the foaming and gelation processes occur simultaneously to create an interconnected macroporous network with pores ranging from 100 to 600 μm.6 The interconnected pores allow for fast absorption of fluids by capillary force and, thus, enable a rapid cell uptake. However, hydrophilic PEGDA hydrogels are considered resistant to cell binding due to poor protein adsorption to the surface.8 The viability of cells encapsulated within unmodified non-porous PEGDA hydrogels drops to 15% within one week of culture.8 We have recently reported that cells can be incorporated into the hydrogel matrix of superporous hydrogels despite the harsh foaming and polymerization conditions.9 These conditions do limit which cell types can be incorporated into the matrix of the SPHs, but leave open the possibility for dual cell type incorporation. Based upon this, there is potential to form these hydrogels in situ with cell encapsulation within the matrix followed by loading of cells within the pores.
Interestingly, our previous findings revealed that PEGDA SPHs, even in the absence of cell adhesive peptides, provide anchorage to hMSCs and support long-term survival suggesting that the porous environment per se imparts properties that could endorse cellular responses. Macroporous degradable PEG hydrogels have been shown to promote hMSC osteoblast differentiation by facilitating cell communication through autocrine and paracrine signaling via the interconnected pore network.10 Importantly, PEGDA SPH synthesized with an anionic monomer, i.e. acrylic acid, eliminated cell adhesion without altering the macroscopic structure, indicating that the observed cell binding phenomenon is not entirely due to the hydrogel architecture.11
Initial anchorage of cells to implant materials occurs via binding to proteins that are either immobilized on the substrate or adsorbed from the serum in culture. Cell-surface integrins subsequently bind to amino-acid sequences, e.g. Arg-Gly-Asp (RGD), in the adsorbed protein molecules.12 Anchorage-dependent cells also deposit their own ECM molecules after seeding on matrices that do not possess any natural binding motifs to prevent anoikis, a form of apoptosis induced by the lack of sufficient cell-ECM contact.13, 14 In 2D monolayer cultures, undifferentiated hMSCs secrete collagen I and IV, and adhesive proteins such as fibronectin and laminin.15-17 The secreted ECM provides environmental cues for cell survival, proliferation and differentiation. In vivo, cell adhesion molecules play an important role in holding the stem cells within their specific niche and thereby allowing cell-cell and cell-ECM interactions. In the current study, we cultured hMSCs in basal medium for a period of 3 weeks within the PEGDA SPHs and monitored the expression of ECM molecules by immunofluorescence and real-time qPCR to reveal cell-scaffold interactions that may influence attachment.
Since the ECM not only promotes hMSCs vitality but also helps to maintain their stem state,18 we investigated if the hMSCs stay undifferentiated after being cultured within the SPH for an extended period of 3 weeks and if they are able to undergo controlled differentiation after osteogenic, chondrogenic and adipogenic induction. The maintenance of hMSCs in their undifferentiated state allows for site-specific cell responses, and thus could be advantageous in many tissue engineering applications where optimal tissue integration is required. To this end, the objective of the present study was to investigate whether hMSCs are able to alter the cell-repellant PEGDA environment to a milieu conducive to cell growth and multi-lineage differentiation by secreting their own adhesive proteins within the SPH scaffolds.
Materials and Methods
Materials
PEGDA (3,400 MW) was obtained from Laysan Bio, Inc. (Arab, AL). Pluronic® F127 was obtained from Sigma-Aldrich, Inc. (St. Louis, MO). N,N,N’,N'-tetramethylethylenediamine (TEMED, 99%, Acros Organics), ammonium persulfate (APS, 98+%, Acros Organics), citric acid anhydrous, sodium bicarbonate and paraformaldehyde (96%, Acros Organics) were purchased from Fisher Scientific (Fair Lawn, NJ). Chemicals were used as received without any further purification. Dulbecco's Modified Eagle's Medium, trypsin-EDTA, penicillin and streptomycin were from Mediatech, Inc. (Cellgro®, Manassas, VA). Foundation™ fetal bovine serum (FBS) was purchased from Gemini Bio-Products (West Sacramento, CA). Primers were ordered from Integrated DNA Technologies (Skokie, IL) (Table 1).
Table 1.
Genes and primers used for real-time RT-qPCR.
| Gene | Full Name | Seqences 5′-> 3′ | Accession number/ Reference |
|---|---|---|---|
| ADIPOQ | Adiponectin | For: AGG GTG AGA AAG GAG ATC C Rev: GGC ATG TTG GGG ATA GTA A |
NM_004797 |
| ACAN | Aggrecan | For: TTC AGT GGC CTA CCA AGT GGC ATA Rev: AGC CTG GGT TAC AGA TTC CAC CAA |
NM_001135 |
| ALPL | Alkaline Phosphatase | For: ATT TCT CTT GGG CAG GCA GAG AGT Rev: ATC CAG AAT GTT CCA CGG AGG CTT |
NM_000478 |
| BGLAP | Bone gamma- carboxy- glutamate (Osteocalcin) | For: CAG CGA GGT AGT GAA GAG AC Rev: TGA AAG CCG ATG TGG TCA G |
NM_199173 |
| COL1A1 | Collagen type I | For: CGC TAC TAC CGG GCT GAT GAT Rev: ATC TTG AGG TCA CGG CAG GTG |
NM_000088 |
| COL2A1 | Collagen type II | For: GTT GCA AAC CCA AAG GAC CCA AGT Rev: ACA TCA GGT CAG GTC AGC CAT TCA |
NM_001844 |
| COL4A1 | Collagen type | For: ACT CTT TTG TGA TGC ACA CCA | NM_00184521 |
| IV | Rev: AAG CTG TAA GCG TTT GCG TA | ||
| FN1 | Fibronectin | For: GGT GAC ACT TAT GAG CGT CCT AAA Rev: AAC ATG TAA CCA CCA GTC TCA TGT G |
NM_054034, RTPrimer DB, ID:109222 |
| LAMA5 | Laminin | For: CCC ACC GAG GAC CTT TAC TGC Rev: GGT GTG CCT TGT TGC TGT TGG |
NM_00556023 |
| LEP | Leptin | For: CTG ATG CTT TGC TTC AAA TCC A Rev: GCT TTC AGC CCT TTG CGT T |
NM_00023024 |
| PCNA | Proliferating cell nuclear antigen | For: AGG CAC TCA AGG ACC TCA TCA Rev: GAG TCC ATG CTC TGC AGG TTT |
NM_002592 |
| RPL13α | Ribosomal protein L13 α | For: CAT AGG AAG CTG GGA GCA AG Rev: GCC CTC CAA TCA GTC TTC TG |
NM_01242320 |
Fabrication of superporous hydrogels
Scaffolds with pore sizes ranging from 100 to 600 μm were prepared by a gas foaming method as previously reported.6 Briefly, aqueous PEGDA solution (15 (w/v) %), foam stabilizer (Pluronic® F-127; 0.6 (w/v) %), and the initiator pair N,N,N’,N’-tetramethylethylenediamine (TEMED; 0.9 (v/v) %) and ammonium persulfate (APS; 0.7 (w/v) %) were added sequentially to a glass vial to a final volume of 1 mL. Saturated citric acid solution (2 (w/v) %) was used to adjust the pH to 3.70. The solution was heated at 37-40°C for 2 minutes. Sodium bicarbonate (200 mg) was stirred into the precursor solution evolving CO2 by reacting with the citric acid. Polymerization occurred simultaneously as the pH increased to a pH of approximately 7. Polymerization was allowed to proceed for 30 min. SPHs were then removed from the vial and washed three times in double-deionized water to remove traces of unpolymerized monomers and salt before sterilizing the gels in 80% ethanol overnight, followed by dehydration in absolute ethanol for 4 hours. SPHs were dried in a food dehydrator at a temperature of 55°C for approximately 2 hours. Dried hydrogels were cut to give scaffolds with a diameter of 5 mm and a thickness of 3 mm.
Human mesenchymal stem cell isolation, seeding and cultivation
Human bone marrow aspirates were obtained from AllCells, LLC (Emeryville, CA) and isolated by density gradient centrifugation utilizing Ficoll-Paque™ PLUS solution and RosetteSep® Human Mesenchymal Stem Cell Enrichment Cocktail (Stem Cell Technologies, Vancouver, Canada) according to the manufacturer's protocol. For each experiment, hMSCs from 3 different donors, up to passage 4, were used. Cells were harvested using 0.25% trypsin with 1.0 M EDTA, centrifuged and expanded in basal medium which consists of high glucose Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 unit/mL penicillin and 100 unit/mL streptomycin. DMEM without FBS was used for serum-free culture. For cell seeding, dehydrated SPH cylinders were placed into 48 well plates. Cell suspension (50 μL) containing 2.5 × 105 cells (unless stated otherwise) was added drop-wise to each SPH. The hydrogels were placed in the incubator for 30 min to allow equilibrium swelling and initial cell attachment. Thereafter, 1.0 mL of basal medium was added to each of the wells, and the plates were incubated at 37°C in 5% CO2. Medium was changed every third day. Cell-seeded constructs were cultured for 3 weeks in basal medium before differentiation medium was added for additional 4 weeks of culture. Adipogenic differentiation was initiated by culturing the cell-seeded constructs in adipogenic induction medium (AI) for 3 days, following treatment with adipogenic maintenance medium (AM) for the next 3 days. The alternation between AI and AM treatment was continued for the 4 weeks of culture. Chondrogenic differentiation was induced by serum free chondrogenic induction medium (Lonza, Walkersville, MD) containing dexamethasone and TGF-β3. Osteogenic differentiation medium (Lonza, Walkersville, MD) consisted of basal medium with 100 nM dexamethasone, 10 mM β-glycerophosphate and 0.05 mM ascorbic acid 2-phosphate. Controls were maintained in basal medium for the entire culture period of 7 weeks.
Cell viability
Cell viability was analyzed immediately after seeding (day 0), and 1, 7, 14, 21 and 28 days after seeding hMSCs at a density of 5 × 103 cells/cm2 on 48 well plates (3750 cells/well) and within PEGDA SPHs (2.5 × 105 cells/SPH). To examine the proliferative activity in serum-free culture, serum-containing medium was switched to serum-free medium 24 hours after seeding. The MTS assay measures cell viability as a function of mitochondrial activity. MTS [3-(4,5-dimethylthiazol-2yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] is reduced into a colored formazan product by metabolically active cells. In brief, 200 μL of 1X DPBS and 40 μL of Cell-Titer 96® Aqueous One Solution reagent (Promega, Madison, WI) were added to each well. The plates were covered and incubated for 3 h at 37 °C. Supernatant (100 μL) was transferred to a new plate and absorbance was measured at 492 nm (Labsystems Multiskan Plate Reader). The controls for background absorbance were unseeded SPHs.
For live-dead staining, the wells were rinsed with 1X DPBS to remove serum and medium components. A solution comprising 2.5 μL calcein acetoxymethyl ester (4 mM) and 2.5 μL ethidium homodimer (2 mM) in 5 mL of 1X DPBS was added to the SPHs and allowed to incubate for 25 min at 37°C. SPHs were rinsed twice with 1X DPBS and fluorescent images were taken with an Olympus IX70 inverted microscope.
Quantitative real-time PCR
At specific timepoints, i.e. 24 hours, 3 and 7 weeks after seeding hMSCs at a density of 5 × 103 cells/m2 on 6 well plates (5 × 104 cells/well) and within PEGDA SPHs (7.5 × 105 cells/SPH), total RNA was extracted using the TRIzol® reagent (Invitrogen) in combination with the PureLink™ RNA Mini Kit (Invitrogen) according to manufacturer's instructions. RNA (180 ng) was reverse transcribed with the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). The PCR reactions were performed on an Applied Biosystems StepOnePlus™ PCR machine using 5 μL SYBR® Green PCR Master Mix (Applied Biosystems), 2 μL sequence specific primers (0.5 mM, Table 1) and 3 μL cDNA under the following conditions: 95°C for 10 min followed by 40 cycles of 15 s of denaturation at 95°C and 60 s of annealing and elongation at 60°C. A melting curve analysis was performed after each run to confirm product specificity. The delta-delta-Ct method19 was employed to determine the relative gene expression level of the gene of interest normalized to the expression of the endogenous control ribosomal protein L13 α (RPL13α).20
Flow cytometry
HMSCs were cultured on 6 well plates (5 × 104 cells/well) or within SPHs (7.5 × 105 cells/SPH) for 21 days in basal medium. To remove hMSCs from the scaffolds, the cell-seeded constructs were incubated with trypsin-EDTA (0.25% trypsin, 2.21 mM EDTA) for 5 min under slight shaking. After centrifugation, cells were incubated on ice for 30 min in 100 μL of FACS buffer (1X DPBS + 5% FBS + 0.05% 3M sodium azide) with anti-human CD105-FITC, CD90-FITC, CD73-APC or CD44-PE. Cells were washed twice with FACS buffer and finally diluted into 100 μL of FACS buffer. Fluorochrome- and isotype-matched antibodies were used as controls. All antibodies were obtained from Biolegend (San Diego, CA) and used at manufacturer's recommended concentrations. Analysis was performed by collecting 15, 000 events on a Beckman Coulter Cyan ADP.
Immunohistofluorescence
Immunostaining was carried out to detect and visualize the expression of collagen type I and IV, laminin, and fibronectin. Briefly, the samples were rinsed with 1X PBS to remove medium components. Following fixation with 3.7% paraformaldehyde for 15 min, the samples were treated with blocking solution (1% BSA in 1X PBS). After 30 min incubation on a shaker, respective primary antibodies (5 μg/ml anti collagen type I; 4 μg/ml anti collagen type IV; 2 μg/ml anti-laminin, 2 μg/ml anti-fibronectin) were added to the blocking solution and samples were further incubated for 3 hours at room temperature (RT). Mouse monoclonal IgG antibodies (Santa Cruz Biotech, Santa Cruz, CA) were used for all ECM molecules. Samples were washed with 1X PBS and incubated with a FITC-labeled goat anti-mouse secondary antibody (5 μg/ml; Molecular Probes, Carlsbad, CA) for 20 min at RT. The nucleus in the 2D samples was counterstained with H33258 (0.5 mg/ml; Molecular Probes, Carlsbad, CA) for 15 min at RT. Appropriate mouse control IgG antibodies were used at dilutions corresponding to the primary antibody (data not shown). Images were taken with an Olympus IX70 inverted microscope and processed using IPLab™ software.
Histochemical Confirmation of Differentiation
Sudan III staining was applied to stain lipid vacuoles to evaluate adipogenic differentiation. The cell-seeded constructs were rinsed with 1X PBS (pH 7.4) and incubated with Sudan III solution (0.3% w/v of Sudan III in 70% ethanol) for 3 minutes. After several washes with double deionized water (DDIW), harris hematoxylin solution was added and incubated for 1 minute. The wells were rinsed with DDIW until the water ran clear. Images were taken under bright field with an Olympus IX70 inverted microscope.
Safranin O staining was used for the detection of glycosaminoglycans to evaluate chondrogenic differentiation. In brief, cell-seeded constructs were washed with 1X PBS (pH 7.4) and fixed with 3.7% paraformaldehyde solution at room temperature. After rinsing with 1X PBS, 0.01 % aqueous fast green was added to the sample and incubated for 3 minutes. The sample was washed twice with 1% acetic acid and incubated with aqueous Safranin O solution (0.1 %) for 5 minutes. The wells were rinsed with DDIW till the water ran clear. Images were taken under bright field with an Olympus IX70 inverted microscope.
To evaluate osteogenic differentiation calcium was determined by complexation of calcium by the phenolsulphonephtalein dye.25 Briefly, the lyophilized hydrogels were homogenized with 1.0 mL of 0.5 N HCL and mixed overnight at 4°C. The supernatant was used for calcium assays, as per manufacturer's protocol (QuantiChrom™ Calcium assay kit, BioAssay Systems).
Statistical analysis
Statistical analysis was performed using a one-way analysis of variance (ANOVA) followed by a Tukey post-hoc test (Origin 8.1) if the means were significantly different at a p-value of 0.05. All data are presented as mean plus or minus standard deviations.
Results
HMSC Viability and Proliferation
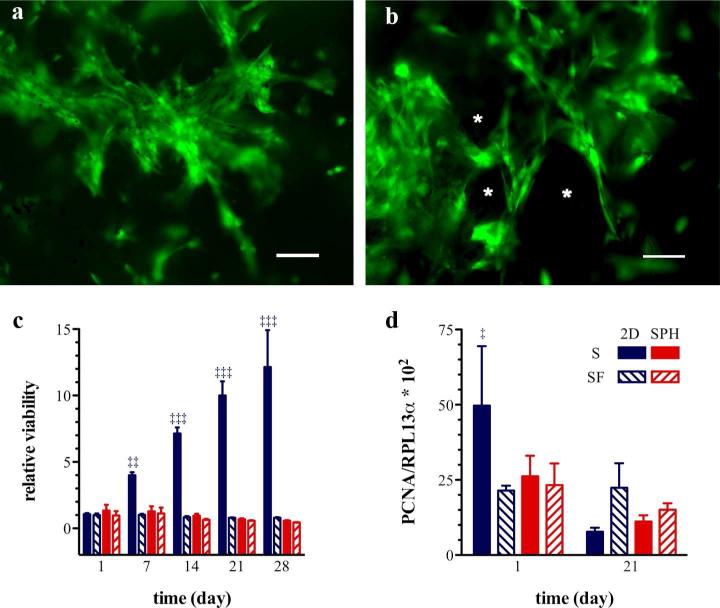
Viable cells with an elongated spindle-shaped, fibroblast-like morphology could be observed within the SPHs during the entire test period of 7 weeks (Figure 1a,b). Serum is necessary for hMSC expansion under 2D conditions on tissue culture plastic, but hMSCs cultured within SPHs proliferate similarly in the presence or absence of serum in basal medium, as assessed by the MTS assay (Figure 1c) and mRNA expression levels of proliferating cell nuclear antigen (PCNA) (Figure 1d).The PCNA mRNA levels of hMSCs cultured for 3 weeks within SPHs remained constant under both, serum-supplemented and serum-free conditions, while PCNA mRNA levels from hMSCs expanded under 2D conditions was initially high and decreased significantly from day 1 to day 21 to a level similar to that of SPH samples. The differences between the MTS assay results and the gene expression patterns of PCNA may arise form the fact that cells grow to a confluent state on the tissue culture plate and stop proliferating at later time points which results in decreased mRNA expression of PCNA. Additionally, it appears that there is less signaling to proliferate within the SPHs leading to a stable number of cells within the structures.
Figure 1. Human mesenchymal stem cells remain viable and proliferate within PEGDA superporous hydrogels.
Representative pseudocolored epifluorescent micrographs of viable hMSCs around the pores (*) of PEGDA SPHs, (a) 3 and (b) 7 weeks post seeding. Viable cells are stained with calcein acetoxymethyl ester dye (green) and dead cells appear red due to staining with ethidium homodimer. The scale bar is 100 μm. (c) MSC viability within PEGDA superporous hydrogels (SPH/red) and on tissue culture plastic as a monolayer (2D/blue) under serum-containing (S/solid bars) and serum-free conditions (SF/diagonal lines within bars) was assessed by the MTS assay. Cells were incubated for one day in the presence of serum, at t = 0 day, and maintained for additional time (as listed on x-axis) in either serum-containing (S) or serum-free conditions (SF). The symbols (‡‡) and (‡‡‡) denote a significant higher cell viability under 2D culture conditions in the presence of serum compared to all other groups at p < 0.01 and p < 0.001 respectively. (d) The proliferative activity of hMSCs was evaluated by the gene expression of PCNA. The symbol (‡) denotes a significant difference in the PCNA mRNA expression at day 1 under 2D culture conditions in the presence of serum compared to all other groups at p < 0.05 (n=3-4; mean ± standard deviation).
Influence of serum on human mesenchymal stem cell attachment within PEGDA superporous hydrogels
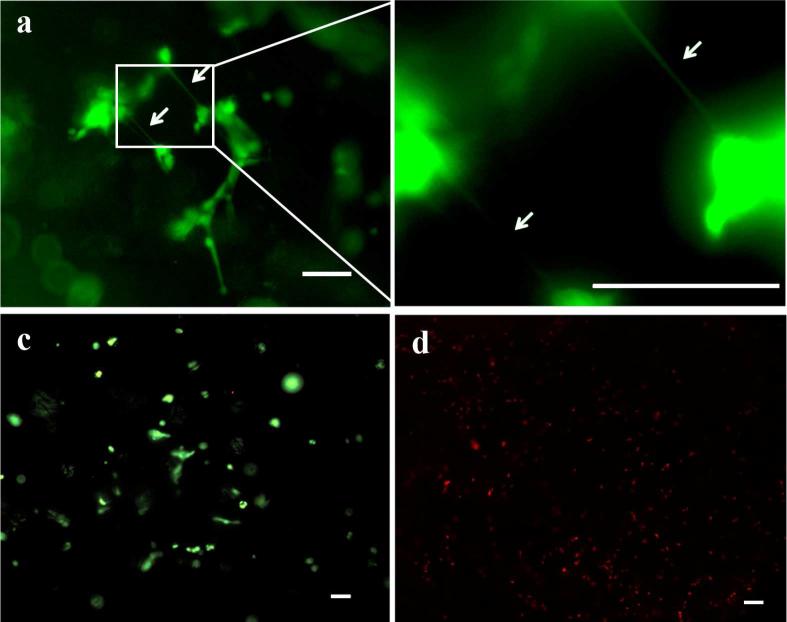
In order to delineate the role of serum proteins in aiding hMSC attachment and survival within PEGDA SPHs, hMSCs were cultured in complete medium and serum-free medium. HMSCs seeded in SPHs and cultured in complete DMEM were viable and displayed an extended morphology (Figure 2a-b). Within 24 hours of seeding, the cells were extending between clusters suggesting cell-cell interactions. When the cell-seeded constructs were transferred to serum free culture for the next 24 hours, the hMSCs retained their viability (Figure 2c). In contrast, when the cell-seeded constructs were directly cultured in serum-free conditions, no viable cells could be found within the SPHs at the end of 24 hours (Figure 2d). These results indicate that the presence of serum proteins is required for initial anchorage of hMSCs within the SPHs but not for the hMSC survival after 24 hours.
Figure 2. Serum proteins are required for initial anchorage of human mesenchymal stem cells to PEGDA superporous hydrogels.
Representative live (green)-dead (red) pseudocolor stained images for hMSCs seeded within PEGDA SPHs and cultured in complete DMEM for 24 hours. (a) Cells with extended morphology toward nearby cells as marked by arrows could be observed. (b) Magnified region from (a) to show the cell-cell projections where the arrows are in approximately the same location. (c) When the culture was deprived of serum for the following 24 hours (total 48 hour culture), the cells remained viable. (d) However, when the hMSCs were seeded in PEGDA SPHs and cultured directly in serum-free medium for 24 hours, the cells did not survive (red staining), as confirmed by the absence of live cells (no green) within the live-dead staining image. The scale bar is 100 μm in each image.
Matrix production within superporous hydrogels
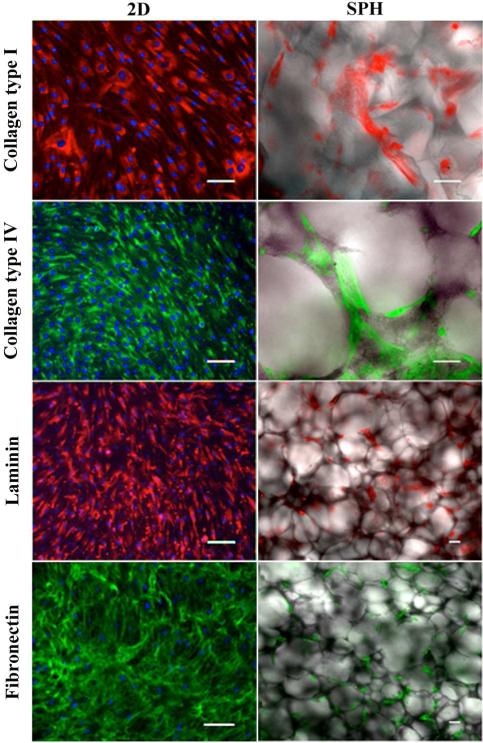
The deposition of ECM molecules by hMSCs cultured within PEGDA SPHs and on 2D tissue culture plastic was examined by immunofluorescence staining (Figure 3) and real-time qPCR (Figure 4). At the end of 3 weeks, hMSCs were embedded in a dense matrix of collagen type I, collagen type IV, laminin, and fibronectin in both culture systems (Figure 3). In 2D culture, the cell-deposited ECM was localized around the nuclei and displayed a fibrous pattern. In the PEGDA SPHs, hMSC-secreted ECM molecules were mainly localized around the porous network with similar fibrillar structures (Figure 3). Since assessment of the protein expression with immunofluorescence staining in the SPH is not quantitative, we conducted real-time qPCR to quantify gene expression of ECM molecules.
Figure 3. Human mesenchymal stem cells synthesize their own extracellular matrix proteins when cultured within PEGDA superporous hydrogels.
Representative pseudocolored immunohistofluorescence micrographs of collagen type I, collagen type IV, fibronectin and laminin expressed by hMSCs cultured within SPHs and on tissue culture plastic (TCP) for 21 days. The expression of the four ECM proteins was positive in both culture systems. H33258 (blue) was used as a nuclear stain in the monolayer group. Due to strong background staining no H33258 staining was conducted in the SPH group. Scale bar is 100 μm in each image.
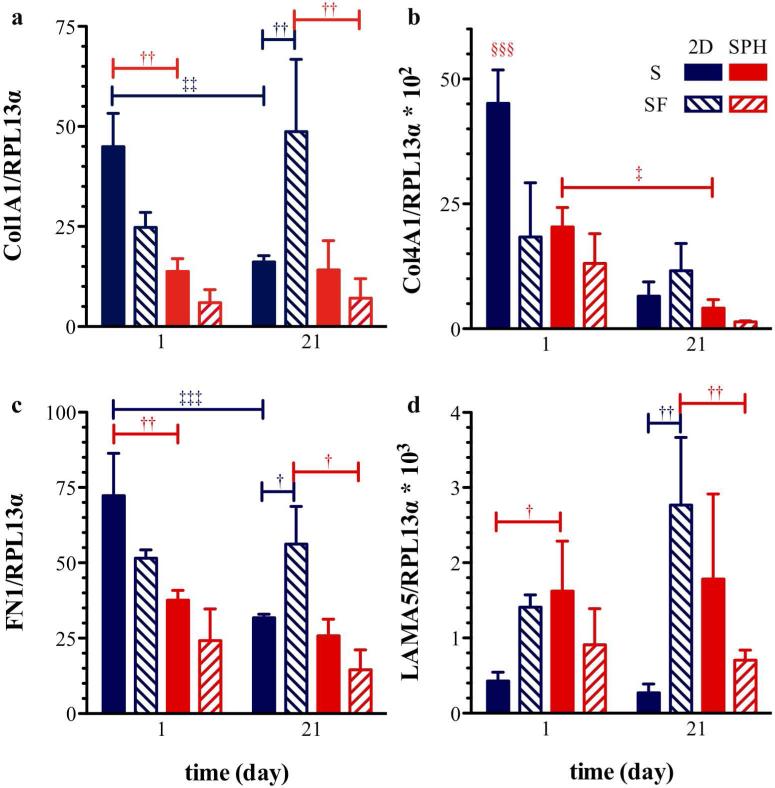
Figure 4. Expression profiles of genes encoding for extracellular matrix proteins.
Expression of (a) collagen type I, (b) collagen type IV, (c) fibronectin, and (d) laminin in hMSCs cultured for 1 and 21 days within PEGDA superporous hydrogels (SPH/red) and on tissue culture plastic as a monolayer (2D/blue) under serum-containing (S/solid bars) and serum-free conditions (SF/diagonal lines within bars); mRNA levels were normalized to the expression of the endogenous control ribosomal protein L13 α (RPL13α). Collagen 1A1 (Col1A1), collagen 4A1 (Col4A1) and fibronectin (FN1) expression was similar to 2D monolayer within the PEGDA SPH system after 21 days of culture in serum-containing basal medium. Values are presented as mean ± standard deviation (n=3-4). Statistical significance is indicated in the figure as † for differences within a specific day, ‡ for differences between day 1 and 21, and § for difference from all other groups on day 1 and the number of symbols indicating level of significance with one, two and three symbols indicating p < 0.05, p < 0.01, and p < 0.001, respectively.
Collagen type I, collagen type IV and fibronectin mRNA expression levels of hMSCs seeded within PEGDA SPHs were similar to the expression levels of hMSCs expanded in 2D monolayer for 21 days in serum-containing basal medium. At the earliest time point, laminin expression was significantly upregulated (p = 0.03) for hMSCs cultured in SPHs compared to on TCP in serum-supplemented media and slightly upregulated, but not statistically different (p= 0.08), in serum-supplemented media at day 21.
Interestingly, the absence of serum did not have any significant effects on the gene expression of ECM molecules within the SPH culture system. As well, the mRNA levels of ECM molecules remained relatively constant over the 21 days of culture within the PEGDA SPHs. Whereas the mRNA levels of collagen type I, collagen type IV and fibronectin decreased from day 1 to day 21 in 2D monolayer in the presence of serum (0.001 < p < 0.05). The decrease in ECM molecule gene expression can be explained by the fact that hMSCs grew confluent within 21 days of culture on the 2D plate and do not need to synthesize more ECM for cell attachment. Similar to SPHs, the hMSCs did not proliferate significantly under 2D serum-free conditions (Figure 1c) and the gene expression did not change significantly from day 1 to day 21.
Flow Cytometry
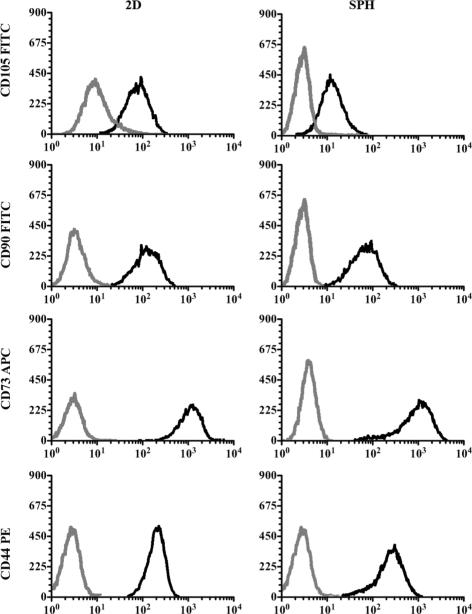
Flow cytometry was employed to assess the surface marker profiles of passage 4 hMSCs cultured for 21 days within SPHs or on 2D TCP (Figure 5). The mean percent expression of CD105 was below 80% whereas CD90, CD73 and CD44 expression levels were over 95% in both culture systems (Table 3). Since the mesenchymal surface marker expression was not significantly altered after the transfer to the 3D system, we conclude that PEGDA SPH culture supports the undifferentiated state of hMSCs and eventually preserves their tri lineage differentiation potential. Flow cytometry of hMSCs from passage 2 revealed similar CD105 expression levels of 78.4% ± 1.2%. Comparable results for the CD105 expression (88.1% ± 7.4%) within unsorted bone marrow derived hMSCs have been reported.26
Figure 5. Expression of human mesenchymal stem cell surface markers of cells cultured within PEGDA superporous hydrogels or on tissue culture plastic (TCP) for 21 days.
Representative flow cytometry histograms from three independent experiments with three different donors are shown. Analysis revealed no statistically significant differences in the expression of CD105, CD90, CD73 and CD44 in hMSCs cultured within SPHs (right) or in 2D (left). Gray lines represent the fluorochrome- and isotype-matched control and black lines the corresponding CD marker-specific antibody.
Table 3.
Flow cytometry comparison of hMSC surface marker expression of cells cultured within SPHs or on 2D monolayer for 21 days.
| Surface marker | 2D (%)† | SPH (%)† |
|---|---|---|
| CD105 | 77.5 ± 5.0 | 72.5 ± 6.9 |
| CD90 | 99.2 ± 1.0 | 97.5 ± 2.6 |
| CD73 | 99.2 ± 0.8 | 98.2 ± 2.1 |
| CD44 | 99.5 ± 0.3 | 98.8 ± 0.3 |
The values listed in the table are presented as the mean values of 3 different donors expressing the indicated cell surface protein ± standard deviation. There is statistical difference between the CD markers (p < 0.001), but no difference between the 2D and SPH samples (p = 0.14).
Multipotency of human mesenchymal stem cells within superporous hydrogels
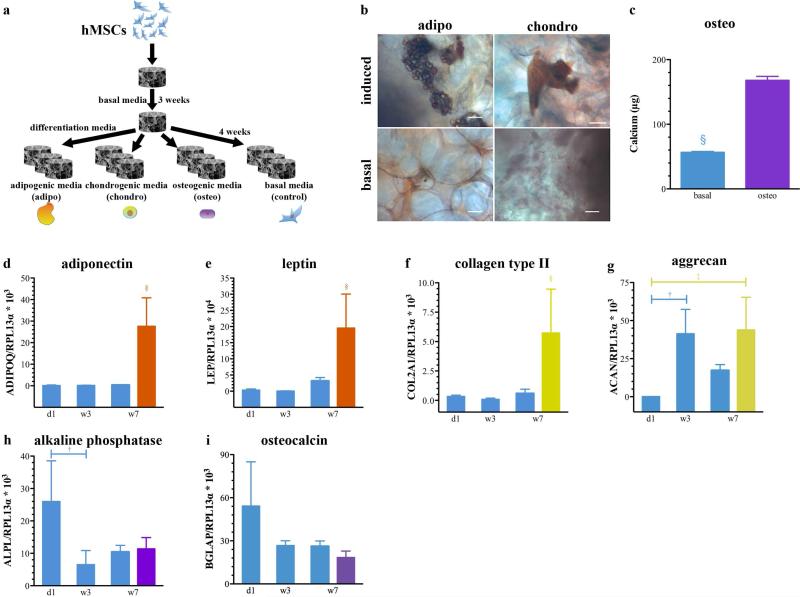
To examine whether hMSCs are able to retain their multi-lineage differentiation potential after cultivation in the SPHs, we cultured the cell-seeded constructs (2.5 × 105 cells/SPH) for 3 weeks in basal medium, followed by treatment with adipogenic, chondrogenic and osteogenic differentiation medium for a period of 4 weeks. Controls were maintained in basal medium for the entire culture time of 7 weeks (Figure 6a). Lipid vacuoles that stained bright red with the lipophilic dye, Sudan III, could be observed within PEGDA SPHs upon exposure to adipogenic medium. No lipid vacuoles could be detected within the non-induced controls (Figure 6b). The presence of proteoglycans was confirmed by Safranin O staining and could only be detected in the chondrogenic induction group (Figure 6b). Cell-seeded constructs cultured in osteogenic medium showed significantly elevated calcium levels compared to the non induced control groups (Figure 6c).
Figure 6. Human mesenchymal stem cells retain their multi-lineage differentiation capability within superporous hydrogels.
(a) Schematic of treatment regimen for cell-seeded constructs cultured for 3 weeks in basal medium and additional 4 weeks in adipogenic, chondrogenic, and osteogenic differentiation medium. Controls were maintained in basal medium for the entire culture time of 7 weeks. (b) Upon adipogenic induction, lipid vacuoles stained red with Sudan III could be observed throughout the SPH. No lipid vacuoles could be detected within the non-induced controls. (b) The induction of chondrogenesis was confirmed by the presence of proteoglycans (red) visualized by a Safranin O stain. No positive staining for proteoglycans could be detected within the basal group. Scale bars are 100 μm. (c) Osteogenic differentiation of hMSCs within SPHs was evaluated by assaying the calcium content within the mineralized matrix. Relative expression of fat-related genes (d-e), cartilage-related genes (f-g) and bone-related genes (h-i) in hMSCs cultured within PEGDA SPHs for 1 day (d1), 3 weeks (w3) and 7 weeks (w7) under basal (blue) culture conditions or for 3 weeks under basal conditions with additional 4 weeks in adipogenic (w7 orange), chondrogenic (w7 yellow) and osteogenic (w7 purple) induction medium. Statistical significance is indicated in the figure as † for differences within the basal groups, ‡ for differences between basal and differentiation media, and § for difference from all other groups at p < 0.05. Values are presented as mean ± standard deviation (n=3, 3 donors).
The relative expression of fat-related genes, adiponectin (ADIPOQ) and leptin (LEP) (Figure 6d-e), cartilage-related genes, collagen type II (COL2A1) and aggrecan (ACAN) (Figure 6f-g), and bone-related genes, alkaline phosphatase (ALPL) and osteocalcin (BGLAP) (Figure 6h-i) was examined by real-time qPCR. The gene expression of adiponectin, leptin and collagen type II exhibited similar patterns: significant higher mRNA levels in the induction groups at week 7 compared to the control groups at day 1, week 3 and week 7. Aggrecan mRNA expression levels were significantly elevated after 3 and 7 weeks of SPH culture compared to day 1 but no significant differences between the control groups and the chondrogenic induction group was observed for week 7. The expression of bone markers, alkaline phosphatase and osteocalcin, showed a decreasing trend over the 7 weeks of SPH culture and no significant differences between the control groups and the osteogenic induction group was seen which contradicts the data observed for calcification (Figure 6c).
Discussion
The goal of this study was to investigate cell-matrix and cell-polymer interactions of human mesenchymal stem cells (hMSCs) cultured within PEGDA superporous hydrogels (SPHs). Since PEG surfaces are thought to be devoid of cell-matrix interactions, we hypothesized that the physicochemical characteristics of the SPH and its macroporous architecture affect cellular responses. Our results demonstrated that macroporous PEGDA hydrogels provide a microenvironment that supports stem cell-derived extracellular matrix (ECM) development. The secreted ECM together with the hydrogel surrounding promoted stem cell maintenance and multi-lineage differentiation.
Over a period of 7 weeks viable cells were detected throughout the PEGDA SPHs (Figure 1). The majority of hMSCs exhibited a spindle-shaped, fibroblast-like morphology. For hMSCs, a thin stellate or spindle-shaped morphology is suggestive of an undifferentiated state.27 In contrast, when encapsulated within nonporous 3D PEGDA hydrogels that are not modified with RGD, hMSCs are forced into a rounded morphology with limited capability to spread, and consequently undergo apoptosis.8, 28 In addition, hMSCs do not survive on top of unmodified, nonporous PEGDA hydrogels, which could serve as a control for the porous architecture of the SPH with similar surface chemistry.6 Thus, it is proposed that the interconnected porous architecture of the PEGDA SPHs promotes long-term survival of hMSCs. Furthermore, there are also chemical and physical components to the ability to populate the scaffolds and create a niche as hMSCs grown in SPHs with similar mechanical properties and morphology were not able to survive for even 24 hours without incorporation of collagen.11 SPHs containing acrylic acid in their formulation were “repulsive” to cell attachment indicating significantly altered protein and cell interactions with the hydrogel.
It is suggested that 3D scaffold culture turns cell proliferation down.29 Duggal et al. showed that hMSCs encapsulated within 2% RGD-alginate gels did not proliferate although they remained viable over the 21 days study period.29 Gene expression of PCNA, encoding for proliferating cell nuclear antigen that is expressed in the nuclei of cells during DNA synthesis, remained constant in hMSCs cultured for 3 weeks within PEGDA SPHs in the presence and absence of serum (Figure 1c). The slow proliferation rate of hMSCs in SPHs compared to their extensive growth under 2D conditions is actually more similar to their in vivo behavior where adult stem cells produce progeny at lower dividing capacity.30
Whereas serum is needed for hMSC expansion under 2D conditions on tissue culture plastic (TCP) (Figure 1c), hMSCs cultured within SPHs proliferate similarly in the presence or absence of serum in the basal medium (Figure 1c), which could have the additional benefit for human cells of avoiding the use of animal serum for culture. We hypothesized that the cells must be secreting their own ECM within the SPHs that may control cell adhesion, proliferation and differentiation.
Cell adhesion to polymer surfaces is mediated by proteins that have been immobilized onto the substrate, adsorbed from the surrounding medium or secreted by the cells themselves. Initial attachment of hMSCs to PEGDA SPHs is mediated by serum proteins (Figure 2) that have been adsorbed onto the scaffold surface or through serum protein mediated cell-cell interactions that allow ECM deposition. HMSCs cultured in serum containing medium are able to anchor to the SPHs and thus overcome initial pro apoptotic signals that lead to anoikis of anchorage-dependent cells.13, 14 However, this effect was not seen when the hMSCs were cultured on the nonporous hydrogels. Besides initial anchorage dependent upon serum proteins, integrin-mediated signaling via attachment to ECM is required to maintain cell viability.12
Undifferentiated hMSCs secrete various ECM proteins that are part of the specialized niche that controls stem cell behavior.31 Immunofluorescence staining revealed expression of collagen type I, collagen type IV, fibronectin and laminin within hMSC-seeded PEGDA SPHs by the end of three weeks (Figure 3). No fluorescence signal could be detected in acellular PEGDA SPHs cultured for 3 weeks in basal media indicating that insufficient ECM ligands have been adsorbed from the serum in the culture media (data not shown) suggesting that the detected ECM molecules originate from the cells within the scaffold. Endothelial cells seeded onto RGD-modified PEGDA hydrogel surfaces were able to attach and secrete ECM even after the RGD recognition sequences had been hydrolyzed from the system.32 The cell sheet did not detach from the hydrogels indicating that, after initial attachment due to the RGD ligands, the cells had secreted ECM that was interacting strongly with the PEGDA surface.32 Rather than direct attachment to the entangled PEGDA chains, it is hypothesized that, the secreted fibronectin filaments and collagen fibrils diffuse within the polymer chains and form an inter-penetrating network with the PEGDA hydrogels, which have a mesh size on the order of 6 nm.33 Although the staining clearly showed ECM, it is unclear how this ECM interacts with the PEGDA hydrogels if at all.
To monitor ECM development, we examined gene expression of hMSCs cultured within PEGDA SPHs and on 2D surfaces. Collagen type I, collagen type IV and fibronectin mRNA expression levels of hMSCs seeded within PEGDA SPHs were similar to the expression levels of hMSCs expanded in 2D monolayers for 21 days in basal medium (Figure 4a-c). Laminin (LAMA5) expression was slightly upregulated in the SPH at day 1 compared to 2D TCP (p=0.03) but no significant difference was observed at day 21 (p=0.08) (Figure 4d). Interestingly, the absence of serum did not have any significant effects on the gene expression of ECM molecules within the SPH culture system. We believe that the cell-secreted ECM supports long-term survival of hMSCs within the PEGDA SPHs and aids in creating a niche environment conducive to stem cell maintenance and multi-lineage differentiation after induction with appropriate cues. The deviation in the ECM expression between 2D and SPH culture may arise from the fact that cells sense different architectural and topographical cues. However, alterations in surface chemistry, polystyrene tissue culture plastic versus poly(ethylene glycol) diacrylate hydrogel substrata, might have also played a role. As stated earlier, we are unable to grow cells directly on 2D PEGDA hydrogels, so comparison is not possible. But, it is clear that there are differences in the amount of mRNA being produced for each of the proteins examined, with laminin in particular being elevated in the SPH cultured hMSCs.
Laminin, known for its role in embryogenesis, has been shown to support human embryonic stem cells (hESCs) proliferation and stem cell maintenance.34 In adhesion assays with ECM protein-coated tissue culture plates, more than 60% of the hMSCs bind to fibronectin, collagen type I, and collagen type IV, and about 30% bind to laminin.16 Laminin is bound by several integrin receptors, all of which are found in hMSCs, suggesting that laminin might play an important role in adhesion of hMSCs to PEGDA SPHs and their end fate. Integrin binding and expression relates to differential expression of ECM molecules35 and the differential binding of serum proteins in the SPH is expected to result in the downstream ECM component differences and possibly different populations of hMSCs being retained.
The cell-secreted ECM resembled the composition of marrow-derived ECM. It has been shown that marrow stem cell-derived ECM restrains osteoblast differentiation and promotes increased replication of multipotent colony forming units (MCFUs).31 The ECM regulates the balance between stem cell replication and differentiation in response to appropriate stimuli. After three weeks of culture within PEGDA SPHs, hMSCs expressed CD105, CD90, CD73 and CD44 to a similar level than their counterparts grown on 2D TCP suggesting that the cells retained their multi-lineage differentiation potential (Figure 5). MSCs are defined by the expression of those cell surface markers together with the ability to differentiate into downstream lineages such as adipocytes, chondrocytes and osteoblasts.4, 36 The CD105 expression is lost when hMSCs start differentiating towards lineages of the mesenchyme. The expression of CD105 in umbilical cord blood derived hMSCs was significantly decreased in differentiated osteoblasts, chondrocytes and adipocytes from 99.4% ± 0.1% to 3.5% ± 1.4%, 3.5% ± 2.3% and 16.7% ± 3.6%, respectively.26 It must be noted, that there are currently no cell surface markers that are specifically and uniquely characteristic of hMSCs, for example CD105 is also associated with vascular endothelial cells.37
Variations in culture methods and differentiation stage of cells can lead to diverging results in surface marker characteristics among the reported studies. For each of the donors, similar marker distribution was identified in the SPHs, suggested that cell populations obtained from within the hydrogels were consistent between experiments. We have done preliminary studies to determine that the cells recovered were as representative as possible, but the possibility that some cells are retained and that these may be one subpopulation can not be ignored. In our experiments CD105 expression within unsorted bone marrow derived hMSCs was about 78.4% ± 1.2% in passage 2 cells. We used passage 4 cells for comparing the surface marker expression of hMSCs cultured within PEGDA SPHs and on 2D TCP for 21 days. Since there was no significant difference in the CD105 expression between 3D scaffold (72.5% ± 6.9%) and 2D monolayer culture (77.5% ± 5%), it suggested that the hMSCs did not undergo substantial differentiation within the PEGDA SPHs and maintain their multi-lineage potential, but it is imperative to show this multipotent differentiation.
The true assessment of the multipotent state is the ability of the hMSCs to differentiate into specific cell types.4, 6, 36 The multi-lineage differentiation capability of hMSCs seeded within PEGDA SPHs was confirmed by their ability to express adipogenic, chondrogenic and osteogenic markers (Figure 6). Although there was no significant increase in the gene expression of osteogenic markers (Figure 6h,i), we observed significantly increased Ca2+-levels after osteogenic induction. The discrepancies between mineralization results and the expression of osteogenic genes have been shown before on 2D TCP.38 Since there is no clear evidence that all cells differentiate and future investigations into the efficiency of differentiation are warranted. The unaltered expression of stem cell surface markers along with the multi-lineage differentiation capability confirms that the cells remain multipotent mesenchymal stem cells within PEGDA SPHs under long-term culture conditions suggesting that the SPH has appropriate chemical (protein absorption) and physical characteristics for stem cell growth and maintenance.
The MSC niche in vivo, the bone marrow31, is a soft tissue that promotes stem cell maintenance. PEGDA hydrogels resemble soft tissues in hydrophilicity and generally in mechanics. Preliminary data suggests a compressive modulus of about 0.15 MPa. We believe that SPH structure, chemistry, and mechanics along with the specific microenvironment created by the stem cells themselves may provide biophysical cues that enable hMSCs to retain their multilineage potency within the SPH. We are the first ones that demonstrated the presence and the development of cell matrix interactions within unmodified macroporous PEGDA hydrogels.
Conclusions
Tissues are constructed of cells that are embedded in a dense ECM. Thus, a scaffold that enables ECM secretion recreates a major component of the native environment of cells in vitro. Within the PEGDA SPHs, initial hMSCs attachment is serum-dependent while survival is serum independent. Additionally, hMSCs secrete ECM that provides environmental cues for cell survival, proliferation and differentiation. In addition to enabling stem cell viability, the PEGDA SPHs supported MSC survival in an undifferentiated state while retaining the multi-lineage potential of the cells. Thus, the PEGDA SPH provides an opportunity for three-dimensional culture of viable and functional hMSCs under controlled conditions for stem cell maintenance and differentiation. These scaffolds have great potential not only in regenerative medicine but also as model 3-D systems for studying cell behavior in response to various stimuli.
Acknowledgments
This investigation was funded in part by the Bundesministerium für Bildung und Forschung (TH) and the National Institutes of Health through grants NS055095 (RAG) and HL090523 (BR). The authors thank Ernie Gemeinhart, Dr. Howard Greissler, members of the laboratory of Dr. Tonetti, and the UIC-RRC Flow Cytometry Facility for significant and helpful discussion. This investigation was conducted, in part, in a facility constructed with support from Research Facilities Improvement Program Grant C06 RR15482 from the National Center for Research Resources, NIH.
REFERENCES
- 1.Kuhn NZ, Tuan RS. J. Cell. Physiol. 2010;222:268–277. doi: 10.1002/jcp.21940. [DOI] [PubMed] [Google Scholar]
- 2.Djouad F, Bouffi C, Ghannam S, Noel D, Jorgensen C. Nat. Rev. Rheumatol. 2009;5:392–399. doi: 10.1038/nrrheum.2009.104. [DOI] [PubMed] [Google Scholar]
- 3.Caplan AI. J. Pathol. 2009;217:318–324. doi: 10.1002/path.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 5.Caplan AI, Dennis JE. J. Cell. Biochem. 2006;98:1076–1084. doi: 10.1002/jcb.20886. [DOI] [PubMed] [Google Scholar]
- 6.Keskar V, Marion NW, Mao JJ, Gemeinhart RA. Tissue Eng., Part A. 2009;15A:1695–1707. doi: 10.1089/ten.tea.2008.0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keskar V, Gandhi M, Gemeinhart E, Gemeinhart R. J. Tissue Eng. Regen. Med. 2009;3:486–490. doi: 10.1002/term.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nuttelman CR, Tripodi MC, Anseth KS. Matrix Biol. 2005;24:208–218. doi: 10.1016/j.matbio.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 9.Desai ES, Tang MY, Ross AE, Gemeinhart RA. Biomed. Mater. 2012:7. doi: 10.1088/1748-6041/7/2/024108. in press, DOI: 10.1088/1748-6041/1087/1082/024108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Betz MW, Yeatts AB, Richbourg WJ, Caccamese JF, Coletti DP, Falco EE, Fisher JP. Biomacromolecules. 2010;11:1160–1168. doi: 10.1021/bm100061z. [DOI] [PubMed] [Google Scholar]
- 11.Kadakia A, Keskar V, Titushkin I, Djalilian A, Gemeinhart RA, Cho MR. Crit. Rev. Biomed. Eng. 2008;36:441–471. doi: 10.1615/critrevbiomedeng.v36.i5-6.50. [DOI] [PubMed] [Google Scholar]
- 12.Frisch SM, Vuori K, Ruoslahti E, Chan-Hui PY. J. Cell Biol. 1996;134:793–799. doi: 10.1083/jcb.134.3.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiarugi P, Giannoni E. Biochem. Pharmacol. 2008;76:1352–1364. doi: 10.1016/j.bcp.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 14.Frisch SM, Francis H. J. Cell Biol. 1994;124:619–626. doi: 10.1083/jcb.124.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chichester CO, Fernandez M, Minguell JJ. Cell Adhes. Commun. 1993;1:93–99. doi: 10.3109/15419069309095685. [DOI] [PubMed] [Google Scholar]
- 16.Conget PA, Minguell JJ. J. Cell. Physiol. 1999;181:67–73. doi: 10.1002/(SICI)1097-4652(199910)181:1<67::AID-JCP7>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 17.Prockop DJ. Science. 1997;276:71–74. doi: 10.1126/science.276.5309.71. [DOI] [PubMed] [Google Scholar]
- 18.Matsubara T, Tsutsumi S, Pan H, Hiraoka H, Oda R, Nishimura M, Kawaguchi H, Nakamura K, Kato Y. Biochem. Biophys. Res. Commun. 2004;313:503–508. doi: 10.1016/j.bbrc.2003.11.143. [DOI] [PubMed] [Google Scholar]
- 19.Schmittgen TD, Livak K. J. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 20.Curtis KM, Gomez LA, Rios C, Garbayo E, Raval AP, Perez Pinzon MA, Schiller PC. BMC Mol. Biol. 2010;11:61. doi: 10.1186/1471-2199-11-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stockert J, Adhikary T, Kaddatz K, Finkernagel F, Meissner W, Muller Brusselbach S, Muller R. Nucleic Acids Res. 2011;39:119–131. doi: 10.1093/nar/gkq773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pattyn F, Robbrecht P, De Paepe A, Speleman F, Vandesompele J. Nucleic Acids Res. 2006;34:D684–D688. doi: 10.1093/nar/gkj155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Landeghem L, Mahe MM, Teusan R, Leger J, Guisle I, Houlgatte R, Neunlist M. BMC Genomics. 2009;10:507. doi: 10.1186/1471-2164-10-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Donzelli E, Lucchini C, Ballarini E, Scuteri A, Carini F, Tredici G, Miloso MJ. Mol. Cell. Biol. 2011;3:123–131. doi: 10.1093/jmcb/mjq050. [DOI] [PubMed] [Google Scholar]
- 25.Woo J, Cannon DC. Metabolic Intermediates and Inorganic Ions. In: Henry JB, McPherson RA, editors. Clinical Diagnosis and Management by Laboratory Methods. 17th ed. Philadelphia, PA; W. B. Saunders: 1991. pp. 140–143. [Google Scholar]
- 26.Kern S, Eichler H, Stoeve J, Kluter H, Bieback K. Stem Cells. 2006;24:1294–1301. doi: 10.1634/stemcells.2005-0342. [DOI] [PubMed] [Google Scholar]
- 27.Sekiya I, Larson BL, Smith JR, Pochampally R, Cui JG, Prockop DJ. Stem Cells. 2002;20:530–541. doi: 10.1634/stemcells.20-6-530. [DOI] [PubMed] [Google Scholar]
- 28.Pierschbacher MD, Ruoslahti E. J. Biol. Chem. 1987;262:17294–17298. [PubMed] [Google Scholar]
- 29.Duggal S, Fronsdal KB, Szoke K, Shahdadfar A, Melvik JE, Brinchmann JE. Tissue Eng., Part A. 2009;15:1763–1773. doi: 10.1089/ten.tea.2008.0306. [DOI] [PubMed] [Google Scholar]
- 30.Scadden DT. Nature. 2006;441:1075–1079. doi: 10.1038/nature04957. [DOI] [PubMed] [Google Scholar]
- 31.Chen XD, Dusevich V, Feng JQ, Manolagas SC, Jilka RLJ. Bone Miner. Res. 2007;22:1943–1956. doi: 10.1359/jbmr.070725. [DOI] [PubMed] [Google Scholar]
- 32.Elbert DL, Hubbell JA. Biomacromolecules. 2001;2:430–441. doi: 10.1021/bm0056299. [DOI] [PubMed] [Google Scholar]
- 33.King PJ, Bryant T, Minas TJ. Knee Surg. 2002;15:177–184. [PubMed] [Google Scholar]
- 34.Miyazaki T, Futaki S, Hasegawa K, Kawasaki M, Sanzen N, Hayashi M, Kawase E, Sekiguchi K, Nakatsuji N, Suemori H. Biochem. Biophys. Res. Commun. 2008;375:27–32. doi: 10.1016/j.bbrc.2008.07.111. [DOI] [PubMed] [Google Scholar]
- 35.Grayson WL, Ma T, Bunnell B. Biotechnol. Prog. 2004;20:905–912. doi: 10.1021/bp034296z. [DOI] [PubMed] [Google Scholar]
- 36.Jones EA, Kinsey SE, English A, Jones RA, Straszynski L, Meredith DM, Markham AF, Jack A, Emery P, McGonagle D. Arthritis Rheum. 2002;46:3349–3360. doi: 10.1002/art.10696. [DOI] [PubMed] [Google Scholar]
- 37.Cheifetz S, Bellon T, Cales C, Vera S, Bernabeu C, Massague J, Letarte M. J. Biol. Chem. 1992;267:19027–19030. [PubMed] [Google Scholar]
- 38.Shafiee A, Seyedjafari E, Soleimani M, Ahmadbeigi N, Dinarvand P, Ghaemi N. Biotechnol. Lett. 2011;33:1257–1264. doi: 10.1007/s10529-011-0541-8. [DOI] [PubMed] [Google Scholar]