Abstract
Objective
Selenium neutralizes interleukin-1β (IL-1β) induced inflammatory responses in chondrocytes. We investigated potential mechanisms for this through in vitro knockdown of three major selenoproteins, Iodothyronine Deiodinase-2 (DIO2), Glutathione Peroxidase-1 (GPX1), and Thioredoxin Reductase-1 (TR1) in primary human chondrocytes.
Methods
Primary human chondrocytes were transfected with scrambled small interfering RNA (siRNA) or siRNA specific for DIO2, GPX1 and TR1. After 48 hours, transfected cells were cultured in serum free media for 48 hours, with or without 10 pg/ml IL-1β for the final 24 hours. The efficiency of siRNAs was confirmed by quantitative Real Time-Polymerase Chain Reaction (qRT-PCR) and Western blot analysis. The gene expression, by qRT-PCR, of cyclooxygenase-2 (COX2), IL-1β, and Liver X receptor (LXR) alpha and beta was evaluated to determine the impact of selenoprotein knockdown on inflammatory responses in chondrocytes.
Results
The mRNA expression of DIO2, GPX1, and TR1 was significantly decreased by the specific siRNAs (reduced 56%, p=0.0004; 96%, p<0.0001; and 66%, p<0.0001 respectively). Suppression of DIO2, but not GPX1 or TR1, significantly increased (~2-fold) both basal (p=0.0005) and IL-1β induced (p <0.0001) COX2 gene expression. Similarly, suppression of DIO2 significantly increased (~9-fold) IL-1β induced IL-1β gene expression (p=0.0056) and resulted in a 32% (p=0.0044) decrease in LXRα gene expression but no effect on LXRβ.
Conclusions
Suppression of the selenoprotein DIO2 resulted in strong pro-inflammatory effects with increased expression of inflammatory mediators, IL-1β and COX2, and decreased expression of LXRα suggesting that this may be the upstream target through which the anti-inflammatory effects of DIO2 are mediated.
Keywords: Chondrocyte, osteoarthritis, inflammation, selenoprotein, cyclooxygenase, Interleukin-1, Iodothyronine Deiodinase-2
Introduction
Profound selenium deficiency is associated with the severe osteoarthropathy known as Kashin-Beck Disease (KBD) that affects around 7 million individuals in China and neighboring regions [1]. In the US, a low but non-deficiency level of selenium has been shown to be associated with OA presence and severity in a large population study [2-3]. These observations suggest a requirement for selenium for cartilage health and OA prevention. Recently, selenium has been shown to be anti-inflammatory by altering cytokine-inducible nitric oxide synthase (iNOS) and cyclooxygenase-2 (COX2) gene expression in response to lipopolysaccharide (LPS) stimulation in cultured macrophages [4-5]. Our recent study demonstrated that pretreatment of chondrocytes with selenomethionine (SeMet) attenuated production of IL-1β induced nitric oxide (NO) and prostaglandin E2 (PGE2), synthesized via the COX and prostaglandin synthase pathways [6]. Given the fact that selenium is incorporated as selenocysteine at the active site of a wide range of selenoproteins, we hypothesized that one or more selenoproteins may be responsible for the anti-inflammatory effects of selenium.
To date, twenty-five mammalian selenoproteins have been identified including three large subfamilies: glutathione peroxidases (GPXs), thioredoxin reductases (TRs), and iodothyronine deiodinases (DIOs) [7]. GPXs and TRs function as antioxidative enzymes to catalyze the reduction of intracellular peroxide and regulate the redox balance in cells [8]. The DIOs regulate the bioactivity of thyroid hormone by controlling levels of thyroxine (T4) and the active hormone, 3,3',5-triiodo L-thyronine (T3) [9]. DIO2 is responsible for local (chondrocyte) conversion of T4 to the active form of the hormone, T3. Several studies now implicate specific selenoproteins from all three of these subfamilies in OA. A genetic variant of GPX1 and reduced plasma GPX activity are associated with increased risk of developing Kashin-Beck disease, a severe form of OA endemic to China [10]. Gene expression of GPX1 and thioredoxin-interacting protein (TXNIP) are downregulated in cartilage lesions with moderate to severe late-stage OA [11]. A variant of DIO2 is associated with risk for developing OA [12] and has been shown to increase the vulnerability of cartilage to OA in association with nonoptimal femoral head bone shapes rather than directly influencing the formation of these shapes [13].Taken together, this evidence shows modulation of major selenoproteins in OA and suggests that they may be important modifiers of the joint to inflammatory responses and thereby osteoarthritis susceptibility. Our goal in this study was to elucidate the biological effects of three specific major selenoproteins through analysis of the consequences of DIO2, GPX1 and TR1 knock-down with small interfering RNAs (siRNAs) on inflammatory responses in primary human chondrocytes.
Materials and Methods
Chondrocyte Isolation and Culture
Anonymous surgical waste articular cartilage samples, taken at the time of joint replacement, were used for this project. Tissue was collected under the approval of the Duke IRB who determined that this protocol met the definition of research not involving human subjects as described in 45CFR46.102(f) and satisfied the Privacy Rule as described in 45CFR164.514. Articular cartilage samples were obtained from 6 patients undergoing total knee replacement surgery [mean age, 60.3 +/- 10.3 years]. Cartilage was harvested from nonlesional areas, further minced, and subjected to pronase and collagenase digestion to isolate primary human chondrocytes, similar to previously published methods [14]. Isolated chondrocytes within the first two passages were used for all experiments.
RNA Interference of DIO2, GPX1 and TR1
siRNA transfection was performed using the program U20 of the Amaxa Nucleofector (Gaithersburg, MD), with either the Amaxa Primary Human Chondrocyte Nucleofector Kit or Mirus Ingenio™ Electroporation Kit (Madison, MD) according to the manufacturers’ protocols. Chondrocytes were transfected with the following siRNAs: 3 μg of Silencer Negative Control No.1 siRNA (Ambion, Applied Biosystem) that served as a scrambled transfection control; 3 μg of human DIO2 specific siRNA (s4106); 1 μg of human GPX1 specific siRNA (s804); or 3 μg of human TR1 specific siRNA (s755).
After transfection, cells were cultured for 48 hours in DMEM/F12 media supplemented with 10% fetal bovine serum to allow gene suppression and turnover of the preexisting targeted proteins. Transfected cells were subsequently cultured in the absence of serum in DMEM media for 24 hours, then treated for another 24 hours with or without 10 pg/ml IL-1β (R & D systems, Minneapolis, MN) in serum free media. The IL-1β concentration of 10 pg/ml was chosen based on evidence for physiological relevance (equivalent to concentrations in human OA synovial fluid) and previously established experiments [6, 15-16]. Thus, the total period of knockdown was 96 hours (72 hours prior to the addition of IL-1β). Cells were treated with RNeasy Lysis buffer (Qiagen Valencia, CA) to isolate RNA for gene expression studies.
RNA Isolation and Real Time RT-PCR
Cell lysates, prepared by RNeasy Lysis buffer from each experimental condition were homogenized by passage through a QIAshredder spin column (Qiagen, Valencia, CA). The total DNA and RNA fractions were further isolated using the AllPrep DNA/RNA/Protein Mini Kit (Qiagen, Valencia, CA) according to the manufacturer's protocol. The isolated total RNA was reverse transcribed into cDNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) for Real Time RT-PCR analysis. The ABI Prism 7000 sequence detection system and relative quantification software (Applied Biosystems, Foster City, CA) were used for real-time analyses. The amplification for real-time RT-PCR used the following Applied Biosystems primer and probe sets: 18S rRNA endogenous control; Hs00255341_ml (DIO2); Hs00829989_gH (GPX1); Hs00182418_ml (TR1); Hs01573474_g1 (COX2); Hs01555410_m1 (IL-1β); Hs00172855_ml (LXRα) and Hs00173195_ml (LXRβ). The real-time reactions were performed in triplicate in a final volume of 25 μl.
mRNA Quantification and Statistical Analysis
Raw mRNA expression values were computed by 2-ΔCt formula [17] with values normalized to 18S rRNA, where ΔCt represents the difference in Ct (threshold cycle) number of the 18S rRNA gene and target genes. Results were derived from a total of 4 independent experiments for COX2, IL-1β, LXRα and LXRβ gene expression, performed in triplicate, using a total of 4 separate primary chondrocyte cell lines. The relative fold changes in mRNA expression levels of target genes were calculated by the 2 -ΔΔCt formula [17], between cells transfected with selenoprotein siRNA and the cells transfected with scrambled siRNA in different treatments. For the purposes of graphical presentation, the relative mRNA level in scrambled transfected cells without treatment (control group) was set at 100%.
Raw mRNA expression data were evaluated by paired t-test of the log transformed 2-ΔCt values comparing subgroups (n=4 in each group from 4 separate cell lines): (1) the scrambled transfected group without IL-1β treatment and individual selenoprotein siRNA transfected groups without IL-1β treatment, and (2) the scrambled transfected group with IL-1β treatment and individual selenoprotein siRNA transfected groups with IL-1β treatment. P values less than 0.05 were considered significant after correction for multiple comparisons (Bonferroni).
Western Blot Quantification of Protein Knock-Down
Whole cell lysates were separated by SDS-PAGE and transferred to nitrocellulose for immunoblotting. Membranes were blocked with 5% BSA in TBS/0.1% Tween 20 (TBS-T). Polyclonal primary antibodies against DIO2, GPX1 and TR1 were obtained from Abcam (Cambridge, MA). A monoclonal antibody against α-tubulin (Sigma) was used as a normalization control at 1:10,000 dilution. Anti-rabbit and anti-mouse IgG-HRP (Jackson ImmunoResearch, West Grove, PA) secondary antibodies were used at a 1:5,000 dilution. The resulting films were scanned using CanoScan LiDE 70 (Canon, Lake Success, NY) and the band intensities were quantified using Adobe Photoshop CS and Image J (National Institutes of Health, Bethesda, MD).
Results
Suppression of Selenoproteins by siRNA
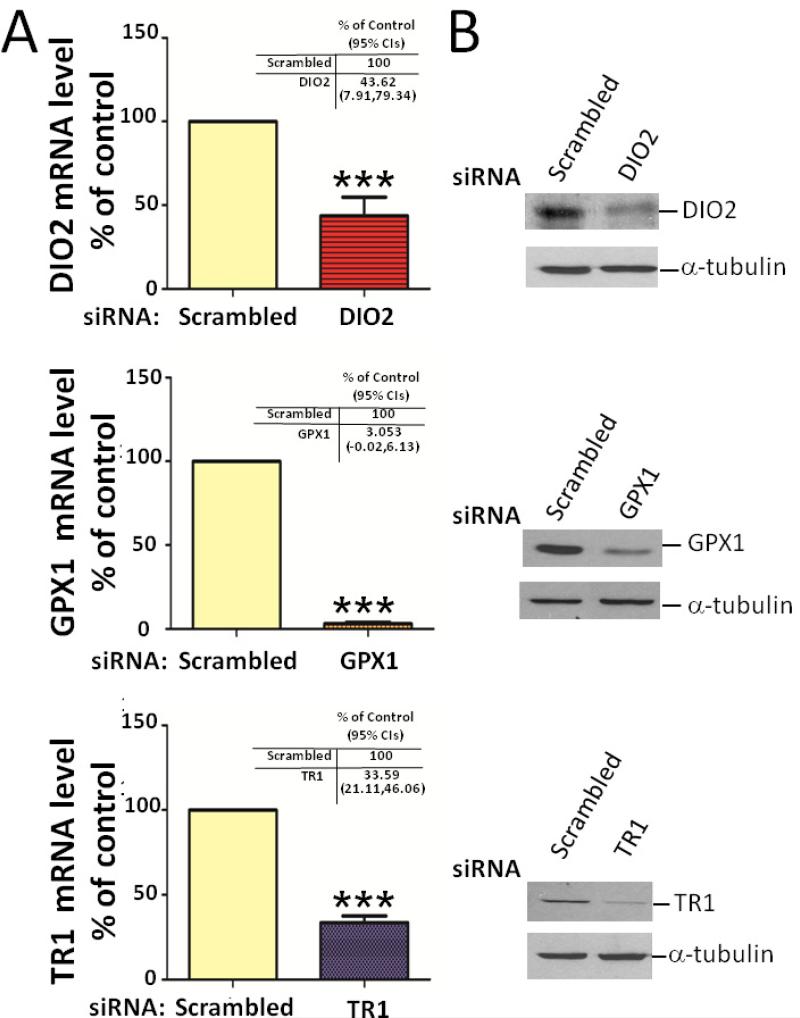
To examine the effects of DIO2, GPX1 and TR1 on IL-1β responses in primary human chondrocytes, we suppressed the expression of DIO2, GPX1, and TR1 with sequence specific siRNAs (Figure 1). Compared with the scrambled siRNA control, the expression of the three selenoproteins was significantly reduced by the specific siRNAs: DIO2 siRNA reduced DIO2 mRNA expression by 56% (p=0.0004) (Figure 1A); GPX1 siRNA reduced GPX1 mRNA expression by 96% (p<0.0001); and TR1 siRNA reduced TR1 mRNA expression by 66% (p<0.0001). By Western blot, protein levels were reduced 50% by the DIO2 siRNA, 80% by the GPX1 siRNA, and 80% by TR1 siRNA (Figure 1B).
Figure 1. Demonstration of siRNA suppression of DIO2, GPX1 and TR1 steady state mRNA gene and protein expression.
Primary human chondrocytes were transfected with a scrambled control small interfering RNA (siRNA) or siRNA specific for DIO2, GPX1 or TR1. At 48 hours post-transfection, siRNA transfected cells were cultured for 48 hours in serum-free medium. (A) DIO2, GPX1 and TR1 steady-state mRNA levels were determined by RT-PCR normalized to 18S rRNA in siRNA-transfected cells (average of triplicates for four independent experiments). Values are the mean and SEM levels of mRNA for DIO2, GPX1 or TR1 expressed as a percentage of the scrambled control. The mRNA level in cells transfected with the scrambled siRNA was set at 100%. *** P<0.001 versus scrambled control (scrambled). The mean fold change as % control and 95% CIs corresponding to each gene target are provided in the tables in each panel. (B) DIO2, GPX1 and TR1 proteins were isolated. Equal amounts of total cell lysate were separated by SDS-PAGE and analyzed by Western blot; α-tubulin (bottom row) was used as a control for normalization.
COX2 Gene Expression in the Setting of Suppressed Selenoprotein Expression
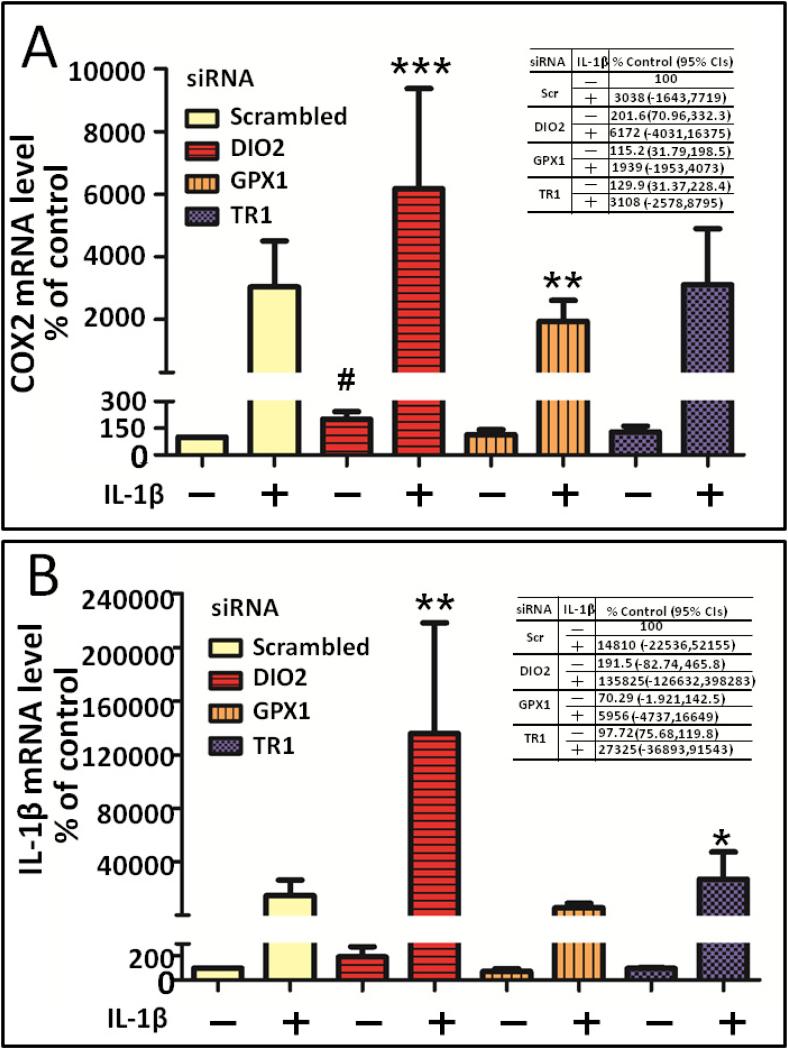
The suppression of DIO2 had a pro-inflammatory effect; it resulted in a 2-fold increase in COX2 basal (absence of IL-1β) mRNA expression (p=0.0005) (Figure 2A). The suppression of DIO2 also resulted in a significant 2-fold increase in IL-1β induced COX2 steady state mRNA expression (P<0.0001) compared to the IL-1β induced COX2 level of gene expression in the presence of the scrambled siRNA control. In contrast, the suppression of GPX1 and TR1 had no significant effect on basal level expression of COX2. Depletion of GPX1 slightly but significantly decreased IL-1β induced COX2 gene expression (p=0.007).
Figure 2. Effect of depleting selenoproteins on COX2 and IL-1β gene expression in primary chondrocytes.
Primary human chondrocytes were transfected with a scrambled control small interfering RNA (siRNA) or siRNA specific for DIO2, GPX1 and TR1. At 48 hours post-transfection, siRNA transfected cells were cultured for 24 hours in serum free medium and followed by 24 hour treatment with or without 10 pg/ml IL-β. Gene expression for COX2 and IL-1β was determined by RT-PCR normalized to 18S rRNA (average of triplicates for four independent experiments). The effect of depleting individual selenoproteins on COX2 (A) and IL-1β (B) gene expression is shown in the absence or presence of IL-1β (10 pg/ml) treatment. Values shown are mean and SEM of COX2 or IL-1β gene expression as a percentage of the scrambled control without IL-1β (set at 100%) from four independent experiments. # P<0.001 versus scrambled without IL-1β; ***P<0.001 versus scrambled with IL-1β stimulation; **P<0.01 versus scrambled with IL-1β stimulation *P<0.05 versus scrambled with IL-1β stimulation. The mean fold change as % control and 95% CIs corresponding to each experimental condition are provided in the tables in each panel.
IL-1β Gene Expression in the Setting of Suppressed Selenoprotein Expression
Based on a report of an inverse relationship of DIOs and IL-1β gene expression in skeletal muscle [18], we next evaluated the effects of selenoprotein depletion on IL-1β mRNA expression in primary human chondrocytes. Similar to the effect on COX2 gene expression, the suppression of DIO2 had a pro-inflammatory effect; it resulted in a 2-fold increase in IL-1β basal steady state mRNA expression but the result did not meet statistical significance (p=0.2837) (Figure 2B). However, the suppression of DIO2 resulted in a 9-fold increase in IL-1β induced IL-1β steady state mRNA expression (p=0.0056) over that induced by IL-1β stimulation in the presence of the siRNA scrambled control. The suppression of GPX1 and TR1 had no significant effect on basal level expression of IL-1β. The suppression of TR1 had a slight but statistically significant pro-inflammatory effect with an increase in the IL-1β induced IL-1β mRNA expression (p=0.0140) beyond the level achieved by IL-1β in the presence of the scrambled siRNA control.
LXR Gene Expression in the Setting of Suppressed DIO2 Expression
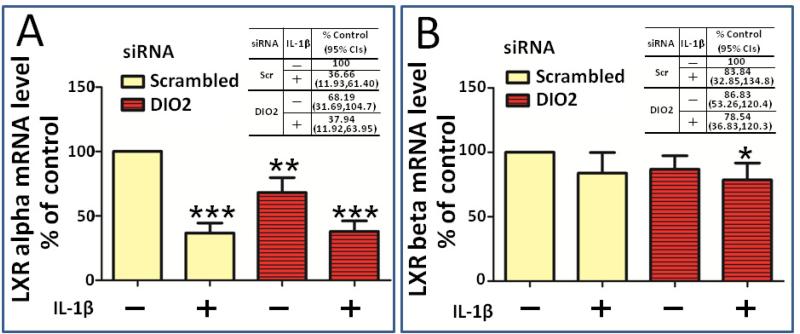
Based on the known regulation of IL-1β and PGE2 production, and regulation of LXRs by thyroid hormone in liver cells [19-20], we next tested the hypothesis that DIO2 modifies inflammatory responses in chondrocytes through regulation of LXR gene expression in chondrocytes. We therefore examined LXRα and LXRβ gene expression under DIO2 depleted conditions. With respect to LXRα, the suppression of DIO2 also resulted in a pro-inflammatory effect with a significant decrease (32%, p=0.0044) in LXRα basal mRNA expression compared to the scrambled control (Figure 3A). IL-1β treatment significantly decreased the LXRα mRNA expression (63%, p=0.0004) and the combination of IL-1β and suppression of DIO2 did not further repress LXRα expression. In contrast to the effect on LXRα gene expression, there were no changes in LXRβ gene expression in response to depletion of DIO2 or IL-1β treatment (Figure 3B).
Figure 3. Effect of depleting DIO2 on LXRα and LXRβ gene expression in primary chondrocytes.
Primary human chondrocytes were transfected with a scrambled control small interfering RNA (siRNA) or siRNA specific for DIO2, GPX1 and TR1. At 48 hours post-transfection, siRNA transfected cells were cultured for 24 hours in serum free medium and followed by 24 hour treatment with or without 10 pg/ml IL-β. Gene expression for LXRα and LXRβ was determined by RT-PCR normalized to 18S rRNA (average of triplicates for four independent experiments). The effect of depleting DIO2 on LXRα (A) and LXRβ (B) gene expression is shown in the absence or presence of IL-1β (10 pg/ml) treatment. Values shown are mean and SEM of LXRα and LXRβ gene expression as a percentage of the scrambled control without IL-1β (set at 100%) from four independent experiments. *** P<0.001 versus scrambled without IL-1β; ** P<0.01 versus scrambled without IL-1β stimulation; *P<0.05 versus scrambled without IL-1β stimulation. The mean fold change as % control and 95% CIs corresponding to each experimental condition are provided in the tables in each panel.
Discussion
Our study highlights a potential new role of DIO2 in modulating the inflammatory response in chondrocytes. Suppression of DIO2 by siRNA knock-down resulted in consistent pro-inflammatory effects in primary human chondrocytes, namely downregulation of LXRα, and upregulation of both IL-1β and COX2 gene expression. These results suggest that DIO2 plays an important anti-inflammatory role in cartilage through local chondrocyte production of thyroid hormone T3 that maintains or activates LXRα, which in turn suppresses IL-1β and COX2 gene expression. In this manner, DIO2 would function as an important countermeasure to inflammatory responses in chondrocytes.
This interpretation is consistent with a variety of evidence from several past studies. For one, this is consistent with a previous study that demonstrated that DIO2 mRNA level was inversely correlated with IL-1β mRNA in skeletal muscle [18]. Our findings are also consistent with data showing that thyroid hormone (T3) upregulates LXRα but not LXRβ gene expression in mouse liver cells [20]. Activation of LXR decreases basal and IL-1β induced PGE2 production in human cartilage explants [19]. This supports our conclusion that DIO2 regulation of LXRα in this study likely mediated the alteration of COX2 gene expression. IL-1β also greatly downregulates LXRα, but not LXRβ gene expression in human OA cartilage [21]. Given that stimulation of LXR transcriptional activity can counteract the catabolic effects of IL-1 [21], the activation or maintenance of LXRα activity by thyroid hormone (produced by DIO2), appears to be one of the mechanisms by which DIO2 exerts a protective anti-inflammatory effect in chondrocytes. This is therefore one pathway and selenoprotein through which selenium may exert an anti-inflammatory effect. The general lack of major modulation of the inflammatory responses due to GPX1 and TR1 knock-down underscores the specificity of these observations of pro-inflammatory effects due to lack of DIO2 and the corollary, that specific anti-inflammatory effects are attributable to DIO2.
Although we only achieved modest knock-down of DIO2, we observed strong effects. Like the GPXs and TRs, there is some redundancy of function within the DIO family of genes consisting of DIO1, DIO2 and DIO3. DIO1 and DIO2 are responsible for conversion of the prohormone T4, to its active form, T3 through deiodination. DIO3 inactivates T4 and T3 through deiodination, while DIO1 also catalyzes deiodination of T3 to inactive 3,3’-diiodothyronine (T2). Only DIO2 and DIO3 are known to be expressed in chondrocytes [22]. To date, genetic polymorphisms in both DIO2 and DIO3 have been implicated in OA susceptibility [12, 23] although the mechanisms are not known. DIO2 expression is highly upregulated in cartilage of late stage OA [24]. It has been demonstrated previously that DIO2 gene expression was potently activated in response to lipopolysaccharide stimulation via the NFKβ pathway in a human mesothelioma cell line [25]. Therefore, the elevated DIO2 expression in OA cartilage could be a consequence of an activated NFKβ pathway activity among other pro-inflammatory pathways during OA disease progression [26]. The findings in this study support the supposition that the local availability of thyroid hormone plays a protective role and that optimal DIO2 activity is essential for maintenance of normal chondrocyte health through production of adequate local T3 concentrations. It is possible however that both depletion (in our study) or overproduction of DIO2 (in another study [24]) may lead to aberrant local levels of T3 and contribute to joint pathology. Further elucidation of the mechanism by which DIO2 or T3 modulate the inflammatory response would be beneficial in determining the optimal DIO2 activity and T3 concentrations for joint health.
There are several limitations of this research. It was interesting that profound depletion of GPX1 (both mRNA and protein) did not have a significant impact on COX2 or IL-1β gene expression. This may however be due to redundancy of this important family of antioxidants. At least 3 other GPX enzymes (GPX2, GPX3 and GPX4) are known to be expressed in cartilage and chondrocytes ([11] and unpublished observation). Similarly, the failure to observe major effects from the TR1 knock-down may be due to redundancy of this family with TRs like TR2 known to be expressed in chondrocytes [27]. Thus, it may be necessary to simultaneously knock-down several GPXs or TRs or require multiple siRNAs for a particular target to more fully suppress the expression and activity of these selenoproteins. It may also require a longer period of knock-down than 72 hours prior to cytokine addition to more fully evaluate the role of these selenoproteins in joint tissue inflammation and joint degradation; although, several recent siRNA knockdown studies in other cell systems demonstrated appreciable reductions of enzymatic activity after 48 hours transient transfection with siRNA (GPX1 or TR1) as well as reduced expression of a T3-driven promoter after 48 hours transfection into a cell line stably transfected with shRNA (DIO2) [28-30].)
Although we observed both mRNA and protein levels of selenoproteins were decreased in siRNA transfected cells, we did not measure their corresponding enzymatic activities. However, prior siRNA studies of these selenoproteins demonstrated a strong concordance of reductions in mRNA steady state and protein levels with reductions in enzymatic activity levels [28-30]. Furthermore, we previously demonstrated that decreases in COX2 steady state transcript levels corresponded to decreases in PGE2, its enzymatic product [6].
Finally, although we focused on the effects of depleting DIO2, GPX1 and TR1, we cannot exclude the possibility that depletion of these selenoproteins may impact the expression of other selenoproteins. For instance, profound depletion of GPX1 may trigger compensatory gene expression of other antioxidative enzymes.
In conclusion, we have demonstrated that depletion of DIO2 increased IL-1β induced COX2 gene expression in chondrocytes. We also showed that depleting DIO2 increased IL-1β induced IL-1β gene expression, which suggests that DIO2 may negatively modulate the IL-1β response through regulation of IL-1β gene expression itself. Finally, we demonstrated that depleting DIO2 decreased the anti-inflammatory LXRα gene expression and this may contribute to the increased IL-1β and COX2 gene expression under DIO2 depleted conditions. These data are consistent with a view that DIO2 in chondrocytes may constitutively maintain or activate LXRα gene expression with downstream effects of decreased IL-1β and COX2 expression and PGE2 production.
Acknowledgements and Funding
This study was funded by the Center for Biomolecular and Tissue Engineering International Fellowship at Duke (T32 GM00855-15 & 16) (to AWMC), an Osteoarthritis Research Society International (OARSI) Scholarship (to AWMC), NIH/NIA Claude D. Pepper OAIC 5P30 AG028716 and NIH/NIAMS grant UO1 AR050898
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authors’ Contributions
AWMC participated in the conception and design of the study, isolation of primary chondrocytes, performance of gene expression studies, and statistical analysis and drafted the manuscript. MB coordinated and organized surgical waste tissue collection and revised the manuscript. VBK conceived of the study, supervised the project, and participated in statistical analysis and manuscript preparation and editing. All authors read and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
References
- 1.Allander E. Kashin-Beck disease. An analysis of research and public health activities based on a bibliography 1849-1992. Scand J Rheumatol Suppl. 1994;99:1–36. doi: 10.3109/03009749409117126. [DOI] [PubMed] [Google Scholar]
- 2.Jordan J, Fang F, Arab L, Morris J, Renner J, Ngwenyama R, et al. Low selenium levels are associated with increased odds of radiographic knee and multijoint OA. Osteoarthritis & Cartilage. 2005;13:S35. [Google Scholar]
- 3.Jordan J, Fang F, Schwartz T, Morris J, Renner J, Chen J, et al. Low Selenium Levels Are Assoicated With Increased Odds Of Radiographic Hip Osteoarthritis In African American And White Women. Osteoarthritis & Cartilage. 2007;15:C33. [Google Scholar]
- 4.Vunta H, Belda BJ, Arner RJ, Channa Reddy C, Vanden Heuvel JP, Sandeep Prabhu K. Selenium attenuates pro-inflammatory gene expression in macrophages. Mol Nutr Food Res. 2008;52:1316–1323. doi: 10.1002/mnfr.200700346. [DOI] [PubMed] [Google Scholar]
- 5.Prabhu KS, Zamamiri-Davis F, Stewart JB, Thompson JT, Sordillo LM, Reddy CC. Selenium deficiency increases the expression of inducible nitric oxide synthase in RAW 264.7 macrophages: role of nuclear factor-kappaB in up-regulation. Biochem J. 2002;366:203–209. doi: 10.1042/BJ20020256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng AW, Stabler TV, Bolognesi M, Kraus VB. Selenomethionine inhibits IL-1beta inducible nitric oxide synthase (iNOS) and cyclooxygenase 2 (COX2) expression in primary human chondrocytes. Osteoarthritis Cartilage. 2011;19:118–125. doi: 10.1016/j.joca.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigo R, et al. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 8.Burk RF. Protection against free radical injury by selenoenzymes. Pharmacol Ther. 1990;45:383–385. doi: 10.1016/0163-7258(90)90073-b. [DOI] [PubMed] [Google Scholar]
- 9.Bianco AC, Salvatore D, Gereben B, Berry MJ, Larsen PR. Biochemistry, cellular and molecular biology, and physiological roles of the iodothyronine selenodeiodinases. Endocr Rev. 2002;23:38–89. doi: 10.1210/edrv.23.1.0455. [DOI] [PubMed] [Google Scholar]
- 10.Xiong YM, Mo XY, Zou XZ, Song RX, Sun WY, Lu W, et al. Association study between polymorphisms in selenoprotein genes and susceptibility to Kashin-Beck disease. Osteoarthritis Cartilage. 2010;18:817–824. doi: 10.1016/j.joca.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 11.Aigner T, Fundel K, Saas J, Gebhard PM, Haag J, Weiss T, et al. Large-scale gene expression profiling reveals major pathogenetic pathways of cartilage degeneration in osteoarthritis. Arthritis Rheum. 2006;54:3533–3544. doi: 10.1002/art.22174. [DOI] [PubMed] [Google Scholar]
- 12.Meulenbelt I, Min JL, Bos S, Riyazi N, Houwing-Duistermaat JJ, van der Wijk HJ, et al. Identification of DIO2 as a new susceptibility locus for symptomatic osteoarthritis. Hum Mol Genet. 2008;17:1867–1875. doi: 10.1093/hmg/ddn082. [DOI] [PubMed] [Google Scholar]
- 13.Waarsing JH, Kloppenburg M, Slagboom PE, Kroon HM, Houwing-Duistermaat JJ, Weinans H, et al. Osteoarthritis susceptibility genes influence the association between hip morphology and osteoarthritis. Arthritis Rheum. 2011;63:1349–1354. doi: 10.1002/art.30288. [DOI] [PubMed] [Google Scholar]
- 14.Kuettner KE, Pauli BU, Gall G, Memoli VA, Schenk RK. Synthesis of cartilage matrix by mammalian chondrocytes in vitro. I. Isolation, culture characteristics, and morphology. J Cell Biol. 1982;93:743–750. doi: 10.1083/jcb.93.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Westacott CI, Whicher JT, Barnes IC, Thompson D, Swan AJ, Dieppe PA. Synovial fluid concentration of five different cytokines in rheumatic diseases. Ann Rheum Dis. 1990;49:676–681. doi: 10.1136/ard.49.9.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kahle P, Saal JG, Schaudt K, Zacher J, Fritz P, Pawelec G. Determination of cytokines in synovial fluids: correlation with diagnosis and histomorphological characteristics of synovial tissue. Ann Rheum Dis. 1992;51:731–734. doi: 10.1136/ard.51.6.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Kwakkel J, van Beeren HC, Ackermans MT, Platvoet-Ter Schiphorst MC, Fliers E, Wiersinga WM, et al. Skeletal muscle deiodinase type 2 regulation during illness in mice. J Endocrinol. 2009;203:263–270. doi: 10.1677/JOE-09-0118. [DOI] [PubMed] [Google Scholar]
- 19.Li N, Rivera-Bermudez MA, Zhang M, Tejada J, Glasson SS, Collins-Racie LA, et al. LXR modulation blocks prostaglandin E2 production and matrix degradation in cartilage and alleviates pain in a rat osteoarthritis model. Proc Natl Acad Sci U S A. 2010;107:3734–3739. doi: 10.1073/pnas.0911377107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hashimoto K, Matsumoto S, Yamada M, Satoh T, Mori M. Liver X receptor-alpha gene expression is positively regulated by thyroid hormone. Endocrinology. 2007;148:4667–4675. doi: 10.1210/en.2007-0150. [DOI] [PubMed] [Google Scholar]
- 21.Collins-Racie LA, Yang Z, Arai M, Li N, Majumdar MK, Nagpal S, et al. Global analysis of nuclear receptor expression and dysregulation in human osteoarthritic articular cartilage: reduced LXR signaling contributes to catabolic metabolism typical of osteoarthritis. Osteoarthritis Cartilage. 2009;17:832–842. doi: 10.1016/j.joca.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 22.Robson H, Shalet S, Williams G. Growth plate chondrocytes express type II and III but not type I deiodinase. Hormones Res. 2002;58(Suppl 2):166. [Google Scholar]
- 23.Meulenbelt I, Bos SD, Chapman K, van der Breggen R, Houwing-Duistermaat JJ, Kremer D, et al. Meta-analyses of genes modulating intracellular T3 bio-availability reveal a possible role for the DIO3 gene in osteoarthritis susceptibility. Ann Rheum Dis. 2011;70:164–167. doi: 10.1136/ard.2010.133660. [DOI] [PubMed] [Google Scholar]
- 24.Ijiri K, Zerbini LF, Peng H, Otu HH, Tsuchimochi K, Otero M, et al. Differential expression of GADD45beta in normal and osteoarthritic cartilage: potential role in homeostasis of articular chondrocytes. Arthritis Rheum. 2008;58:2075–2087. doi: 10.1002/art.23504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeold A, Doleschall M, Haffner MC, Capelo LP, Menyhert J, Liposits Z, et al. Characterization of the nuclear factor-kappa B responsiveness of the human dio2 gene. Endocrinology. 2006;147:4419–4429. doi: 10.1210/en.2005-1608. [DOI] [PubMed] [Google Scholar]
- 26.Roman-Blas JA, Jimenez SA. NF-kappaB as a potential therapeutic target in osteoarthritis and rheumatoid arthritis. Osteoarthritis Cartilage. 2006;14:839–848. doi: 10.1016/j.joca.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 27.Kruger C, Kappen C. Microarray analysis of defective cartilage in Hoxc8- and Hoxd4-transgenic mice. Cartilage. doi: 10.1177/1947603510363005. first published online March 22 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watson WH, Heilman JM, Hughes LL, Spielberger JC. Thioredoxin reductase-1 knock down does not result in thioredoxin-1 oxidation. Biochem Biophys Res Commun. 2008;368:832–836. doi: 10.1016/j.bbrc.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lubos E, Kelly NJ, Oldebeken SR, Leopold JA, Zhang YY, Loscalzo J, et al. Glutathione peroxidase-1 deficiency augments proinflammatory cytokine-induced redox signaling and human endothelial cell activation. J Biol Chem. 2011;286:35407–35417. doi: 10.1074/jbc.M110.205708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dentice M, Marsili A, Ambrosio R, Guardiola O, Sibilio A, Paik JH, et al. The FoxO3/type 2 deiodinase pathway is required for normal mouse myogenesis and muscle regeneration. J Clin Invest. 2010;120:4021–4030. doi: 10.1172/JCI43670. [DOI] [PMC free article] [PubMed] [Google Scholar]