Abstract
Cell proliferation is crucial to tissue growth and form during embryogenesis, yet dynamic tracking of cell cycle progression and cell position present a challenging roadblock. We have developed a fluorescent cell cycle indicator and single cell analysis method, called CycleTrak, which allow better spatiotemporal resolution and quantification of cell cycle phase and cell position than current methods. Our method was developed on the basis of the existing FUCCI method. CycleTrak uses a single lentiviral vector that integrates mKO2-hCdt1 (30/120), and a nuclear-localized eGFP reporter. The single vector and nuclear localized fluorescence signals simplify delivery into cells and allow for rapid, automated cell tracking and cell cycle phase readout in single and subpopulations of cells. We validated CycleTrak performance in metastatic melanoma cells and identified novel cell cycle dynamics in vitro and in vivo after transplantation and 3D confocal time-lapse imaging in a living chick embryo.
Keywords: cell cycle, cell tracking, CycleTrak, fluorescence, embryo, melanoma
INTRODUCTION
The cell cycle consists of 4 phases (G1, S, G2, M) during which a cell goes through periods of growth, chromosome replication and mitosis. Intrinsic properties of the cell and signals from the local microenvironment are thought to influence when the cell enters each phase of the cycle (Ho and Dowdy, 2002). The inability of a cell to correctly regulate its entry into different phases of the cell cycle increases genomic instability and may lead to programmed cell death or cancer (Nurse, 2000). Dynamic tracking of cell cycle progression and cell position present a challenging roadblock. The first step in determining the cell cycle profile of a cell is the accurate classification of each of its phases. Second, measuring the dynamics of cell cycle profile and linking this to cell position requires the spatiotemporal readout of cell cycle events. Thus, the development of tools to accurately determine cell cycle phase and cell position would offer a powerful approach and may significantly impact studies of cell cycle regulation during events in embryo and adult morphogenesis, tissue regeneration, and cancer.
Cell cycle phase analysis is typically limited to static, fixed-tissue methods such as 5-bromo-2'-deoxyuridine (BrdU) or thymidine incorporation, and staining for cell cycle markers such as Ki67, PCNA, or phopho-H3. These methods provide a snapshot of the cell cycle at specific time points, but cannot directly measure cell cycle length or provide dynamic, temporal information. The recent design of a Fluorescent, Ubiquitin based, Cell Cycle Indicator system (Fucci) as a dual construct, fluorescent reporter system allows for the visualization of the spatiotemporal dynamics of cell cycle progression (Sakaue-Sawano et al., 2008a). The Fucci system has two components: mAG-hGem (1/110), which produces a green fluorescent signal during G2/S (and very little in M) and mKO2-hCdt1 (30/120) which produces an orange fluorescent signal during G1. Because each protein in the Fucci system is rapidly degraded after their respective phase of the cell cycle is complete, decreasing fluorescent intensities of mAG-hGem (1/110) and mKO2-hCdt1 (30/120) mark the transitions between particular cell cycle phases.
Since its original design, modifications to Fucci have included altering the fluorescent proteins attached to hGem or hCdt1, localizing the hCdt1-dependent reporter to the cytoplasm rather than to the nucleus, and designing Fucci reporters based on the zebrafish versions of Cdt1 and Geminin (Sakaue-Sawano et al., 2008b; Sugiyama et al., 2009). However, none of the Fucci designs provide a fluorescent readout through the late M/early G1 phases or describe a quantitative framework that would allow for automated cell cycle measurements. Also, the system may be cumbersome to apply since it requires the introduction of two separate constructs. These limitations of Fucci may lead to inefficient labeling of all cells in culture or in an embryo, and an inability to both track cell position during mitosis and to measure cell cycle phase progression from time-lapse imaging data.
To overcome these limitations, we have developed CycleTrak, a novel, single-plasmid system that provides a fluorescent readout of G1 and continuous tracking of cell position through all the phases of the cell cycle. As part of the CycleTrak system, we also developed semi-automated analysis tools to interrogate dynamic information on cell cycle progression from time-lapse images and to more accurately determine cell cycle progression with single cell precision. To validate our system, we used CycleTrak to analyze cell cycle dynamics in both HeLa cells and a highly aggressive metastatic melanoma cell line (Welch et al., 1991). We found that CycleTrak provided the same dynamic cell cycle phase resolution as Fucci, and uniquely allows the use of automated cell tracking programs to follow cell position and cell cycle progression in fluorescently labeled cells over time. Also, the semi-automated quantitative analysis tools of CycleTrak allowed us to track cell cycle dynamics in both individual and populations of cells simultaneously. These features are unique to CycleTrak and, particularly in an embryonic environment, provide distinct advantages over cell cycle analysis using Fucci.
MATERIALS AND METHODS
Cell Culture
HeLa.S-Fucci are a variant of the human cervical carcinoma cell line HeLa.S that express Fucci-Orange-G1 and Fucci-Green-S/G2/M (Sakaue-Sawano et al., 2008a), and were kindly provided by the Cell Bank of the RIKEN BioResource Center (cat. #RCB2812). HeLa.S-Fucci cells were grown in DMEM with 10% fetal bovine serum (FBS) at 37°C in 5% CO2.
C8161 cells, a highly metastatic melanoma, were a kind gift from Mary Hendrix (Children’s Memorial Research Center, Northwestern University) and were grown in RPMI with 10% FBS.
Construction and Confirmation of Single-Vector Fucci Reporters
To allow for simultaneous expression of both Fucci proteins, Fucci-Orange-G1 and Fucci-Green-S/G2/M were PCR amplified, linked via the porcine teschovirus-1 2A peptide sequence (P2A) (Szymczak-Workman et al., 2007) and cloned into the EcoRI and XbaI sites of pcDNA3.1(+) (Invitrogen). In order to allow for automated tracking of cells through time, H2B-eGFP was PCR amplified and inserted into the BspEI and XbaI sites of the dual Fucci vector to generate a plasmid expressing both Fucci-Orange-G1 and a nuclear-localized H2B-eGFP (Fig. 1b). For infection of the C8161 cells, we transferred the Fucci-Orange-G1/2A/H2B-eGFP cassette into the PstI and Acc65I restriction sites of pLenti6 (Invitrogen) downstream of the PGK promoter.
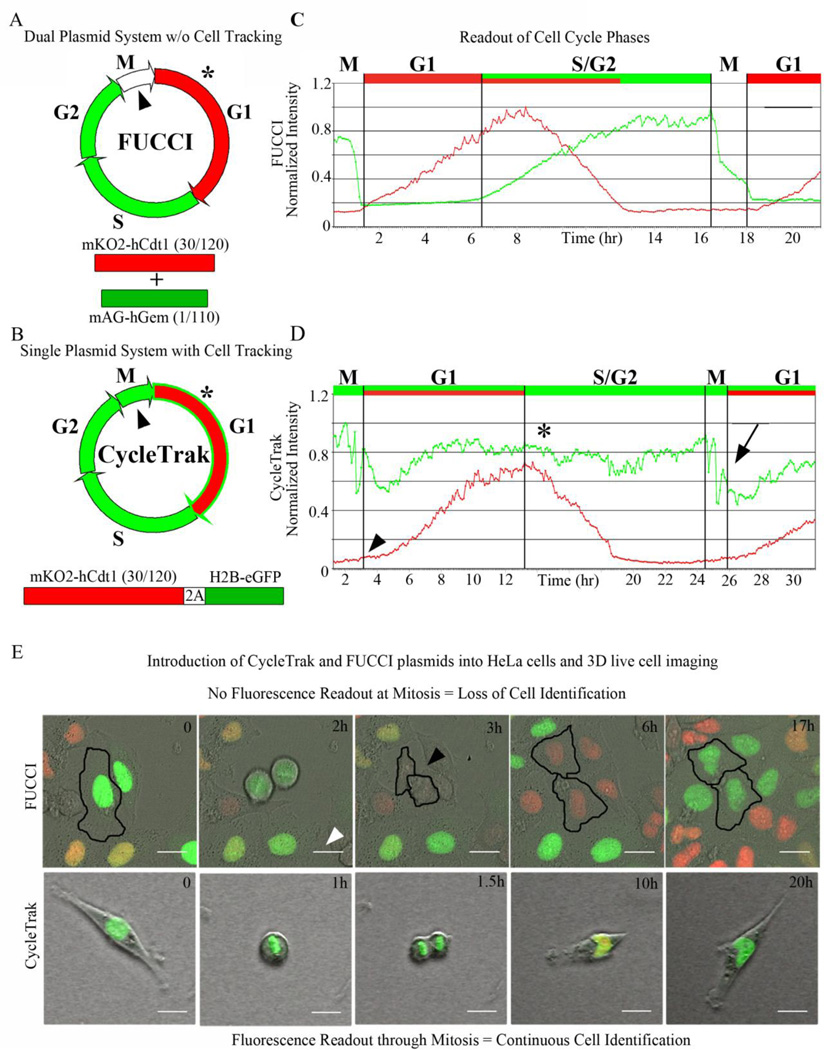
Figure 1. Comparison of cell cycle dynamics using Fucci versus CycleTrak systems.
(A) Two-plasmid Fucci system with M phase (arrowhead) and beginning of G1 (asterisk), when little or no fluorescence signal is visible. (B) Single-plasmid CycleTrak system displays red and green fluorescence signals during G1 (asterisk) and green fluorescence signal (H2B-eGFP) throughout the remaining cell cycles, including M phase (arrowhead). (C) A graph of the normalized intensities of red and green fluorescence of Fucci-labeled HeLa cells. mKO2-hCdt1 (30/120) fluorescence (red) increases at the onset of G1 and begins to degrade at the onset of S phase, concomitant with an increase in hGem-mAG fluorescence (green). Green fluorescence decreases with the onset of mitosis, leaving a brief period in late M/early G1 during which no fluorescence is visible. (D) A graph of the normalized intensities of red and green fluorescence of CycleTrak-labeled metastatic melanoma cells. CycleTrak-labeled cells are green through all phases of the cell cycle, including mitosis (arrow). The red signal continues to fluctuate due to changes in the cell cycle, with an increase in mKO2-hCdt1 (30/120) intensity at the beginning of G1 (arrowhead), followed by a decrease in the mKO2-hCdt1 (30/120) intensity at the onset of S phase (asterisk). (E) Representative images from confocal time-lapse movies of Fucci-labeled and CycleTrak-labeled cells in vitro. In Fucci-labeled HeLa cells (upper panels), red fluorescence increases in G1, decreases at the onset of S and is absent in G2 and M. The loss of fluorescence signal immediately following mitosis (arrowhead) makes it difficult to track cells during this phase. In CycleTrak-labeled metastatic melanoma cells (lower panels), the green fluorescence signal is present in all phases of the cell cycle, allowing for cell tracking throughout the entire cell cycle.
Lentiviral Preparation and Infection
Lentiviral particles were prepared using the ViraPower Lentiviral Packaging mix, Lipofectamine 2000, and the 293FT cell line according to the manufacturer’s directions (Invitrogen). Supernatant was collected at 48 and 72 hours post-transfection, and was concentrated using PEG precipitation. Briefly, the supernatant was collected, spun at 3,000 rpm for 5 minutes and then filtered using a 0.45um filter. For the precipitation, 3 ml of filter-sterilized 40% PEG-8,000 in PBS and 180 ul FBS was added to each 9 ml of supernatant. The virus was allowed to precipitate for at least 72 hours prior to pelleting at 1,500g for 10 minutes. The supernatant was removed and the tube was spun down again at 1,500g for 1 minute to remove residual PEG solution. The viral pellet was resuspended in culture media by gently pipetting up and down, before being stored at −80°C prior to use. Supernatant was concentrated 1:500 to 1:1,000, resulting in viral titers as high as 108 titerable units (TU)/ml.
For infection, C8161 cells were seeded into 24-well plates at 1×104 cells/well, and allowed to adhere overnight. The next day, the cells were infected at various MOIs in culture media in the presence of 6 ug/ml polybrene (Sigma-Aldrich) overnight, before undergoing a media change. The cells were then expanded prior to in vitro time-lapse imaging experiments or in vivo transplant experiments.
Chick Embryo Transplant Model
Metastatic melanoma cells (C8161) were trypsinized and resuspended at 4×106 cells/ml. A 20 ul drop with approximately 80,000 cells/drop was placed on a petri dish lid and grown for 40 hours. A typical subset of cells (in a cluster) was cut by glass needle from a larger cell cluster grown in a hanging drop as described above. This reduced the size of the cell cluster to fit into a gap in the dorsal neural tube of the e1.5 chick embryo made by separating the dorsal midline tissue of the neural tube by glass needle and inserting the tumor cell cluster inside. Each tumor cluster contained approximately 300 cells.
Time-Lapse Imaging
For in vitro imaging, cells were seeded in a glass-bottom petri dish (MatTek Corp.). Images were taken on a Zeiss 510 inverted confocal microscope using a 488nm laser for GFP and a 543 nm or a 560 nm laser for mKO2, and were collected at 1024×1024 pixels every 6 minutes. In vivo imaging was performed as previously described (Kulesa et al., 2010). Briefly, chick embryos (after tumor cell transplantation) were fitted with a Teflon window and placed on an upright Zeiss 710 confocal microscope. Embryos were imaged in ovo and z-stack images were collected with 9 slices (for a total of 62um) every 10 minutes.
Cell Tracking
Cell nuclei were tracked using Imaris (Bitplane Inc.) using the spots function. Spot size was set to the average size of a nucleus (10um) and cells were tracked for the entire time they were visible in the field of view. Tracks were verified by hand to be sure that one track represented the same cell for the duration of the track. Spots’ intensity data was exported and analysis was completed using a MATLAB (Mathworks Inc.) function. Though Imaris was used, the MATLAB analysis can be applied to any data for translational use with other tracking software.
Intensity Analysis and Automated Assignment of Phase
Cells often have a wide variety of mKO2-hCdt1 (30/120) fluorescence intensities from cell to cell and also from cycle to cycle within a single cell so simple threshold analysis was insufficient. Local minima were found in the fluorescence time trace to approximate the time of M phase, and then verified by checking that the intensity was less than 15% of the total range of mKO2 intensities. A temporary threshold was created, multiplying each local minima by a user defined parameter. When the mKO2 intensity crossed this threshold, a start of G1 was marked for each minima found. The completion of G1 was marked at the site of local maxima. As soon as the mKO2 intensity started to decrease, S/G2 began and this phase continued until M phase was reached.
M phase demarcation used the H2B-eGFP intensity by exploiting the fact that as a cell “rounded up” to divide, the cell either pulled up off the glass in vitro causing a decrease in mean intensity or in vivo, the condensed chromatin caused an increase in mean intensity. We therefore looked for changes in H2B-eGFP intensity greater than a certain number standard deviations from the mean. We found 2.5 worked well for our cells. This potential M phase was verified by first checking that the cell had not already been defined in G1 and second that a G1 phase started within 20% of the estimated total cell cycle time after our potential M phase beginning. If the conditions were met, the start of M phase was defined and all time points between here and the start of G1 were defined as M. All remaining time points in the trace were defined as S/G2.
Verification of Analysis Method
Fifty tracked cells were analyzed by hand for the time at which each phase of the cell cycle began. The start of M phase was defined by a retraction of the cell’s extended processes and the end upon the reemergence of the cellular extensions. G1 was defined as beginning with the first detection of red fluorescence and ending at the maximal red intensity. The results were compared to the same 50 cells analyzed by the MATLAB code and the results agreed within 15 minutes of the manually annotated data.
RESULTS
Development of CycleTrak as a single plasmid system that allows for cell cycle phase and cell position readout
The original Fucci constructs designed by the Miyawaki lab used two plasmids and exploited the cyclical expression and degradation of Cdt1 and Geminin as a means of tracking cell cycle progression (Sakaue-Sawano et al., 2008a). We harnessed the most informative component of Fucci, mKO2-hCdt1 (30/120) which specifically labels cells in G1, and coupled it with a nuclear-localized eGFP that expresses throughout the entire cell cycle (compare Fig. 1A,B and C,D; see Sup Movies 1 and 3). These proteins are separated by a 2A self-cleaving peptide sequence, allowing them to be expressed simultaneously and in equimolar amounts from the same promoter (de Felipe et al., 2006). The nuclear-localized eGFP is constantly expressed so that single cell identification and tracking may be accurately automated, even during mitosis (Fig 1E; see Sup Movies 2 and 3).
We verified that our single-plasmid system did not affect cell cycling and that we were able to discretely distinguish S/G2, M and G1 phases (Fig. 1D; see Sup Movies 1–3). We showed that the fluorescence readout using CycleTrak was distinct from that of Fucci, demonstrating that CycleTrak-labeled cells were visible throughout all phases of the cell cycle (Fig. 1E and Sup Movie 3) and were efficiently tracked (Sup Movie 2). For clarity of visualizing cell cycle progression in individual cells, we chose to present subregions where cells were more sparsely distributed.
Generation of the CycleTrak cell tracking and quantitative analysis software
We exploited the phase-specific fluorescent readouts of the CycleTrak plasmid to create a MATLAB function that automatically assigned cell cycle phase for single cells, based on fluorescence intensity. Using CycleTrak, an increase in mKO2-hCdt1 (30/120) intensity marked the onset of G1 phase (Fig. 1D, arrowhead). Thus, we were able to distinguish M phase by a rapid change in the average eGFP fluorescence intensity as a cell began chromosomal segregation (Fig. 1D, arrow; see Sup Movie 1). S/G2 phase was marked by the onset of degradation of mKO2 fluorescence intensity and constant eGFP fluorescence signal (Fig. 1D, asterisk). Thus, our single-plasmid CycleTrak system and quantitative tools provided a seamless analysis of cell cycle progression.
Since every cell may display different overall fluorescence intensity, this could present a roadblock to proper cell cycle analysis. Our analysis was based on relative changes in intensity over time. That is, our method for cell identification and tracking was not affected by cell to cell variability in fluorescence intensity, any more than other variables such as size and shape. Our method for segmenting out phases of the cell cycle is capable of handling fluorescence intensity changes with each cycle. Thus, our method based on relative changes in intensity is robust and overcomes this roadblock such that future studies will not need to generate a stable cell line in order to use the CycleTrak system.
We used a commercial software package (Imaris, Bitplane) to identify and track cells; however, any tracking software can be used to then interface with our MATLAB routines for analysis. We decided to use commercial software as the basis to integrate our cell cycle analysis algorithm. That is, instead of writing an entirely new cell tracking program that would not be user-friendly, cell type specific, or easily distributable to a wide audience within the community, we took advantage of a well-established commercial cell tracking program that allowed integration of externally-generated algorithms. Using a basic package to track cells showed that our system was easy to use. Additionally, users will not be required to use specialty software that is unfamiliar or that may not work optimally with their cell lines or embryos. Thus, our algorithm for cell cycle analysis will integrate with other cell tracking software that is user selected.
Validation of the CycleTrak system in vitro and identification of cell cycle heterogeneities in cultured human metastatic melanoma cells
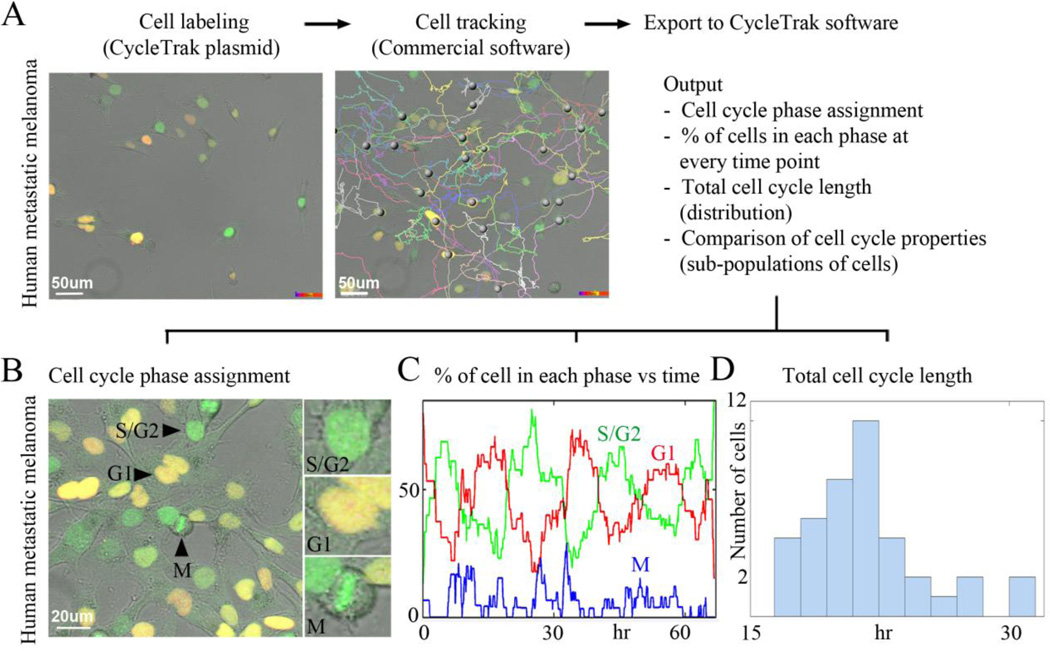
One of the major readouts of cancer progression is tumor growth. Studies of cell growth kinetics in tumors have shown that the duration of the cell cycle and the proportion of actively dividing cells are crucial to tumor growth in vivo (Ho and Dowdy, 2002). However, estimations of tumor growth as assessed by typical static methods are prone to error due to limitations in dynamic cell cycle analysis. We applied CycleTrak to fluorescently label, accurately track and then analyze cell cycle progression in individual human metastatic melanoma cells (Fig. 2; see Supplementary Movies 1 and 2). Our CycleTrak analysis of metastatic melanoma cells (n=55 cells) in vitro estimated the lengths (± standard deviation) of M, G1, and S/G2 as 1±0.9, 9±3.7 and 11±3.4 hours, respectively. This predicted a total cell cycle length of approximately 21 hrs±3.4, which agreed with estimates from manual measurements of the data and with reports from the literature (Zhu et al., 2004). We plotted the distribution of the total cell cycle time for all cells that completed at least one full cycle and found a wide variability within the population of cells (Fig. 2D). This variability would not have been detected without the ability to dynamically track cell cycle progression in individual cells. The variability in cell cycle was likely due to inherent heterogeneities within the population of our melanoma cells (see Sup Movie 1). Thus, using CycleTrak, it was possible to examine both population and individual cell cycle responses to an application of a drug or a change in microenvironment.
Figure 2. Visualization and analysis of human cancer cell cycle dynamics in vitro using CycleTrak.
(A) Unsynchronized human metastatic melanoma cells imaged in culture using confocal time-lapse microscopy for approximately 66 hours. A sampling of cell trajectories is shown. (B) A typical image from a confocal time-lapse session showing human metastatic melanoma cells in different cell cycle phases. (C) The graph shows the percentage of cells assigned to each cell cycle phase at every time point. (D) Histogram of total cell cycle length for all cells that completed at least one full cycle. Note the wide degree of variability in cell cycle length at the single cell level.
Novel in vivo cell cycle dynamics of human metastatic melanoma cells in a chick embryo transplant model as revealed by the CycleTrak system
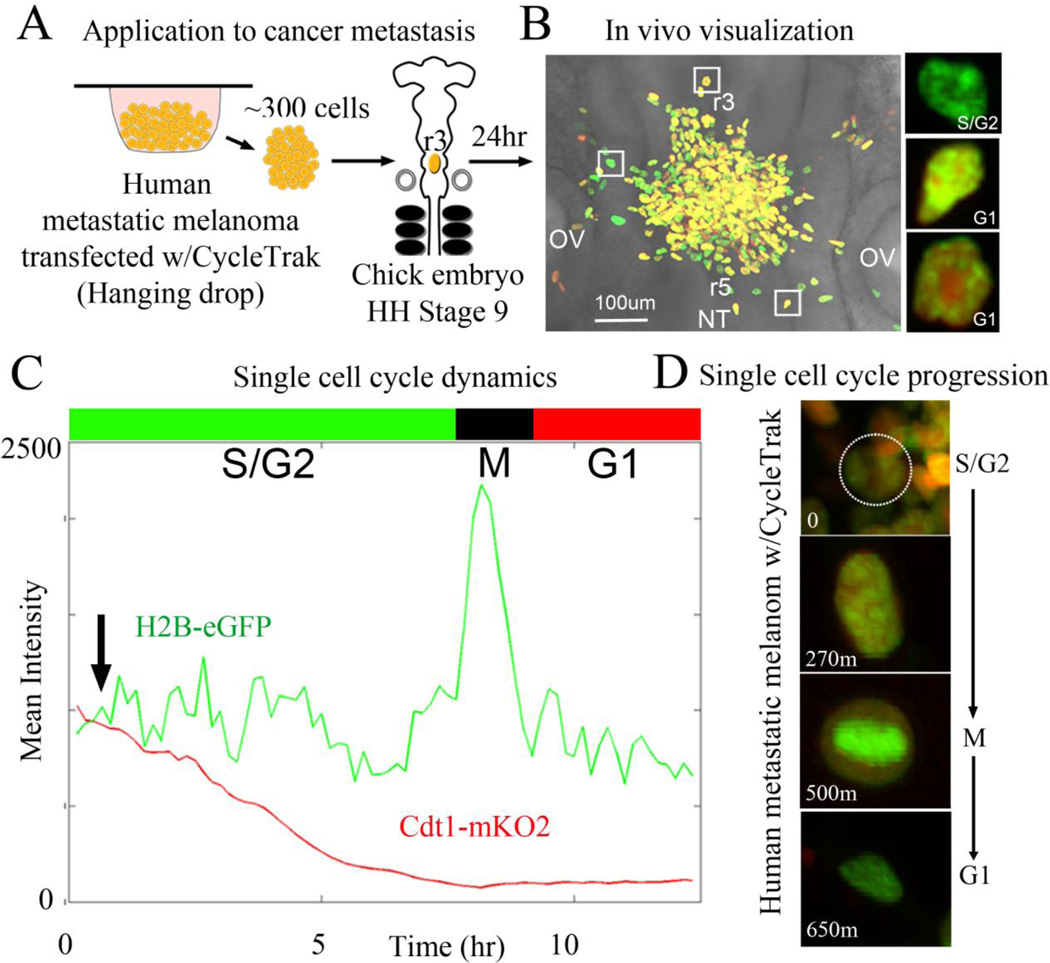
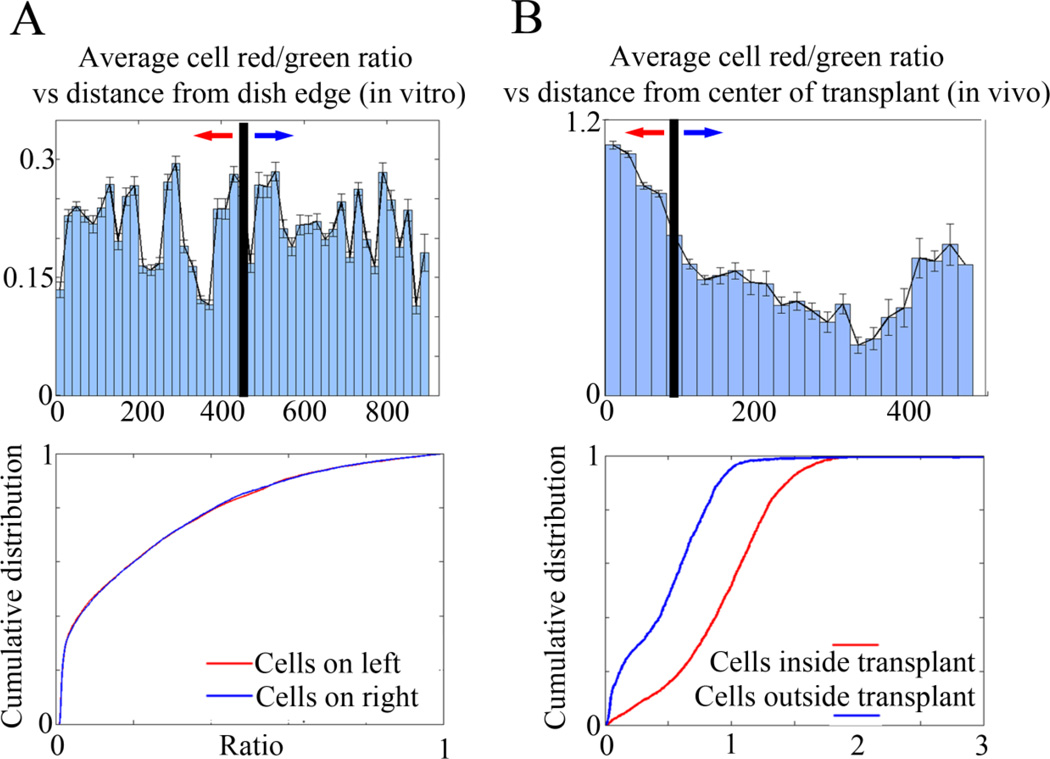
To study the cell proliferation dynamics of human metastatic melanoma cells in vivo, we took advantage of our chick embryo transplant model system for studying single human tumor cell behaviors in vivo. We have previously shown that human metastatic melanoma cells transplanted into the chick embryonic neural crest microenvironment invaded and moved along stereotypical host neural crest cell migratory pathways (Kulesa et al., 2006). Here, we transplanted small subpopulations of human metastatic melanoma cells (~300 cells), fluorescently-labeled with our single-plasmid CycleTrak system, into the dorsal neural tube of early-stage chick embryos (Fig. 3A; see Sup Movie 4). Our in vivo experiments revealed 3 main conclusions. First, cells that remained at the transplant site were more likely to be found in G1 (Fig. 3B, Fig. 4B). Second, upon exit from the transplant site and migration along host neural crest cell migratory pathways, tumor cells resumed cycling and continued to cycle during their invasion throughout the chick embryo (Fig. 3B–D, compare Fig. 4A,B; see Sup Movie 4). Third, our cell population analysis of transplanted tumor cells resulted in only a 4% phase classification error compared to measurements by hand, and allowed for examination of cell-to-cell variation (Fig. 3C). In comparison, other models that used the Fucci system to acquire whole-field information based on the fluorescence intensity obtained only gross changes in the percent of cells in a particular cell cycle phase (Sakaue-Sawano et al., 2011). In addition, only one other method describes single cell tracking using cell cycle-specific probes, and the non-automated results included only a small fraction of the population (Hahn et al., 2009).
Figure 3. Visualization and analysis of human cancer cell cycle dynamics in vivo using CycleTrak.
(A) Schematic of our in vivo chick embryo transplant model system. (B) Representative transplant after 24 hours of culture time with metastatic melanoma cells exhibiting all phases of the cell cycle within the embryonic neural crest microenvironment. Square boxes highlight examples of 3 melanoma cells in either G1 or S/G2 cell cycle phases. (C) Graph of the red and green fluorescent intensity traced over time for a typical, individual melanoma cell captured during an in ovo confocal time-lapse imaging session. Phases of the cell cycle as determined by CycleTrak are shown above the graph. The cell first exits the clump 30 minutes after the start of the time-lapse (arrow). (D) Cell from (B) at four different time points showing different phases of the cell cycle within the embryo. At time = 0, the cell is located within the transplant site. Over the course of the time-lapse session, the cell migrates away from the transplant site and into the embryonic environment. The cell begins in G1 phase and starts to cycle as it migrates, shortly after exiting the transplant.
Figure 4. Comparison of in vitro and in vivo cell cycle dynamics of human metastatic melanoma cells using CycleTrak.
(A) Upper panel - The average mKO2/eGFP intensity ratio (R/G) for all cells in a representative in vitro confocal time-lapse imaging session as a function of distance from the edge of the field of view. Lower panel - Distribution of R/G ratios of the left and right half of the field of view showing no significant difference (n=~4000 cells, p=0.47 by K-S test). (B) Upper panel - The average R/G ratio for all cells in the in vivo transplant time-lapse as a function of distance from the center of the transplant site. The transplant is approximately 90um in diameter. Lower panel - Distribution of R/G ratios showing a significant difference (n=4559 cells, p=1.1×10e-213 by K-S test) between the ratios of cells still located within the transplant site and the ratios of cells that have migrated into the embryonic environment.
DISCUSSION
The ability to accurately determine cell cycle phase and cell position in single and subpopulations of cells in vitro and in vivo at high spatiotemporal resolution using our newly developed CycleTrak system overcomes a major roadblock in cell dynamics analysis. CycleTrak allows cell cycle analysis in living tissue, unlike current static methods, such as BrdU and immunostaining, that require fixation. In addition, Fucci and other dynamic cell cycle-specific probes only provide general observations about large scale proliferative events. This is because of a lack of accurate cell tracking capability through late M/early G1 (Sakaue-Sawano et al., 2008a) or G1 to S/G2 (Hahn et al., 2009). This makes analysis of large numbers of cells during dynamic processes very labor intensive and time consuming. Although there is a slight decrease in CycleTrak’s ability to resolve early G1 from G2 in analysis of static images (because of constant green fluorescence signal and the lack of red fluorescence), CycleTrak overcomes the current roadblock of Fucci and other cell cycle reporters by allowing the tracking and visualization of all phases of the cell cycle through time. Thus, CycleTrak is unique in its ability to interrogate cell proliferation dynamics in single cells, or in small subpopulations of cells, particularly within a developing embryo.
Because of the ability to automate cell tracking and the quantitation of fluorescence signals, CycleTrak may be especially well suited for analyzing large-scale 3D data sets. Emerging imaging technologies, such as sheet plane illumination microscopy (SPIM) (Keller et al., 2010; Truong et al., 2011), have been elegantly applied to visualize Drosophila and zebrafish embryogenesis and offer one example. High content screening may also benefit from using the CycleTrak system, since multiple wells may be rapidly screened for changes in cell cycle progression in the presence of various siRNA or chemical compound treatments. Other application areas for CycleTrak include cancer growth and metastasis and stem cell biology, where the accurate readout of cell cycle progression and cell position would shed light on whether changes in cell cycle progression predict metastatic behavior and niche size, respectively.
The goal of this study was to generate a novel fluorescent reporter system and analytical tools for automated analysis of cell cycle progression. We have shown proof of principle of this system in vitro in human cell lines and in vivo by transplantation of human cancer cells into a chick embryo metastasis model. We believe the generation of transgenic animal models based on our CycleTrak reporter system will be exciting and will become a valuable resource for the developmental biology community.
In summary, we have created a single plasmid vector and semi-automated tools, called CycleTrak. CycleTrak allows for higher fidelity visualization and analysis of cell cycle phase and cell position than current methods. As a single plasmid system, CycleTrak enables a more effective delivery method capable of providing a fluorescent readout of G1 and cell nucleus position. The cell labeling strategy and software tools of CycleTrak allow for enhanced studies of cell cycle regulation owing to a more accurate means to identify and determine cell cycle phase and cell position than previous methods. This is largely due to the cell cycle readout of CycleTrak being based on the presence or absence of nuclear localized fluorescence signal, rather than on localization of signal (Hahn et al., 2009) and fluorescence through all cell cycle phases, unlike Fucci (Sakaue-Sawano et al., 2008a). We have validated CycleTrak and applied it in practice to identify heterogeneities in human cancer cell cycle progression in cultured metastatic melanoma cells and to follow in vivo single cell cycle progression after cell transplantation into a chick embryo model. We suggest that CycleTrak offers a powerful approach distinct from current techniques and is broadly applicable to cell proliferation questions in development, cancer, and regeneration.
Highlights.
We have developed a novel fluorescent reporter for dynamic cell cycle analysis
CycleTrak consists of a single plasmid vector and semi-automated quantitative tools
CycleTrak provides a simultaneous readout of G1 and cell nucleus position
We validated CycleTrak in vitro and an in vivo chick embryo model of metastasis
Advances current methods by allowing dynamic cell cycle readout and tracking
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Prof. Atsushi Miyawaki for his advice during the early phase of this project. PMK would like to acknowledge partial funding from NIH grant 1R01HD057922 and the Stowers Institute for Medical Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- de Felipe P, Luke GA, Hughes LE, Gani D, Halpin C, Ryan MD. E unum pluribus: multiple proteins from a self-processing polyprotein. Trends Biotechnol. 2006;24:68–75. doi: 10.1016/j.tibtech.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Hahn AT, Jones JT, Meyer T. Quantitative analysis of cell cycle phase durations and PC12 differentiation using fluorescent biosensors. Cell Cycle. 2009;8:1044–1052. doi: 10.4161/cc.8.7.8042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho A, Dowdy SF. Regulation of G(1) cell-cycle progression by oncogenes and tumor suppressor genes. Curr Opin Genet Dev. 2002;12:47–52. doi: 10.1016/s0959-437x(01)00263-5. [DOI] [PubMed] [Google Scholar]
- Keller PJ, Schmidt AD, Santella A, Khairy K, Bao Z, Wittbrodt J, Stelzer EH. Fast, high-contrast imaging of animal development with scanned light sheet-based structured-illumination microscopy. Nat Methods. 2010;7:637–642. doi: 10.1038/nmeth.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulesa PM, Bailey CM, Cooper C, Fraser SE. In ovo live imaging of avian embryos. Cold Spring Harb Protoc 2010, pdb prot5446. 2010 doi: 10.1101/pdb.prot5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulesa PM, Kasemeier-Kulesa JC, Teddy JM, Margaryan NV, Seftor EA, Seftor RE, Hendrix MJ. Reprogramming metastatic melanoma cells to assume a neural crest cell-like phenotype in an embryonic microenvironment. Proc Natl Acad Sci U S A. 2006;103:3752–3757. doi: 10.1073/pnas.0506977103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse P. A long twentieth century of the cell cycle and beyond. Cell. 2000;100:71–78. doi: 10.1016/s0092-8674(00)81684-0. [DOI] [PubMed] [Google Scholar]
- Sakaue-Sawano A, Kobayashi T, Ohtawa K, Miyawaki A. Drug-induced cell cycle modulation leading to cell-cycle arrest, nuclear mis-segregation, or endoreplication. BMC Cell Biol. 2011;12:2. doi: 10.1186/1471-2121-12-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, Imamura T, Ogawa M, Masai H, Miyawaki A. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008a;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- Sakaue-Sawano A, Ohtawa K, Hama H, Kawano M, Ogawa M, Miyawaki A. Tracing the silhouette of individual cells in S/G2/M phases with fluorescence. Chem Biol. 2008b;15:1243–1248. doi: 10.1016/j.chembiol.2008.10.015. [DOI] [PubMed] [Google Scholar]
- Sugiyama M, Sakaue-Sawano A, Iimura T, Fukami K, Kitaguchi T, Kawakami K, Okamoto H, Higashijima S, Miyawaki A. Illuminating cell-cycle progression in the developing zebrafish embryo. Proc Natl Acad Sci U S A. 2009;106:20812–20817. doi: 10.1073/pnas.0906464106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymczak-Workman AL, Vignali KM, Vignali DA. Generation of 2A Peptide-linked Multicistronic Vectors. In: Friedmann T, Rossi J, editors. Gene Transfer: Delivery and Expression of DNA and RNA. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 2007. pp. 137–147. [Google Scholar]
- Truong TV, Supatto W, Koos DS, Choi JM, Fraser SE. Deep and fast live imaging with two-photon scanned light-sheet microscopy. Nat Methods. 2011;8:757–760. doi: 10.1038/nmeth.1652. [DOI] [PubMed] [Google Scholar]
- Welch DR, Bisi JE, Miller BE, Conaway D, Seftor EA, Yohem KH, Gilmore LB, Seftor RE, Nakajima M, Hendrix MJ. Characterization of a highly invasive and spontaneously metastatic human malignant melanoma cell line. Int J Cancer. 1991;47:227–237. doi: 10.1002/ijc.2910470211. [DOI] [PubMed] [Google Scholar]
- Zhu N, Lalla R, Eves P, Brown TL, King A, Kemp EH, Haycock JW, MacNeil S. Melanoma cell migration is upregulated by tumour necrosis factor-alpha and suppressed by alpha-melanocyte-stimulating hormone. British journal of cancer. 2004;90:1457–1463. doi: 10.1038/sj.bjc.6601698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.