Abstract
The mouse posterior primitive streak at neural plate/headfold stages (NP/HF, ~7.5dpc–8dpc) represents an optimal window from which hemangioblasts can be isolated. We performed immunohistochemistry on this domain using established monoclonal antibodies for proteins that affect blood and endothelial fates. We demonstrate that HoxB4 and GATA1 are the first set of markers that segregate independently to endothelial or blood populations during NP/HF stages of mouse embryonic development. In a subset of cells, both proteins are co-expressed and immunoreactivities appear mutually excluded within nuclear spaces. We searched for this particular state at later sites of hematopoietic stem cell emergence, viz., the aorta-gonadmesonephros (AGM) and the fetal liver at 10.5–11.5dpc, and found that only a rare number of cells displayed this character. Based on this spatial-temporal argument, we propose that the earliest blood progenitors emerge either directly from the epiblast or through segregation within the allantoic core domain (ACD) through reduction of cell adhesion and pSmad1/5 nuclear signaling, followed by a stochastic decision toward a blood or endothelial fate that involves GATA1 and HoxB4, respectively. A third form in which binding distributions are balanced may represent a common condition shared by hemangioblasts and HSCs. We developed a heuristic model of hemangioblast maturation, in part, to be explicit about our assumptions.
Keywords: Hemangioblast, HSC, Posterior Primitive Streak, GATA1, HoxB4, cross antagonism
Introduction
Understanding the complex sequence of interactions that lead to robust determination of distinct cell types during embryonic development will directly enhance our ability to generate sustainable populations for regenerative therapies. A critically important but unresolved problem is to clarify the origin (Danchakoff, 1916; Sabin, 1922) and natural paths (Chan and Yoder, 2004; Dzierzak and Speck, 2008) of the earliest blood cells during embryogenesis.
Combined developmental and clinical investigations have identified hemangioblasts and hematopoietic stem cells (HSCs) as the earliest progenitors that contribute to the complete set of blood types. The term hemangioblast was originally coined by Murray to emphasize the specific potential of angioblasts to contribute to both blood and endothelial cell types (Murray, 1932). Around that period, “angioblast” was used as a morphological term to describe primitive mesenchymal cells emerging from the epiblast that aggregate into cords, which then transform into blood islands through lumenization (Downs, 2003; Sabin, 2002). More recently, genetic analyses have distinguished angioblasts by their restricted potential to contribute only to endothelial and possibly smooth muscle cells (Ema et al., 2003). Complementary examinations of post-gastrulation stage mouse embryos and the related embryoid body system have helped dissect further specific molecular and genetic differences between hemangioblasts and angioblasts (Park et al., 2004; Robb et al., 1995). However, failure to distinguish behavior at the single cell level in toto has obstructed continued progress toward successful derivation and amplification of HSCs from ES (Ying et al., 2008) and EpiSCs (Brons et al., 2007; Tesar et al., 2007).
HSCs are considered to derive from hemangioblasts through an intermediate phenotype called hemogenic endothelium (Eilken et al., 2009; Zovein et al., 2010). HSCs are operationally defined as mesodermal cells that can functionally reconstitute a lethally irradiated host to a particular degree (Medvinsky et al., 2011; Warr et al., 2011), the success of which can be sensitive to donor/recipient interactions (Yoder, 2004). However, due to lack of an explicit (Till et al., 1964) awareness of how particular microenvironments modulate hemangioblasts or HSCs at specific times, a consensus definition sensu stricto has not yet been achieved, leading to relaxed usage to different degrees by specialists in different disciplines. Despite this potential problem, a minimal combination of markers that include SLAM (Signalling Lymphocytic Activation Molecule) factors (Kiel et al., 2005) have been identified to execute up to 37% successful long-term functional reconstitution using supplemented single cells from adult and fetal tissues as early as the 14.5dpc mouse liver (Kim et al., 2006), while other combinations may be used to enhance reconstitution potential to a lesser degree from tissues as early as 10.5dpc (Chen et al., 2011). SLAM factors are transmembrane proteins of the CD2 superfamily of Immunoglobulin receptors that are physically clustered in Chromosome 1 of both mouse and humans (Sidorenko and Clark, 2003). Finally, emergent behaviors that correlate with the rare number of HSCs identified through a limiting dilution assay (Kumaravelu et al., 2002) has been directly visualized using a transgenic reporter driven by the Ly6A promoter (Boisset et al., 2010) within the ventral region of dorsal aorta explants at 10.5dpc, a site of HSC emergence.
The first blood and endothelial cells in the mouse embryo appear at ~6.5dpc with onset of gastrulation (Gilbert, 2008; Stern et al., 2006), a highly conserved (Voiculescu et al., 2007) metazoan morphogenetic event that initiates with formation of the primitive streak (Vasiev et al., 2010). Using single cell labeling and comprehensive mapping of descendants, Lawson et al., first characterized the dynamics of the earliest zones that contribute directly to extraembryonic mesoderm at early streak stages (Lawson et al., 1991). Building on this antero-proximal to postero-lateral epiblast recruitment model, others (Kinder et al., 1999) used multiple retrospective analyses to argue that early primitive erythroid and endothelial lineages are sorted separately and sequentially at early streak to mid-streak stages, respectively. Interpretations of multi-color chimeric studies based on the assumption that cells do not rearrange to a significant degree have supported this temporal segregation model in which epiblast cells control blood or vessel decisions directly rather than through generation of hemangioblasts (Ueno and Weissman, 2006).
Blood and vessel specification at gastrulation stages is dependent on a diffusible interaction with the visceral endoderm (Belaoussoff et al., 1998). Although Indian Hedgehog (Ihh) has been implicated as an inducer (Dyer et al., 2001), deletion of the Ihh-encoding gene does not result in early blood and vessel defects (Byrd et al., 2002). However, the severity of vascular defects displayed by smoothened −/− embryos is phenocopied by removal of both Shh and Ihh, implicating overlapping roles for the two ligands (Astorga and Carlsson, 2007) and an essential requirement for Hedgehog signaling for patterning early embryonic blood and vessel development. Rescue of endothelial defects with exogenous treatment of Bmp ligands in mice lacking Foxf1, a bHLH transcription factor responsive to Hedgehog signaling, support a model in which vascular remodeling is controlled indirectly by Hedgehog signaling through induction of Bmp4 via control of Foxf1 in a tissue-specific manner (Astorga and Carlsson, 2007).
At neural plate/headfold stages (~7.5dpc, please see morphological criteria, such as the shape of the allantois (Downs and Davies, 1993)), Huber et al., demonstrated optimal hemangioblast potential using clonal analysis of cells isolated from Brachyury/Flk1-positive posterior streak tissue (Huber et al., 2004). Based on these data, the investigators proposed that hemangioblasts migrate directly into the yolk sac upon delamination from the posterior streak. Persistent assortment of mesoderm into the yolk sac was previously shown using orthotopic grafts to the region ventral to the amniotic insertion site at headfold stages (Tam and Beddington, 1987). Most recently, transplant of large cell clusters followed by expression mapping showed fluid contribution from posterior and allantoic regions to the extraembryonic yolk sac (Mikedis and Downs, 2011), arguing that mesoderm contribution to the yolk sac exhibits both coherence and exchangeability with neighbors. However, definitive assortment trajectories have not yet been mapped to define the degree to which cell adhesion and segregation operates across the single cell level.
The posterior primitive streak (Downs, 2009) at headfold stages is an ambiguously defined caudal region of active cell sorting that exhibits a remarkable degree of organization (Mitiku and Baker, 2007), as illustrated by simultaneous coordination of blood island formation (Ferkowicz and Yoder, 2005) allantoic bud growth (Downs et al., 2009), primordial germ cell maturation (Fujiwara et al., 2001; McLaren, 2003; Saitou and Yamaji, 2010) and segregation of the embryo proper (Deschamps and van Nes, 2005). Despite this criticality, physical suitability for analyses (Gardner, 1985), and its position as a restricted point of divergence for three different HSC reservoirs (the allantois, yolk sac and future AGM (Wang and Wagers, 2011)), physical difficulty with handling the delicate gastrulation stage mouse embryo has precluded repeatable examinations for improving resolution of cell segregation behaviors.
In this report, we examined HoxB4 and GATA1 at different stages of blood and vessel development beginning at neural plate/headfold stages. We show that the GATA1/HoxB4 combination is the earliest set of markers that distinguish blood and endothelial lineages, respectively. The two functionally distinct transcription factors are co-expressed in the nucleus in a salt and pepper pattern until at least 8.5dpc embryos. Moreover, we demonstrate that the signature is found in rare populations in the 10.5dpc AGM and 11.5dpc fetal liver. Based on these observations, we hypothesize that the HoxB4/GATA1 interaction is an example of transcription factor cross-antagonism (Cantor and Orkin, 2001) specific for early mouse embryonic hemangioblasts and shared by HSCs in the AGM and fetal liver. Cross-antagonism is a concentration-dependent mutually exclusionary lineage specification process involving competitive interactions between two functionally opposed transcription factors, although the specific manner by which the mechanism is executed can vary.
Materials and Methods
Embryo collection
Embryos were collected from ICR or C57BL/6 crosses. Embryos were staged according to morphological criteria (Downs and Davies, 1993). Neural Plate/Headfold (NP/HF) stage embryos were assigned as those embryos that have formed an allantois that had not yet fused with the chorion while displaying thickening of rostral neural tissue and no somites. Further subdivisions were based on shape of allantois. NP/HF, early somite stage (8.5dpc) and 9.5–11.5dpc embryos were collected in a 5% FBS/DPBS dissecting buffer and fixed with 4% PFA overnight at 4 degrees with rocking.
Immunohistochemistry
Embryos were washed thoroughly in PBT.1. Samples were placed in blocking buffer (PBT.1 + 5% Goat and 5% Donkey Serum) for one hour. The blocking buffer was replaced with the following monoclonal antibodies at 1:150 dilution in blocking buffer: Flk1, PECAM-1, CD41 (Pharmingen), GATA1, pSMAD1/5, E-Cadherin, PU.1 (Cell Signaling Technologies), pan-Laminin (Sigma) and HoxB4 (DHSB, http://dshb.biology.uiowa.edu/). After overnight incubation at 4 deg, samples were washed with three changes in blocking buffer and placed in appropriate secondaries at 1:200 dilution overnight in blocking buffer. Secondary antibodies were all purchased from Invitrogen (Alexa-Fluor conjugated, InVitrogen). Samples were washed in PBT.1 three times and placed in an undersaturated DAPI dilution (~7 mg/ml) overnight for counterstaining. DAPI was rinsed in PBT.1 prior to imaging. At least 3 embryos and multiple optical sections of different regions were examined for each antibody. Generally, we examined distributions until they became qualitatively predictable.
Section preparation
Samples were placed in an agarose/sucrose block (2%/1% in PBS) for vibratome sectioning. Samples were cut in 80–100μm sections and only the middle section containing the longest allantoic fragment was examined to facilitate comparative analysis and record developmental stage. Samples were manually mounted by placing the droplet of PBS with the sample onto the coverslip with silicon posts. The buffer was removed with a pipet to promote attachment of sample onto coverslip, then a slide was placed over the sample, being careful not to crush the sample. PBS was replaced and the sample was imaged using a confocal microscope.
Microscopy
Samples were examined using a Zeiss 510META system. We used a 10× Plan-Neofluar 10×/0.3, F-Fluar 40×/1.3 Oil or Alpha-Plan-Apochromat 100×/1.46 Oil DIC lens.
Quantification and Image Processing
Zen software that is a standard feature of the confocal system was used to quantify intensities. Briefly, the widest field was captured using the 40× lens to relate relative intensities between cells located across a single field. Then, higher magnification subfield images were generated and relations in this field were normalized to the wider image. Surface area of intensity was measured from equivalently sized nuclei selected from a Z-series, then plotted using a spreadsheet. No processing other than Photoshop Image Levels, Rotate or Mode Grayscale was used.
Netlogo modeling
The Netlogo software is free to download from the Northwestern University Center for Connected Learning website (Wilensky, 1999), which includes step by step instructions on how to proceed and is actively supported by a large user community.
Results
Posterior epiblast/extraembryonic mesoderm interface changes throughout gastrulation and the exact boundaries remain ambiguous
We developed a simple and reproducible stereological technique (Hilliard and Lawson, 2003) to better resolve spatial boundaries at neural plate/headfold stages (NP/HF). We attempted to spatially localize the earliest hematopoietic and endothelial determination event during NP/HF stages using immunohistochemistry. As a marker of the undifferentiated state, we chose E-Cadherin (ECad), a calcium dependent adhesion molecule that has an essential and specific role in trophectoderm development (Kan et al., 2007). Although it is also expressed in the allantois and visceral endoderm throughout headfold stages (Daane et al., 2011), its clearance specifically in embryonic mesoderm (Burdsal et al., 1993) is considered a critical aspect of the coordinate remodeling necessary for proper segregation and has been used as a marker for the epiblast (Ciruna and Rossant, 2001; Rakeman and Anderson, 2006).
As an indicator of the determined state, we chose Flk1/VEGFR2/KDR, a growth factor receptor necessary for blood and vessel development (Shalaby et al., 1997), and perhaps smooth muscle precursors (Ema et al., 2003; Yamashita et al., 2000). Finally, we examined pSmad1/5 distributions to quantify the nuclear activation state of Bmp-Receptor regulated Smads (BRSmads)(Massague et al., 2005) in Flk1 positive cells. Smad1 (Tremblay et al., 2001) and Smad5 (Chang et al., 1999) are cytoplasmic modulators of the BR-Smad class that affect posterior development (Winnier et al., 1995) at different stages and through multiple mechanisms (de Sousa Lopes et al., 2004; Fujiwara et al., 2001; Lawson et al., 1999), in contrast to R-Smads (Smad2/3), which generally participate in orchestrating embryonic polarity through control of anterior, neural and cardiac fates in a concentration-dependent manner (Kattman et al., 2011; Waldrip et al., 1998).
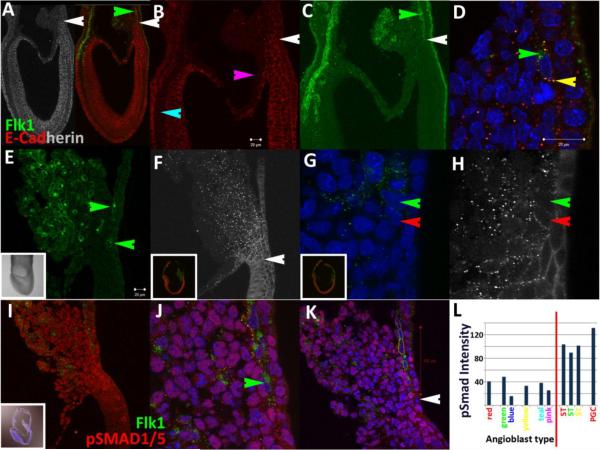
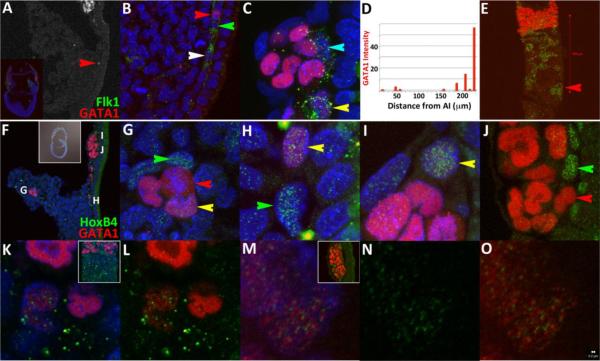
At early bud stages (~7.25dpc), ECad is differentially expressed as dense punctate clusters on membranes throughout various tissues including the embryonic ectoderm and endoderm and reduced in the middle germ layer except in a narrow domain at the posterior end of the embryo (Fig.1A–C; for nomenclature and anatomy of posterior region, please see summary Fig.4). More specifically, the protein is maintained within a pseudo-stratified region ~16 cells-wide across the embryonic/extraembryonic junction (Fig.1A,B). The posterior limit of ECad expression (white arrows, Fig.1A,B) abuts Flk1 expressing cells, creating a sharp boundary that divides the Allantoic Core Domain (ACD) in half across its base (white arrows, Fig.1A–C). The absence of signal in extraembryonic amniotic mesoderm (pink arrow, Fig.1B, (Dobreva et al., 2010)) and head mesenchyme (teal arrow, Fig.1B) serves as internal negative controls for antibody specificity. ECad expression on angioblast membranes is maintained as puncta that overlaps with Flk1 (yellow arrow, Fig.1D) but always excluded from an extranuclear lipid trafficking compartment (green arrow, Fig.1D).
Figure 1.
The interface between pluripotent and determined cell types is dynamic across gastrulation. Flk1 (green) and E-Cadherin (red and grayscale) expression at early bud (A–D) and headfold stages (E–H). White arrows in A–C mark the interface. Green arrows in (C,D) mark Flk1 expression in ECad-negative recycling compartment. Yellow arrow in (D) marks co-localization on membrane. ECad expression is different at mid-headfold stages (F–H).
Nuclear pSmad1/5 levels (I–K) in angioblasts (Flk1 positive, green arrow) at headfold stages is reduced. Quantification (L) of pSmad1/5 nuclear levels of cells marked in (K) with y-axis total intensity and x-axis distance from the amniotic insertion site (white arrow, K). White scale bars in (B,D,E) are 20μm. Generally, most nuclei are ~10μm, although mesothelial cells have larger nuclei, ~20μm (not shown). All inset figures are 1250μm across in this report except Fig.3B,F and 7C.
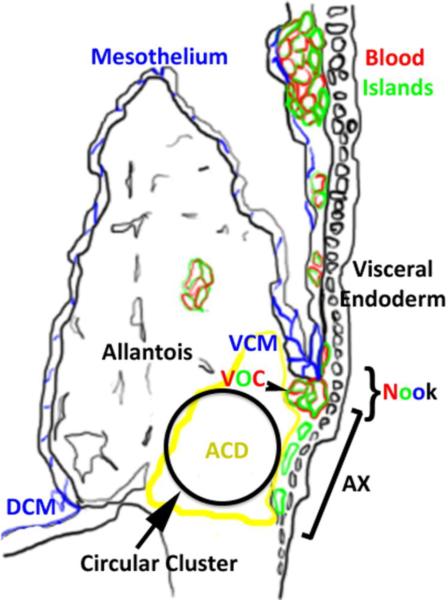
Figure 4.
Summary figure describes new observations in the context of an existing model of the posterior region at headfold stages. The embryonic stage can be estimated based on the shape of the allantois, in this case, late headfold stage. Colors of letters and structures correlate. Blue marks mesothelium, red marks PECAM-1/CD41, green marks Flk1 and black marks visceral endoderm, allantois, amnion and embryo. The circular cluster is variable in size. DCM (dorsal cuboidal mesothelium); VCM (ventral cuboidal mesothelium); VOC (vessel of confluence); ACD (allantoic core domain); AX (allantois associated visceral endoderm or transitional visceral endoderm). The figure is modified from (Daane et al., 2011).
Determining the location of the epiblast/angioblast interface was difficult at headfold stages due to changes in both ECad and Flk1 expression distributions (Fig.1E–H, Fig.2A–D). Flk1 positive cells expanded its territory distally across the entire transitional visceral endoderm interface (AX)(Bonnevie, 1950; Downs et al., 2009) often as individuated cells (green arrows, Fig.1E) but sometimes as a continuous stream (Fig.1J). A qualitative difference in ECad distribution was apparent in cells positioned across the line defined as the amniotic insertion point (white arrow, Fig.1F). Generally, cells proximal to the site of implantation displayed punctate membrane expression while continuous patterns were evident on distal cells. Closer examination of the ACD revealed varying degrees of ECad membrane clustering across subsets of visceral endoderm and stromal cells (Fig.1H, Fig.2B,C) that appeared to mature to continuous junctions by late headfold stages (Fig.2D). Flk1 positive cells maintained reduced ECad levels at headfold stages (compare green and red arrows, Fig.1G,H). These data suggest that regulation of ECad membrane dynamics (Hong et al., 2010) is changing across headfold stages.
Figure 2.
The circular cluster occupies the allantoic core domain after mid-headfold stages. The size of the circular cluster has a diameter of 50μm in (A–C), ambiguous in (D), 70μm in (E–G) and 90μm in (H). Laminin expression in (F,G) marks extracellular matrix and outlines the circular domain.
ECad puncta in cells occupying the proximal allantoic core domain (ACD) (Mikedis and Downs, 2011) were sometimes arranged in circular patterns (Fig.2A–D), suggestive of polarized clustering. Using nuclear arrangements as readout, we examined 21 embryos and found 12 circular clusters that ranged from 50 to 100μm in size. All 12 samples with recognizable arrangements were from mid headfold stages and older, up to early somite stages. Although these data argue that the circular cluster within the ACD matures throughout headfold stages, this qualitative observational argument is expected to be sensitive to mounting orientation and dynamics of underlying adhesive interactions.
To determine if the circular arrangement was associated with other markers of tissue polarity, we stained for Laminin (Hamill et al., 2009), a basement membrane protein that forms an integral component of the extracellular matrix (Mikedis and Downs, 2009; Zamir et al., 2008). Although the round nuclear arrangement was recognizable in a mid headfold stage embryo (Fig.2E), Laminin expression was mostly restricted to tissue interfaces and stromal cells adjacent to the AX. Laminin expression increased by late headfold and the signal surrounded a 70μm circular cluster (Fig.2F,G) that made contact across the amniotic insertion site. A different example of a circular cluster is shown in Fig.2H to illustrate its spatial relationship to hematopoietic determination. Note the positioning of cells expressing sparse punctate levels of CD41, a marker of both primitive and definitive hematopoiesis (Ferkowicz et al., 2003; Mikkola et al., 2003), contacting the base of the circle adjacent to the AX (red arrow, Fig.2H). Combined, these data suggest that Flk1+ cells are determined through a direct interaction with the circular domain that lies across a triangular region within the proximal ACD that has demonstrated potential to contribute to hematopoiesis, the vessel of confluence and yolk sac endothelial cells (Mikedis and Downs, 2011). Since the displayed images are renders of static optical sections, further investigation is warranted to elucidate the transforming volumetric structure and its potential role in affecting posterior patterning.
Bmp signaling is critical for mesoderm induction (Winnier et al., 1995) and subsequent maturation, proliferation and survival of Flk1 positive cells (Park et al., 2004). To detect changes in Bmp Receptor mediated induction associated with cell determination, we examined distributions of the phosphorylated form of Smad1/5. Tissue-level staining patterns at headfold stages resembled published reports (Fig.1I–K)(Fukuda et al., 2006). Namely, expression is higher in the posterior region (Fig.1I) but also present in rostral compartments near the anterior visceral endoderm (data not shown). Within the ACD (Fig.1I–K), nuclear localization is present throughout all cells, albeit at different levels. Notably, there was a significant and specific decrease in nuclear accumulation in all angioblasts adjacent to the AX (green arrow, Fig.1J). Punctate signals that colocalize with Flk1 outside of the nucleus were also apparent. In blood islands, Flk1 positive cells maintained low nuclear pSMAD1/5 levels (data not shown). In the allantois, some Flk1 positive cells near the base of the ACD displayed low pSMAD1/5 nuclear levels similar to cells of the yolk sac but distal cells associated with allantoic blebs (Daane et al., 2011) exhibited higher intensities (Fig.1I,K).
We quantified phosphorylated Smad1/5 distributions in the caudal region (Fig.1L). A set of cells with characteristic round nuclei that resemble migrating PGCs had higher but consistent intensities (~130 RLUs), likely due to induction by extraembryonic mesoderm-derived Bmp4 signals (Fujiwara et al., 2001). Visceral endoderm displayed heterogeneous nuclear levels, stromal cells of the ACD had consistent intensities (~100 RLUs), while angioblasts displayed the lowest average nuclear accumulation (~33 RLUs). These data are consistent with a threshold model for survival, maintenance and segregation of different lineages (Lawson et al., 1999). Most notably, our results show that pSmad1/5 nuclear accumulation was reduced and maintained at low levels in Flk1 positive cells lining the visceral endoderm. These results support arguments that stress the importance of timing in Smad1 modulation across distinct stages (Adelman et al., 2002; Cook et al., 2011; Zafonte et al., 2007).
Blood and vessel determination begins prior to yolk sac blood island formation at headfold stages
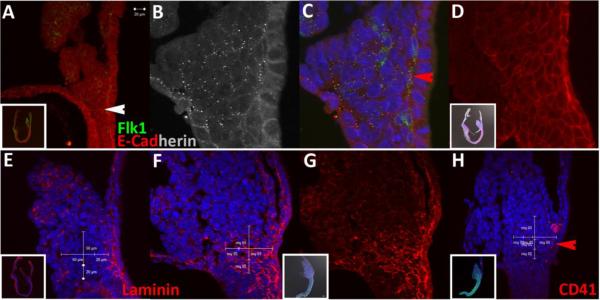
To determine the positioning of blood and vessel segregation at headfold stages, we performed immunohistochemistry on embryos using PECAM-1 and CD41, respectively. PECAM-1 (Newman and Newman, 2003) is an integral membrane glycoprotein that serves as a marker for endothelial cells, while CD41, the alpha subunit of integrin αIIbβ3 (Coller and Shattil, 2008), has been implicated as the first marker for both primitive (Ferkowicz et al., 2003) and definitive (Mikkola et al., 2003) hematopoiesis. Consistent with previous reports (Ema et al., 2006), CD41 and PECAM-1 did not discriminate between blood and endothelial cells at headfold stages. Both monoclonal antibodies reacted with cells throughout blood islands, albeit at different and complicated intensities and subcellular distributions (Fig.3D,H), ie., distributions varied from sample to sample and we were not able to accurately predict the degree of punctate and continuous membrane distributions. Such distributions argue that we did not achieve a sufficient level of sampling and are suggestive of rapid membrane dynamics, which will require increased temporal resolution for improved interpretation. The first consistent site of expression was present in cell clusters located beneath the Ventral Cuboidal Mesothelium (VCM)(red arrows, Fig.3A–C), at the developing site of the vessel of confluence (VOC)(Downs et al., 2009). For example, a rare cluster expressing copious levels of the CD41 aggregation receptor throughout membranes was observed in only one out of five embryos examined. Connected cells extending both proximally and distally from the cluster displayed sparse punctate stains (white arrow, Fig.3C). Other examined embryos exhibited a different type of punctate pattern outside blood islands, which is suggestive of varied dynamic membrane regulation (data not shown). We also observed PECAM-1 expression in disconnected cells surrounding a lumen (Downs, 2003; Zeeb et al., 2010) (green arrows, Fig.3E–G), which supports the view that blood island formation initiates prior to cells reaching the blood island proper.
Figure 3.
Yolk sac blood and endothelial determination initiates near the nook. A and E are posterior views whereas all others are sagittal views. Red arrows in (A–D) mark CD41 expression at the nook in a single embryo. White arrow in (C) marks a sparse punctate cluster in a putative Flk1-positive cell abutting the AX. Green arrows in (E–H) mark PECAM-1 expression at the nook in a single embryo. Therefore, both markers are expressed at the nook during headfold stages. PECAM-1 expressing cells (A,E) are not continuous at this stage. Both CD41 and PECAM-1 expression mark cells at endothelial and blood positions in blood islands (D,H), although expression patterns are not yet predictable and arranged in an ambiguous pattern.
Blood islands can be defined as compositions of blood and endothelial cells derived from lumenized angioblasts (Downs, 2003; Sabin, 2002), or by the now more conventional usage denoting its spatial location, as a belt around the extraembryonic region (Ferkowicz and Yoder, 2005). We decided to adopt the term “nook” to describe the region bounded by the AX, ACD and VCM (the region in which the VOC forms) to emphasize the special characteristics of this environment and its distinct spatial location (Fig.4). Namely, different permeability properties between the ventral and dorsal cuboidal mesothelium have been described (Daane et al., 2011), which could potentially indicate and affect distinct interactions with surrounding cells. Our choice to use the term “nook” was based on its colloquial description as a tucked corner/niche in which stem cells sit (Powell, 2005). We showed that standard markers for blood and endothelial cells are already expressed at the nook, which argues that determination initiates prior to arrival at the blood islands proper. We have summarized our findings based on (Daane et al., 2011) in Fig.4. However, detailed dynamics of angioblast transitions from the epiblast or ACD at headfold stages remain ambiguous and must incorporate live imaging to improve temporal resolution.
Complementary expression of HoxB4 and GATA1 is observed at both the cellular and subcellular levels at headfold stages
Since PECAM-1 and CD41 did not discriminate between blood and endothelial cells at headfold stages, we chose to examine a different combination of markers to distinguish between the blood and vessel populations. GATA1 is required for erythroid development (Orkin and Zon, 1997) and is co-expressed with VE-Cadherin in blood island cells at headfold stages, a subset of which displayed hemangioblast properties (Yokomizo et al., 2007).
At headfold stages, low-level accumulation appears in a subset of Flk1-positive cells distal to the nook (white, green arrows, Fig.5A,B). A population of GATA1 negative angioblasts that display consistent levels just above background was also observed immediately adjacent to GATA1 positive cells at the nook and blood islands (green and teal arrows, Fig.5B,C). We quantified GATA1 nuclear accumulation intensities in a single embryonic section using standard quantification software. In the embryo presented, signals increase monotonically as cells approach blood islands. Total levels in the most immature cell is 2% of a typical signal measured in blood islands, while some Flk1 positive cells expressing GATA1 have been measured as high as 55% intensity compared to cells within blood islands (Fig.5D and data not shown). Assuming constant and unidirectional migration of angioblasts away from the epiblast (please see illustrations in (Coultas et al., 2005)(Huber et al., 2004)) with onset of GATA1 induction at the amniotic insertion site, these data argue that growth of GATA1 accumulation is slow initially and rapid as cells arrive closer to blood islands. However, examinations of other sections did not show monotonic behavior, with high GATA1 positive levels appearing as early as the nook in some instances (red arrow, Fig.5E). Together, these data argue that arrangement of determined hematopoietic populations is influenced by local interactions at headfold stages. To implicate a manner by which this difference may be operating, we chose to examine distribution patterns for HoxB4 (Gould et al., 1998), a homeobox gene (Krumlauf, 1994; Scott, 1992) that suppresses erythroid differentiation when ectopically expressed at high levels (Pilat et al., 2005) and increases repopulation potential when complemented with existing selection strategies (Hills et al., 2010; Kyba et al., 2002). In addition, HoxB4 has been associated with a hematoendothelial phenotype when overexpressed in human embryoid bodies (Unger et al., 2008) and thought to be downregulated upon multipotent progenitor and HSC differentiation based on dynamic expression analysis of FACS sorted fetal liver and embryoid body samples (Pineault et al., 2002).
Figure 5.
HoxB4 and GATA1 segregate at two different levels in headfold stage embryos. GATA1 expression levels are not uniform and are present in some (white, red and yellow arrows, A–C, E) but not all (green and teal arrows) angioblasts. (C) Flk1 is maintained mostly in cells adjacent to VEGF sources (B,C), the visceral endoderm and mesothelium. (D) Quantification of GATA1 levels measured with respect to the amniotic insertion site (AI). Although this particular embryo shows monotonic growth of the signal, a frontal view of a different embryo (E) shows that GATA1 expression is high in some cells at the nook (red arrow). (F–O) HoxB4 and GATA1 distributions at headfold stages. (Green arrows) HoxB4 only, (red arrows) GATA1 only and (yellow arrows) co-expressing cells are found in the (G) allantois, (H) nook and (I) blood islands. (J) Absolute segregation is unambiguous (compare red and green arrows in (J). Nuclear expression of GATA1 and HoxB4 (K–O) is distinct and only occupy non-heterochromatic nuclear space (bright blue DAPI staining). The serial images in (K–O) were taken at 300× resolution with pinhole set at 1μm and rendered using standard visualization software. Red scale bar in (E) is 200μm. White scale bar in (O) is 200nm to show the relationship to size of puncta.
At headfold stages, embryonic expression of HoxB4 is displayed most strongly in mesoderm and ectoderm with rostral boundaries reaching the node ((Forlani et al., 2003) and data not shown). In extraembryonic regions, positive signals were detected at varying levels throughout the ACD, allantois and yolk sac mesoderm, but were selectively high in subsets of cells adjacent to cells expressing GATA1 (Fig.5F–J). This configuration was detected in the allantois (Fig.5G), the nook (Fig.5H) and blood islands (Fig.5I,J). Absolute segregation of HoxB4 and GATA1 to endothelial (green arrows) or blood cells (red arrows), respectively (Fig.5G,J), was apparent adjacent to cells expressing both markers (yellow arrows, Fig.5G–I). Combined, these data argue that GATA1, HoxB4 and GATA1/HoxB4 expression patterns mark three different stable states, viz., blood, endothelial and bipotent celltypes during headfold stages. Moreover, the signals consistently avoided heterochromatin and each other within the nucleus (Fig.5K–O and Supplemental Fig.1).
Dual expression is present in rare populations within 10.5dpc AGM and 11.5dpc fetal liver
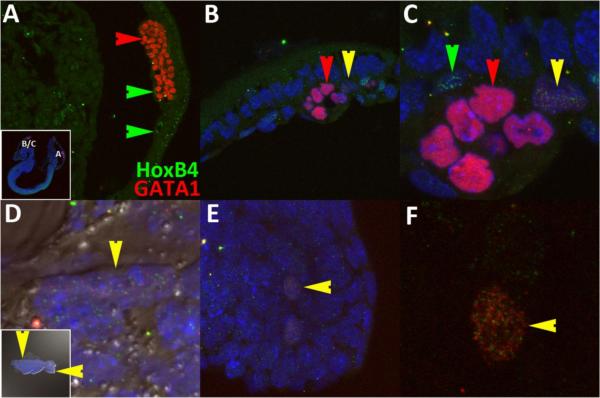
With consideration to theoretical studies (Huang et al., 2007), we postulated that the salt and pepper HoxB4/GATA1 expression signature is a marker of a pluripotent state that may be maintained at later stages. Therefore, if this combined set of characters, defined as observable features at a certain level of integration that is associated with some reference process (Wagner, 2000), is to be used to distinguish HSCs, then the pattern must be present in rare populations within regions of HSC emergence, viz., AGM at 10.5dpc and 11.5dpc fetal liver (Kumaravelu et al., 2002). To determine whether the HoxB4/GATA1 metastable state qualifies as a marker for the HSC, we examined sections for evidence of HSC emergence along possible migratory trajectories. Since these paths have not been defined explicitly, we focused on regions associated with omphalomesenteric artery maturation (OA, also called vitelline artery, VA) (Daane and Downs, 2011; Walls et al., 2008) because the developmental boundaries of the paraortic splanchnopleura, the precursor to the AGM, were too large and ambiguous. The OA matures as extensions from the posterior-lateral regions of the vessel of confluence (VOC) (Daane and Downs, 2011). Hematopoietic cluster formation, which has been associated with HSC emergence, is optimal at 10.5dpc near the OA (Yokomizo and Dzierzak, 2010) and has been associated with HSC emergence. The optimal site of HSC emergence then switches to the fetal liver by ~11.5dpc (Dzierzak and Speck, 2008; Medvinsky et al., 2011).
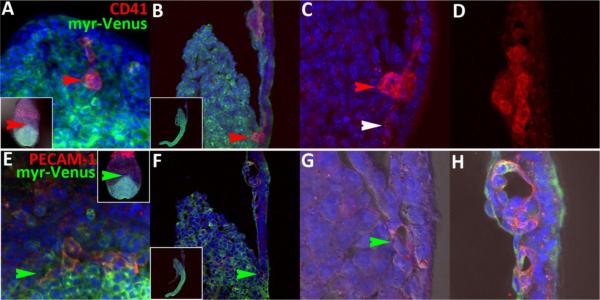
At early somite stages (8.5dpc, Fig.6A–F), medial sections showed that GATA1 and HoxB4 have segregated to either blood (red arrows) or endothelial (green arrows) cells in posterior regions (Fig.6A). However, we were able to detect the combinatorial state in a cell sandwiched between fusing blood islands in rostral blood islands (yellow arrows, Fig.6B,C). Since the OA develops as lateral outgrowths of the VOC, we examined posterior lateral domains. Due to the difficulty of acquiring an appropriate physical section, we mounted the tissue after manual dissection (allantois was removed) and viewed the tissue from the exocoelomic side (inset, Fig.6D). The combinatorial expression state was found in two separated cells at the junction of the body wall and amnion (vertical yellow arrows and not shown, Fig.6D) that will later contribute to the umbilical vein (Walls et al., 2008). In addition, two mesodermal cells located close to one another expressed the signature inside the tailbud (Fig.6E,F). We consistently detected paired states (e.g., Fig.6H) throughout our investigation but do not know the full significance of this observation other than that it suggests overlap in cell cycle regulation.
Figure 6.
HoxB4/GATA1 combinatorial expression is present in different regions regions of 8.5dpc embryos. Lateral views of a sagittal section (A–C) and manual mounted (D–F) 8.5dpc embryo displaying GATA1 only, HoxB4 only and combined expression in yolk sac (A–C), embryo/abembryo junction (vertical yellow arrow, D) and tailbud mesoderm (horizontal yellow arrow, E,F).
No combinatorial states in yolk sacs or embryo at 9.5dpc were found, although it was difficult to get sufficient coverage. More specifically, HoxB4 and GATA1 expression was present in either endothelial or blood cells of the yolk sac, respectively, but no co-expression of GATA1 and HoxB4 puncta in nuclei was detected. In the embryo, many cells in posterior regions co-expressed the markers at low levels throughout the cytoplasm but not in the nucleus (data not shown).
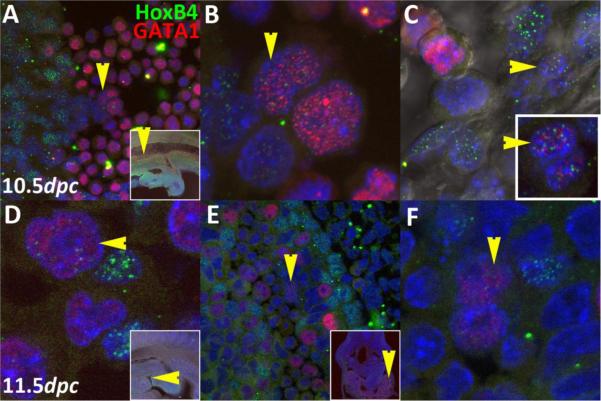
At 10.5dpc (Fig.7A–C), we only detected the combinatorial state in ventral regions of the dorsal aorta. One cluster of four cells was found in an oblique section inside a single hematopoietic cluster ~750μm rostral to the OA (yellow arrows, Fig.7A,B). The cells displayed two different concentrations but were pair-matched. Although downstream events are known to be sensitive to HoxB4 concentration effects (Pilat et al., 2005; Unger et al., 2008), we cannot currently distinguish the limits and significance of the different concentrations in this preliminary report. However, spatial exclusion of GATA1 and HoxB4 puncta could be distinguished in nuclei of even the highest expressing cells. In a different embryo, we detected only one cell ~350μm rostral to the OA (yellow arrows, Fig.7C) that displayed the combinatorial character from examinations of multiple thick sections. This cell was embedded in an endothelial position but its nuclear morphology was closer to that of hematopoietic cells, i.e., it was ellipsoid and folded instead of flattened.
Figure 7.
The combinatorial HoxB4/GATA1 character is present at the two earliest sites with demonstrated HSC potential, viz., 10.5dpc AGM (A–C) and 11.5dpc fetal liver (D–F). Colored arrows represent cells that are either HoxB4 only (green), GATA1 only (red), or both (yellow). Arrow orientation marks the same cell along each row. Anterior is to the left in D and all sagittal sections.
At 11.5dpc (Fig.7D–F), we only detected the combinatorial state in blood forming regions subjacent to the fetal liver cortex. General HoxB4 expression was mostly extinguished from the dorsal aorta by this time, illustrating the transient association between HoxB4 and endothelial cells, but expression was maintained at high levels in fetal liver cortex. We found one cell in one embryonic section (Fig.7D) and two cells close to one another in a different section of a different embryo (Fig.7E,F). On a technical note, detecting these rare populations was particularly challenging at this stage due to presence of a hazy background, which made it difficult to spot the character state unless scanning at higher than 200× (100× lens with 2× confocal software magnification, pinhole set at 1μm optical section). However, the size (~150nm when set under signal saturation), numbers of puncta and their configurations made pluripotent cells distinguishable. To allow the reader to better assess the quality of differences, we generated a supplemental figure (Supplemental Fig.1) of separated channels and selected controls. Based on the above analyses, we argue that the rarity in specific regions of HSC emergence and nowhere else supports our hypothesis that GATA1/HoxB4 combinatorial state is a specific HSC marker.
Using Multiagent-based model of HSC determination at headfold stages as a heuristic
We formalized our observations using Netlogo, a user-friendly open source agent based modeling platform designed in part to facilitate visualization (Ottino, 2010) of complex behaviors typical of general systems (Epstein, 2008; Wilensky, 1999) and to make explicit the hypothetical assumptions that are used for deduction (Woodger, 1930). In addition, the software allows dynamic views and parameter sweeping for economizing executable interrogations of developmental interactions.
Our Netlogo interface is modeled as a frontal view of the inner posterior yolk sac containing patches (immovable spatial positions that reflect product from visceral endoderm and/or mesothelium or matrix-bound secreted products to denote memory of previous migration events) that secrete diffusible chemicals in order to promote cell clustering (Fig.8 and the Netlogo model is available as Supplemental Fig.2). Migrating angioblasts are analogized as agents endowed with the ability to secrete and respond to chemicals (Serini et al., 2003; Wilensky and Resnick, 1999), resulting in probabilistic migration towards the higher chemical gradient.
Figure 8.
Heuristic Netlogo model of Hemangioblast determination. Views of preliminary (A,B) and improved model (C–E). White arrows mark the location of patches that secrete user-controlled chemical levels. White background indicates a visual summary of chemical concentrations. Small red and lime green mark pre-existing blood and endothelial cells, respectively. Targets mark the epiblast or ACD. Therefore, the origin of emerging blood and endothelial cells (big yellow, red and dark green agents) is not clearly defined. The images are export images of two independent simulations. The model is provided as a supplemental figure (Supplemental Fig.2). For more details, please read the Information Tab of the model.
We emphasize that the current model is to be explored and modified by the user as a heuristic for eventual understanding of how both competence and interactions with environmental parameters can affect distribution and determination of hemangioblasts throughout the yolk sac under a given set of assumptions when the full extent of data is not available. Unlike previous reports (Kinder et al., 1999; Ueno and Weissman, 2006), this model assumes migration behaviors as an essential effector of final cell distributions. Moreover, it does not directly incorporate inductive influences (Dyer et al., 2001; Matsuoka et al., 2001) on final cell fate patterning, instead relying heavily on a stochastic, additive process for simplicity.
Formation of the vascular network proved sensitive to prepatterning (Waddington, 2008). That is, specific configurations of chemical sources at the nook, migration zone and blood islands were necessary to generate shapes that resembled a primitive plexus (compare Fig.5E, Fig.8A,B and Fig.8C–E). While VEGF/Flk1 interaction is likely to represent the actual mediators of this process (Carmeliet et al., 1996; Miquerol et al., 1999; Shalaby et al., 1997), our model argues that specific arrangement into a particular configuration is also necessary.
To set up the steady state representative of headfold stages, the model is run for ~1000 ticks/iterations to generate a shape resembling the initial conditions (small lime-green agents, assumed to be Flk1+, Tal1−, Lmo2− (Ema and Rossant, 2003) endothelial cells that mostly emerge at mid-gastrula stages). These iterations were necessary to generate sufficient context to communicate the novel observations reported in the current study. Under these conditions, global flow of cells was toward blood islands but local movement was stochastic. Real migration trajectories have not yet been reported and are a subject of current investigation.
Newly specified hemangioblasts (big yellow clouds, assumed to be Flk1+, Tal1+, Lmo2+) that are considered to correlate with the competence associated with reduced ECad and nuclear Smad1/5 levels (this study) appear from the epiblast/ACD (targets) at a frequency set by the user. New hemangioblasts are initialized with random arbitrary HoxB4 and GATA1 values between 1 and 50 to represent nuclear accumulation. With each iteration/tick, the number is increased for both HoxB4 and GATA1 at stochastic increments between 1 and 5. Although the total amount of GATA1 and HoxB4 proteins is expected to represent a steady state between synthesis and degradation of protein products, little is known of actual kinetics in vivo. The half-life of HoxB4 measured in different FACS sorted populations from different species was ~1 hour (Zhang et al., 2007), while that for GATA1 has proved to be variable, most likely due to differences in inherent competencies of examined cell lines. Moreover, we observed an upper limit for HoxB4 expression in different tissues throughout the embryo, marked by our ability to discern individual punctate distributions and absence from hetereochromatin.
Given the lack of precise details in dynamic protein level regulation, we attempted to capture the essence of the output of the steady state through creation of a “threshold-difference” slider, which allows the user to define the upper limit of the difference at which cells must adopt either a primitive erythroid (red cloud, GATA1+) or endothelial fate (dark green amoeboid cell, HoxB4+). This aspect was included to recognize the implication that promotion of a state is associated with a critical transition, a general feature of complex adaptive systems (Bak and Chen, 1991). The choice to use hematopoietic, endothelial and bipotent fates was based on observations made in this study combined with a previous report that showed three different potentials expressed in subsets of CD45negPFV from embryoid body sorted populations (Wang et al., 2004) and the suggestion that HoxB4 overexpression may induce endothelial fate (Unger et al., 2008).
The user may view the running difference by pausing the simulation and accessing a general feature of the Netlogo interface, “inspect agent”, which will reveal values of the random walk. The general spatial arrangements resembling observed states are used to calibrate HoxB4/GATA1 threshold ranges. We expect the model to be modified and improved over time with integration of real values and observations. In the meantime, our heuristic model attempts to explain dynamic positioning of GATA1/HoxB4 positive cells within the migration zone and posits how graded and binary cell decisions are made in the posterior yolk sac. Although we take several liberties, no similar model currently exists (Roeder and Radtke, 2009) that places a coherent quantitative context for spatial positioning of chemical gradients, cell behaviors and gene expression patterns toward understanding hematopoietic cell fate transitions during mouse headfold stages.
Discussion
On the identification of the pluripotent population
In this report, we attempted to improve resolution of the initial stages of blood and endothelial determination using immunohistochemical analysis of mouse headfold stage embryos. Our strategy was to mark the pluripotent population, either the epiblast or ACD, and determine the parsimonious relationship to blood and endothelial cells based on proximity. Although useful for early bud stages, ECad expression changed over time, which made it difficult to distinguish the spatial origin of Flk1 positive cells after early bud stages. At headfold stages, we observed circular ECad arrangements embedded within the triangle region known to contribute blood cells to the yolk sac (Mikedis and Downs, 2011) and the hematopoietic allantois (Corbel et al., 2007; Zeigler et al., 2006). Specifically, we examined 21 embryos and measured the size of clusters using nuclear arrangements as readout. We were able to detect round clustering behavior in 12 samples, which ranged between 50 and 100μm in diameter. Although the number of cell types that maintain this structure is not entirely clear, blood and endothelial determination were consistently detected at a contact site at the periphery abutting the AX (red arrow, Fig.2C,H). We placed our observations in context of the current map for the posterior region in Fig.4 (Daane et al., 2011).
On HoxB4/GATA1 as a marker of hemangioblasts and HSCs
We identified HoxB4 and GATA1 transcription factors as the first set of markers that becomes restricted to endothelial and blood cells, respectively. Moreover, we identified the nuclear salt and pepper pattern as a combinatorial signature that is shared between hemangioblasts at headfold stages and rare clusters that are found only at later sites of definitive HSC emergence. In consideration with previous reports that showed correlation of a subset of GATA1 positive cells with hemangioblast potential (Yokomizo et al., 2007), it is likely that at least some HoxB4/GATA1 co-expressing cells at headfold stages are hemangioblasts. Although this hypothesis has support, live examinations will be required to connect the immunohistochemical signature to the hemangioblast.
We speculate that the rare populations found at later sites of HSC emergence are HSCs. This hypothesis must be verified using reconstitution or clonal analysis due to the operational definition, although an alternative indirect approach would be to demonstrate correspondence between the HoxB4/GATA1 expression signatures with established HSC characters (Boisset et al., 2010; Kim et al., 2006).
However, there is a deeper issue to associating morphological and genetic markers for defining functional populations. The sensitivity of the operational definition (Morita et al., 2010) to environmental conditions and the tendency of open systems to display similar qualities via different routes have made standards for naming hemangioblasts and HSCs a moving target. Moreover, while the clinicians' reasons (Weissman and Shizuru, 2008) for maintaining rigorous “intentions” (Bertalanffy, 1955) must be respected, the selection and sorting protocols familiar to experimental hematology do not transfer easily to standards utilized by organismal disciplines, the focus of which is to understand how distributed connectivities affect the emergence of behaviors and patterns (Allen, 2004). Therefore, we believe a complementary approach that explicitly represents the combined considerations of experienced stem cell biologists (Dyer et al., 2001; Medvinsky et al., 2011) with clonal (Eilken et al., 2009; Ema et al., 2003; Lancrin et al., 2009) and embryonic (Yokomizo et al., 2007) reports are necessary to reduce conflict as we progress toward a synthesis through interdisciplinary investigations.
As an example, we discuss our observations in context of the recently modified epigenetic landscape (Gilbert, 1991), which considers hematopoiesis as a specific sequence of metastable state transitions driven by transcription factor cross-antagonism (Graf and Enver, 2009). We showed that GATA1 and HoxB4 expression begins to segregate to blood and endothelial cell types prior to arrival at the nook, which is the first site of high CD41 expression (Fig.3A–C). Moreover, PU.1 expression, which labels multipotent progenitor (MPP) cells (Arinobu et al., 2007) initiates at late headfold stages immediately prior to chorio-allantoic fusion and only in blood islands but never at the nook (data not shown). Based on the spacing and timing of marker emergence, this would place the HoxB4/GATA1 cross-antagonism upstream of the GATA1/PU.1 interaction, which implies that HSCs must exist at headfold stages. However, no successful single cell long-term multilineage reconstitution using cells from 7.5dpc embryos has been reported and specific epigenetic differences (Deng and Blobel, 2010; Xu et al., 2011) between hemangioblasts and HSCs are not known. Determining whether these conflicts are enough to demand generation of distinct maps for each independent hematopoietic organ system or whether modification of a single general model is sufficient for comparison across all systems is an open problem. Developing such maps will be useful for economizing strategies to scale-up generation of sustainable therapeutic populations.
On the mechanism of HoxB4/GATA1 cross antagonism
Theoretical studies argue that transcription factor-based metastable states persist only under certain regimes of balanced positive and negative feedback (Huang, 2009). Positive feedback has been experimentally demonstrated for both GATA1 and HoxB4 through binding to its own respective promoter (Gould et al., 1997; Tsai et al., 1991). Negative feedback is expected to be present given the functionally antagonistic roles associated with the respective transcription factors and the persistence of the signature at later stages of development as rare populations. However, defining the specific control mechanism is difficult due to the observed separation of nuclear puncta, which argues that interactions are likely to be indirect.
To our knowledge, all defined transcriptional mechanisms of cross-antagonism, in which abundance of one transcription factor drives simultaneous suppression of the alternative lineage, operate through direct physical interactions in cis or trans to regulate alternative states. This stochastic mechanism appears to be general, as it has proved to operate not only during blood determination but also during the first series of lineage commitments in mouse embryonic patterning (Chen et al., 2009; Niwa et al., 2005; Plusa et al., 2008). For example, it has been shown that determination toward the trophectoderm fate operates through cross antagonism of an ICM promoting transcription factor, Oct3/4, against that of a trophectoderm specific regulator, Cdx2. The mechanism involves generation of a repressor complex comprising both proteins that binds directly to autoregulatory elements, which leads to suppression of the respective cell fates via generation of heterochromatin (Niwa et al., 2005). What is notable about this particular mechanism is that the observed nuclear distribution is exactly opposite to that characterized for the putative HSC metastable condition described in our study.
GATA1 (Wadman et al., 1997) and Hox (Joshi et al., 2007; Mann and Affolter, 1998) proteins act as multimeric complexes, have different optimal cis-binding preferences and share interaction with only one trans factor, the ubiquitous histone acetyltransferase complexes, p300/CBP (Choe et al., 2009; Letting et al., 2003). Therefore, our observations of mutually exclusive HoxB4/GATA1 nuclear patterns detected with confocal laser scanning microscopy (Schermelleh et al., 2008) would support the argument that HATs are recruited to different genomic loci and activate functionally different modules of downstream effectors. Alternatively, gene regulation may involve nucleosome-independent HAT activity. For example, CBP can directly acetylate GATA1 to promote DNA binding (Boyes et al., 1998) and in vitro experiments have shown that CBP binding to Hox proteins reduces acetylase activity while simultaneously sequestering the protein from binding DNA (Shen et al., 2001). Finally, a mechanism that excludes HAT activity altogether but relies only on homeodomain-dependent DNA binding function has been proposed for Hox regulation of the α and γ-globin loci (Shen et al., 2004). It is possible that all three models are operating at the same time to drive opening of distinct genome-wide compartments (Lieberman-Aiden et al., 2009) but understanding the systemic coordination of lineage commitment by HoxB4/GATA1 will require more explicit awareness of affected loci (Jing et al., 2008).
Validating such indirect mechanisms is currently difficult because experimental constraints have not allowed an integrated understanding of local and global forces acting on gene regulation (Belmont et al., 1999; Segal and Widom, 2009). Therefore, the prospect of combining existing medical and engineering technologies (Beck, 1993; Cremer et al., 2004; Kurimoto and Saitou, 2010; Sinclair et al., 2010) to link across this notoriously difficult nano- to micrometer scale (Tsien, 2003) makes the challenge of computing a systemic, predictable awareness of early blood cell determination across the single cell level ultimately surmountable. Understanding such regular events is necessary to enhance awareness of disease mechanisms (Misteli, 2010).
Conclusions
In this report, we identified GATA1/HoxB4 distributions as the earliest combinatorial set of markers that segregate to blood and endothelial cells, respectively. We placed this primary hematopoietic determination event in angioblasts that lie along the transitional visceral endoderm at the time of exit from the posterior streak at headfold stages. We showed that this event is associated with simultaneous increase in Flk1 expression and reduction in ECad and pSmad1/5 nuclear accumulation, which limits the domain influenced by HoxB4 for affecting blood and endothelial cell determination. In consideration with published reports, we speculate that HoxB4/GATA1 cross-antagonism in hemangioblasts operates in part through competitive recruitment of p300/CBP to different genomic loci that stabilizes mutually exclusive states via its effects on nuclear organization. We propose that this signature represents a metastable state that is common to hemangioblasts and HSCs and may be useful as a physiological target for comparison across species and at different stages of ontogenesis.
Our development of simple stereological techniques allows exploration of cell interactions from the epiblast to blood islands during a stage of development that has escaped interrogation due to difficulties with physical manipulation. Although we have discussed specific directions for future research, the range of hierarchies across which this process operates makes the system particularly amenable for examination through multiple perspectives (Robertson and Joyner, 2010; Roeder and Radtke, 2009; Ronneberger et al., 2008). Given the inherent problems associated with integrating across disciplines (Borner et al., 2010), it is necessary to map such information onto a coherent dynamic visual framework (Fox DT, 2009; Rakic, 2009) that can not only support representation of the many simultaneous interactions that are operating in growing animal systems but also improve communication to diverse audiences. For this purpose, we placed our observations onto Waddington's epigenetic landscape, which was envisioned as a revisable but explicit probability map of sequential cell fate bifurcations (Gilbert, 1991). Its utility is embedded in its hierarchical structure, which is shared by all natural complex systems (Koestler, 1969; Riedl, 1978) and may be exploited as a device for comparing the extent of homology across development and evolution (Hemberger et al., 2009; Smith, 2003). For simulation of instances at faster timescales, we utilized a powerful but user-friendly open source executable software platform (Laursen, 2009; Wilensky, 1999) as a heuristic means by which to integrate our current level of understanding and expose open problems for future investigations.
Supplementary Material
Highlights
We resolved the boundaries of mouse hemangioblast maturation at headfold stages.
HoxB4 and GATA1 are the first markers that distinguish vessel and blood lineages.
A third population co-expresses both markers, indicating a metastable state.
We found similar states in rare cells at earliest sites of HSC emergence.
We formalized our observations using a heuristic Netlogo model.
Acknowledgements
We would like to thank Karen Downs for invaluable discussions and support. We thank Susana de Sousa Lopes, Vasil Galat and William Tse for critical reading, attendees at the EMBO workshop on Lineage Commitments at the VIB who provided valuable advice (Downs, 2011), Kinjel Shastri for initiating the Netlogo model, Forrest Stonedahl and the Netlogo user community for general guidance with programming; Sean Flannery for organizing and maintaining mouse colonies, Greg Taborn for mouse and reagent support; Jose Hernandez for animal consultations. The research was supported in part by PHS grant EY020946 from the NIH and the George M. Eisenberg Foundation for Charities.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- Adelman CA, Chattopadhyay S, Bieker JJ. The BMP/BMPR/Smad pathway directs expression of the erythroid-specific EKLF and GATA1 transcription factors during embryoid body differentiation in serum-free media. Development. 2002;129:539–49. doi: 10.1242/dev.129.2.539. [DOI] [PubMed] [Google Scholar]
- Allen GE. A Pact with the Embryo: Viktor Hamburger, Holistic and Mechanistic Philosophy in the Development of Neuroembryology, 1927–1955. Journal of the History of Biology. 2004;37:421–475. doi: 10.1007/s10739-004-4448-2. [DOI] [PubMed] [Google Scholar]
- Arinobu Y, Mizuno S, Chong Y, Shigematsu H, Iino T, Iwasaki H, Graf T, Mayfield R, Chan S, Kastner P, Akashi K. Reciprocal activation of GATA-1 and PU.1 marks initial specification of hematopoietic stem cells into myeloerythroid and myelolymphoid lineages. Cell Stem Cell. 2007;1:416–27. doi: 10.1016/j.stem.2007.07.004. [DOI] [PubMed] [Google Scholar]
- Astorga J, Carlsson P. Hedgehog induction of murine vasculogenesis is mediated by Foxf1 and Bmp4. Development. 2007;134:3753–61. doi: 10.1242/dev.004432. [DOI] [PubMed] [Google Scholar]
- Bak P, Chen K. Self-Organized Criticality. Scientific American. 1991;264:46–53. [Google Scholar]
- Beck RN. Overview of imaging science. Proc Natl Acad Sci U S A. 1993;90:9746–50. doi: 10.1073/pnas.90.21.9746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belaoussoff M, Farrington SM, Baron MH. Hematopoietic induction and respecification of A-P identity by visceral endoderm signaling in the mouse embryo. Development. 1998;125:5009–18. doi: 10.1242/dev.125.24.5009. [DOI] [PubMed] [Google Scholar]
- Belmont AS, Dietzel S, Nye AC, Strukov YG, Tumbar T. Large-scale chromatin structure and function. Curr Opin Cell Biol. 1999;11:307–11. doi: 10.1016/S0955-0674(99)80041-6. [DOI] [PubMed] [Google Scholar]
- Bertalanffy L. v. An Essay on the Relativity of Categories. Philosophy of Science. 1955;22:243–263. [Google Scholar]
- Boisset JC, van Cappellen W, Andrieu-Soler C, Galjart N, Dzierzak E, Robin C. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature. 2010;464:116–20. doi: 10.1038/nature08764. [DOI] [PubMed] [Google Scholar]
- Bonnevie K. New facts on mesoderm formation and proamnion derivatives in the normal mouse embryo. Journal of Morphology. 1950;86:495–545. doi: 10.1002/jmor.1050860303. [DOI] [PubMed] [Google Scholar]
- Borner K, Contractor N, Falk-Krzesinski HJ, Fiore SM, Hall KL, Keyton J, Spring B, Stokols D, Trochim W, Uzzi B. A multi-level systems perspective for the science of team science. Sci Transl Med. 2010;2:49cm24. doi: 10.1126/scitranslmed.3001399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes J, Byfield P, Nakatani Y, Ogryzko V. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 1998;396:594–8. doi: 10.1038/25166. [DOI] [PubMed] [Google Scholar]
- Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, Vallier L. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–5. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- Burdsal CA, Damsky CH, Pedersen RA. The role of E-cadherin and integrins in mesoderm differentiation and migration at the mammalian primitive streak. Development. 1993;118:829–44. doi: 10.1242/dev.118.3.829. [DOI] [PubMed] [Google Scholar]
- Byrd N, Becker S, Maye P, Narasimhaiah R, St-Jacques B, Zhang X, McMahon J, McMahon A, Grabel L. Hedgehog is required for murine yolk sac angiogenesis. Development. 2002;129:361–72. doi: 10.1242/dev.129.2.361. [DOI] [PubMed] [Google Scholar]
- Cantor AB, Orkin SH. Hematopoietic development: a balancing act. Curr Opin Genet Dev. 2001;11:513–9. doi: 10.1016/s0959-437x(00)00226-4. [DOI] [PubMed] [Google Scholar]
- Carmeliet P, Ferreira V, Breier G, Pollefeyt S, Kieckens L, Gertsenstein M, Fahrig M, Vandenhoeck A, Harpal K, Eberhardt C, Declercq C, Pawling J, Moons L, Collen D, Risau W, Nagy A. Abnormal blood vessel development and lethality in embryos lacking a single VEGF allele. Nature. 1996;380:435–9. doi: 10.1038/380435a0. [DOI] [PubMed] [Google Scholar]
- Chan RJ, Yoder MC. The multiple facets of hematopoietic stem cells. Curr Neurovasc Res. 2004;1:197–206. doi: 10.2174/1567202043362324. [DOI] [PubMed] [Google Scholar]
- Chang H, Huylebroeck D, Verschueren K, Guo Q, Matzuk MM, Zwijsen A. Smad5 knockout mice die at mid-gestation due to multiple embryonic and extraembryonic defects. Development. 1999;126:1631–42. doi: 10.1242/dev.126.8.1631. [DOI] [PubMed] [Google Scholar]
- Chen L, Yabuuchi A, Eminli S, Takeuchi A, Lu CW, Hochedlinger K, Daley GQ. Cross-regulation of the Nanog and Cdx2 promoters. Cell Res. 2009;19:1052–61. doi: 10.1038/cr.2009.79. [DOI] [PubMed] [Google Scholar]
- Chen MJ, Li Y, De Obaldia ME, Yang Q, Yzaguirre AD, Yamada-Inagawa T, Vink CS, Bhandoola A, Dzierzak E, Speck NA. Erythroid/Myeloid progenitors and hematopoietic stem cells originate from distinct populations of endothelial cells. Cell Stem Cell. 2011;9:541–52. doi: 10.1016/j.stem.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe SK, Lu P, Nakamura M, Lee J, Sagerstrom CG. Meis cofactors control HDAC and CBP accessibility at Hox-regulated promoters during zebrafish embryogenesis. Dev Cell. 2009;17:561–7. doi: 10.1016/j.devcel.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciruna B, Rossant J. FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Dev Cell. 2001;1:37–49. doi: 10.1016/s1534-5807(01)00017-x. [DOI] [PubMed] [Google Scholar]
- Coller BS, Shattil SJ. The GPIIb/IIIa (integrin alphaIIbbeta3) odyssey: a technology-driven saga of a receptor with twists, turns, and even a bend. Blood. 2008;112:3011–25. doi: 10.1182/blood-2008-06-077891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook BD, Liu S, Evans T. Smad1 signaling restricts hematopoietic potential after promoting hemangioblast commitment. Blood. 2011;117:6489–97. doi: 10.1182/blood-2010-10-312389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbel C, Salaun J, Belo-Diabangouaya P, Dieterlen-Lievre F. Hematopoietic potential of the pre-fusion allantois. Dev Biol. 2007;301:478–88. doi: 10.1016/j.ydbio.2006.08.069. [DOI] [PubMed] [Google Scholar]
- Coultas L, Chawengsaksophak K, Rossant J. Endothelial cells and VEGF in vascular development. Nature. 2005;438:937–45. doi: 10.1038/nature04479. [DOI] [PubMed] [Google Scholar]
- Cremer T, Kupper K, Dietzel S, Fakan S. Higher order chromatin architecture in the cell nucleus: on the way from structure to function. Biol Cell. 2004;96:555–67. doi: 10.1016/j.biolcel.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Daane JM, Downs KM. Hedgehog signaling in the posterior region of the mouse gastrula suggests manifold roles in the fetal-umbilical connection and posterior morphogenesis. Dev Dyn. 2011;240:2175–93. doi: 10.1002/dvdy.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daane JM, Enders AC, Downs KM. Mesothelium of the murine allantois exhibits distinct regional properties. J Morphol. 2011;272:536–56. doi: 10.1002/jmor.10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danchakoff V. Origin of the blood cells. Development of the haematopoetic organs and regeneration of the blood cells from the standpoint of the monophyletic school. The Anatomical Record. 1916;10:397–416. [Google Scholar]
- de Sousa Lopes SM, Roelen BA, Monteiro RM, Emmens R, Lin HY, Li E, Lawson KA, Mummery CL. BMP signaling mediated by ALK2 in the visceral endoderm is necessary for the generation of primordial germ cells in the mouse embryo. Genes Dev. 2004;18:1838–49. doi: 10.1101/gad.294004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Blobel GA. Do chromatin loops provide epigenetic gene expression states? Curr Opin Genet Dev. 2010;20:548–54. doi: 10.1016/j.gde.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps J, van Nes J. Developmental regulation of the Hox genes during axial morphogenesis in the mouse. Development. 2005;132:2931–42. doi: 10.1242/dev.01897. [DOI] [PubMed] [Google Scholar]
- Dobreva MP, Pereira PN, Deprest J, Zwijsen A. On the origin of amniotic stem cells: of mice and men. Int J Dev Biol. 2010;54:761–77. doi: 10.1387/ijdb.092935md. [DOI] [PubMed] [Google Scholar]
- Downs KM. Florence Sabin and the mechanism of blood vessel lumenization during vasculogenesis. Microcirculation. 2003;10:5–25. doi: 10.1038/sj.mn.7800171. [DOI] [PubMed] [Google Scholar]
- Downs KM. The enigmatic primitive streak: prevailing notions and challenges concerning the body axis of mammals. Bioessays. 2009;31:892–902. doi: 10.1002/bies.200900038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs KM. Lineage Commitments: Emphasis On Embryonic-Extraembryonic Interfaces. EMBO Rep. 2011;12:987–90. doi: 10.1038/embor.2011.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs KM, Davies T. Staging of gastrulating mouse embryos by morphological landmarks in the dissecting microscope. Development. 1993;118:1255–66. doi: 10.1242/dev.118.4.1255. [DOI] [PubMed] [Google Scholar]
- Downs KM, Inman KE, Jin DX, Enders AC. The Allantoic Core Domain: new insights into development of the murine allantois and its relation to the primitive streak. Dev Dyn. 2009;238:532–53. doi: 10.1002/dvdy.21862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer MA, Farrington SM, Mohn D, Munday JR, Baron MH. Indian hedgehog activates hematopoiesis and vasculogenesis and can respecify prospective neurectodermal cell fate in the mouse embryo. Development. 2001;128:1717–30. doi: 10.1242/dev.128.10.1717. [DOI] [PubMed] [Google Scholar]
- Dzierzak E, Speck NA. Of lineage and legacy: the development of mammalian hematopoietic stem cells. Nat Immunol. 2008;9:129–36. doi: 10.1038/ni1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilken HM, Nishikawa S, Schroeder T. Continuous single-cell imaging of blood generation from haemogenic endothelium. Nature. 2009;457:896–900. doi: 10.1038/nature07760. [DOI] [PubMed] [Google Scholar]
- Ema M, Faloon P, Zhang WJ, Hirashima M, Reid T, Stanford WL, Orkin S, Choi K, Rossant J. Combinatorial effects of Flk1 and Tal1 on vascular and hematopoietic development in the mouse. Genes Dev. 2003;17:380–93. doi: 10.1101/gad.1049803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ema M, Rossant J. Cell fate decisions in early blood vessel formation. Trends Cardiovasc Med. 2003;13:254–9. doi: 10.1016/s1050-1738(03)00105-1. [DOI] [PubMed] [Google Scholar]
- Ema M, Yokomizo T, Wakamatsu A, Terunuma T, Yamamoto M, Takahashi S. Primitive erythropoiesis from mesodermal precursors expressing VE-cadherin, PECAM-1, Tie2, endoglin, and CD34 in the mouse embryo. Blood. 2006;108:4018–24. doi: 10.1182/blood-2006-03-012872. [DOI] [PubMed] [Google Scholar]
- Epstein J. Why Model? Journal of Artificial Societies and Social Simulation. 2008;11:12. [Google Scholar]
- Ferkowicz MJ, Starr M, Xie X, Li W, Johnson SA, Shelley WC, Morrison PR, Yoder MC. CD41 expression defines the onset of primitive and definitive hematopoiesis in the murine embryo. Development. 2003;130:4393–403. doi: 10.1242/dev.00632. [DOI] [PubMed] [Google Scholar]
- Ferkowicz MJ, Yoder MC. Blood island formation: longstanding observations and modern interpretations. Exp Hematol. 2005;33:1041–7. doi: 10.1016/j.exphem.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Forlani S, Lawson KA, Deschamps J. Acquisition of Hox codes during gastrulation and axial elongation in the mouse embryo. Development. 2003;130:3807–19. doi: 10.1242/dev.00573. [DOI] [PubMed] [Google Scholar]
- Fox DT ML. Nystul T, Spradling AC, Lineage analysis of stem cells. StemBook. 2009 [PubMed] [Google Scholar]
- Fujiwara T, Dunn NR, Hogan BL. Bone morphogenetic protein 4 in the extraembryonic mesoderm is required for allantois development and the localization and survival of primordial germ cells in the mouse. Proc Natl Acad Sci U S A. 2001;98:13739–44. doi: 10.1073/pnas.241508898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda T, Scott G, Komatsu Y, Araya R, Kawano M, Ray MK, Yamada M, Mishina Y. Generation of a mouse with conditionally activated signaling through the BMP receptor, ALK2. Genesis. 2006;44:159–67. doi: 10.1002/dvg.20201. [DOI] [PubMed] [Google Scholar]
- Gardner RL. Clonal analysis of early mammalian development. Philos Trans R Soc Lond B Biol Sci. 1985;312:163–78. doi: 10.1098/rstb.1985.0186. [DOI] [PubMed] [Google Scholar]
- Gilbert SF. Epigenetic Landscaping: Waddington's Use of Cell Fate Bifurcation Diagrams. Biology and Philosophy. 1991;6:135–154. [Google Scholar]
- Gilbert SF. All I really needed to know I learned during gastrulation. CBE Life Sci Educ. 2008;7:12–3. doi: 10.1187/lse.7.1.cbe12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould A, Itasaki N, Krumlauf R. Initiation of rhombomeric Hoxb4 expression requires induction by somites and a retinoid pathway. Neuron. 1998;21:39–51. doi: 10.1016/s0896-6273(00)80513-9. [DOI] [PubMed] [Google Scholar]
- Gould A, Morrison A, Sproat G, White RA, Krumlauf R. Positive cross-regulation and enhancer sharing: two mechanisms for specifying overlapping Hox expression patterns. Genes Dev. 1997;11:900–13. doi: 10.1101/gad.11.7.900. [DOI] [PubMed] [Google Scholar]
- Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462:587–94. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- Hamill KJ, Kligys K, Hopkinson SB, Jones JC. Laminin deposition in the extracellular matrix: a complex picture emerges. J Cell Sci. 2009;122:4409–17. doi: 10.1242/jcs.041095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemberger M, Dean W, Reik W. Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington's canal. Nat Rev Mol Cell Biol. 2009;10:526–37. doi: 10.1038/nrm2727. [DOI] [PubMed] [Google Scholar]
- Hilliard JE, Lawson LR. Stereology and Stochastic Geometry. Kluwer Academic Publishers; 2003. [Google Scholar]
- Hills D, Gribi R, Ure J, Buza-Vidas N, Luc S, Jacobsen SE, Medvinsky A. Hoxb4-YFP reporter mouse model: a novel tool for tracking HSC development and studying the role of Hoxb4 in hematopoiesis. Blood. 2010 doi: 10.1182/blood-2009-12-253989. [DOI] [PubMed] [Google Scholar]
- Hong S, Troyanovsky RB, Troyanovsky SM. Spontaneous assembly and active disassembly balance adherens junction homeostasis. Proc Natl Acad Sci U S A. 2010;107:3528–33. doi: 10.1073/pnas.0911027107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S. Reprogramming cell fates: reconciling rarity with robustness. Bioessays. 2009;31:546–60. doi: 10.1002/bies.200800189. [DOI] [PubMed] [Google Scholar]
- Huang S, Guo YP, May G, Enver T. Bifurcation dynamics in lineage-commitment in bipotent progenitor cells. Dev Biol. 2007;305:695–713. doi: 10.1016/j.ydbio.2007.02.036. [DOI] [PubMed] [Google Scholar]
- Huber TL, Kouskoff V, Fehling HJ, Palis J, Keller G. Haemangioblast commitment is initiated in the primitive streak of the mouse embryo. Nature. 2004;432:625–30. doi: 10.1038/nature03122. [DOI] [PubMed] [Google Scholar]
- Jing H, Vakoc CR, Ying L, Mandat S, Wang H, Zheng X, Blobel GA. Exchange of GATA factors mediates transitions in looped chromatin organization at a developmentally regulated gene locus. Mol Cell. 2008;29:232–42. doi: 10.1016/j.molcel.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi R, Passner JM, Rohs R, Jain R, Sosinsky A, Crickmore MA, Jacob V, Aggarwal AK, Honig B, Mann RS. Functional specificity of a Hox protein mediated by the recognition of minor groove structure. Cell. 2007;131:530–43. doi: 10.1016/j.cell.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan NG, Stemmler MP, Junghans D, Kanzler B, de Vries WN, Dominis M, Kemler R. Gene replacement reveals a specific role for E-cadherin in the formation of a functional trophectoderm. Development. 2007;134:31–41. doi: 10.1242/dev.02722. [DOI] [PubMed] [Google Scholar]
- Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, Ellis J, Keller G. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–40. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Kiel MJ, Yilmaz OH, Iwashita T, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–21. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- Kim I, He S, Yilmaz OH, Kiel MJ, Morrison SJ. Enhanced purification of fetal liver hematopoietic stem cells using SLAM family receptors. Blood. 2006;108:737–44. doi: 10.1182/blood-2005-10-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinder SJ, Tsang TE, Quinlan GA, Hadjantonakis AK, Nagy A, Tam PP. The orderly allocation of mesodermal cells to the extraembryonic structures and the anteroposterior axis during gastrulation of the mouse embryo. Development. 1999;126:4691–701. doi: 10.1242/dev.126.21.4691. [DOI] [PubMed] [Google Scholar]
- Koestler A. Beyond Atomism and Holism- The concept of the Holon. In: Smythies A. K. a. J. R., editor. The Alpbach Symposium 1968: Beyond Reductionism, New perspectives in the life sciences. The Macmillan Company; 1969. pp. 192–232. [Google Scholar]
- Krumlauf R. Cell. Vol. 78. 1994. Hox genes in vertebrate development; pp. 191–201. [DOI] [PubMed] [Google Scholar]
- Kumaravelu P, Hook L, Morrison AM, Ure J, Zhao S, Zuyev S, Ansell J, Medvinsky A. Development. Vol. 129. 2002. Quantitative developmental anatomy of definitive haematopoietic stem cells/long-term repopulating units (HSC/RUs): role of the aorta-gonad-mesonephros (AGM) region and the yolk sac in colonisation of the mouse embryonic liver; pp. 4891–9. [DOI] [PubMed] [Google Scholar]
- Kurimoto K, Saitou M. Curr Opin Genet Dev. Vol. 20. 2010. Single-cell cDNA microarray profiling of complex biological processes of differentiation; pp. 470–7. [DOI] [PubMed] [Google Scholar]
- Kyba M, Perlingeiro RC, Daley GQ. Cell. Vol. 109. 2002. HoxB4 confers definitive lymphoid-myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors; pp. 29–37. [DOI] [PubMed] [Google Scholar]
- Lancrin C, Sroczynska P, Stephenson C, Allen T, Kouskoff V, Lacaud G. Nature. Vol. 457. 2009. The haemangioblast generates haematopoietic cells through a haemogenic endothelium stage; pp. 892–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laursen L. Nature. Vol. 462. 2009. Computational biology: Biological logic; pp. 408–10. [DOI] [PubMed] [Google Scholar]
- Lawson KA, Dunn NR, Roelen BA, Zeinstra LM, Davis AM, Wright CV, Korving JP, Hogan BL. Genes Dev. Vol. 13. 1999. Bmp4 is required for the generation of primordial germ cells in the mouse embryo; pp. 424–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson KA, Meneses JJ, Pedersen RA. Development. Vol. 113. 1991. Clonal analysis of epiblast fate during germ layer formation in the mouse embryo; pp. 891–911. [DOI] [PubMed] [Google Scholar]
- Letting DL, Rakowski C, Weiss MJ, Blobel GA. Mol Cell Biol. Vol. 23. 2003. Formation of a tissue-specific histone acetylation pattern by the hematopoietic transcription factor GATA-1; pp. 1334–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J. Science. Vol. 326. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome; pp. 289–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann RS, Affolter M. Curr Opin Genet Dev. Vol. 8. 1998. Hox proteins meet more partners; pp. 423–9. [DOI] [PubMed] [Google Scholar]
- Massague J, Seoane J, Wotton D. Genes Dev. Vol. 19. 2005. Smad transcription factors; pp. 2783–810. [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Tsuji K, Hisakawa H, Xu M, Ebihara Y, Ishii T, Sugiyama D, Manabe A, Tanaka R, Ikeda Y, Asano S, Nakahata T. Blood. Vol. 98. 2001. Generation of definitive hematopoietic stem cells from murine early yolk sac and paraaortic splanchnopleures by aorta-gonad-mesonephros region-derived stromal cells; pp. 6–12. [DOI] [PubMed] [Google Scholar]
- McLaren A. Primordial germ cells in the mouse. Dev Biol. 2003;262:1–15. doi: 10.1016/s0012-1606(03)00214-8. [DOI] [PubMed] [Google Scholar]
- Medvinsky A, Rybtsov S, Taoudi S. Embryonic origin of the adult hematopoietic system: advances and questions. Development. 2011;138:1017–31. doi: 10.1242/dev.040998. [DOI] [PubMed] [Google Scholar]
- Mikedis MM, Downs KM. Collagen type IV and Perlecan exhibit dynamic localization in the Allantoic Core Domain, a putative stem cell niche in the murine allantois. Dev Dyn. 2009;238:3193–204. doi: 10.1002/dvdy.22129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikedis MM, Downs KM. STELLA-positive subregions of the primitive streak contribute to posterior tissues of the mouse gastrula. Dev Biol. 2011 doi: 10.1016/j.ydbio.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkola HK, Fujiwara Y, Schlaeger TM, Traver D, Orkin SH. Expression of CD41 marks the initiation of definitive hematopoiesis in the mouse embryo. Blood. 2003;101:508–16. doi: 10.1182/blood-2002-06-1699. [DOI] [PubMed] [Google Scholar]
- Miquerol L, Gertsenstein M, Harpal K, Rossant J, Nagy A. Multiple developmental roles of VEGF suggested by a LacZ-tagged allele. Dev Biol. 1999;212:307–22. doi: 10.1006/dbio.1999.9355. [DOI] [PubMed] [Google Scholar]
- Misteli T. Higher-order genome organization in human disease. Cold Spring Harb Perspect Biol. 2010;2:a000794. doi: 10.1101/cshperspect.a000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitiku N, Baker JC. Genomic analysis of gastrulation and organogenesis in the mouse. Dev Cell. 2007;13:897–907. doi: 10.1016/j.devcel.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Morita Y, Ema H, Nakauchi H. Heterogeneity and hierarchy within the most primitive hematopoietic stem cell compartment. J Exp Med. 2010;207:1173–82. doi: 10.1084/jem.20091318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray PDF. Proceedings of the Royal Society of London. Vol. 111. 1932. The Development in vitro of the Blood of the Early Chick Embryo; pp. 497–521. (Series B). [Google Scholar]
- Newman PJ, Newman DK. Signal transduction pathways mediated by PECAM-1: new roles for an old molecule in platelet and vascular cell biology. Arterioscler Thromb Vasc Biol. 2003;23:953–64. doi: 10.1161/01.ATV.0000071347.69358.D9. [DOI] [PubMed] [Google Scholar]
- Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, Rossant J. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123:917–29. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- Orkin SH, Zon LI. Genetics of erythropoiesis: induced mutations in mice and zebrafish. Annu Rev Genet. 1997;31:33–60. doi: 10.1146/annurev.genet.31.1.33. [DOI] [PubMed] [Google Scholar]
- Ottino J. The art of mixing with an admixture of art: Fluids, solids, and visual imagination. Physics of Fluids. 2010;22:12. [Google Scholar]
- Park C, Afrikanova I, Chung YS, Zhang WJ, Arentson E, Fong Gh G, Rosendahl A, Choi K. A hierarchical order of factors in the generation of FLK1- and SCL-expressing hematopoietic and endothelial progenitors from embryonic stem cells. Development. 2004;131:2749–62. doi: 10.1242/dev.01130. [DOI] [PubMed] [Google Scholar]
- Pilat S, Carotta S, Schiedlmeier B, Kamino K, Mairhofer A, Will E, Modlich U, Steinlein P, Ostertag W, Baum C, Beug H, Klump H. HOXB4 enforces equivalent fates of ES-cell-derived and adult hematopoietic cells. Proc Natl Acad Sci U S A. 2005;102:12101–6. doi: 10.1073/pnas.0505624102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineault N, Helgason CD, Lawrence HJ, Humphries RK. Differential expression of Hox, Meis1, and Pbx1 genes in primitive cells throughout murine hematopoietic ontogeny. Exp Hematol. 2002;30:49–57. doi: 10.1016/s0301-472x(01)00757-3. [DOI] [PubMed] [Google Scholar]
- Plusa B, Piliszek A, Frankenberg S, Artus J, Hadjantonakis AK. Distinct sequential cell behaviours direct primitive endoderm formation in the mouse blastocyst. Development. 2008;135:3081–91. doi: 10.1242/dev.021519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell K. Stem-cell niches: it's the ecology, stupid! Nature. 2005;435:268–70. doi: 10.1038/435268a. [DOI] [PubMed] [Google Scholar]
- Rakeman AS, Anderson KV. Axis specification and morphogenesis in the mouse embryo require Nap1, a regulator of WAVE-mediated actin branching. Development. 2006;133:3075–83. doi: 10.1242/dev.02473. [DOI] [PubMed] [Google Scholar]
- Rakic P. Evolution of the neocortex: a perspective from developmental biology. Nat Rev Neurosci. 2009;10:724–35. doi: 10.1038/nrn2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl R. A Wiley-Interscience publication. Wiley, Chichester; New York: 1978. Order in living organisms : a systems analysis of evolution; pp. 58–59. [Google Scholar]
- Robb L, Lyons I, Li R, Hartley L, Kontgen F, Harvey RP, Metcalf D, Begley CG. Absence of yolk sac hematopoiesis from mice with a targeted disruption of the scl gene. Proc Natl Acad Sci U S A. 1995;92:7075–9. doi: 10.1073/pnas.92.15.7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson L, Joyner A. An interview with Alex Joyner and Liz Robertson: development editors at the helm of Developmental Biology Societies. Development. 2010;137:2075–7. doi: 10.1242/dev.054098. [DOI] [PubMed] [Google Scholar]
- Roeder I, Radtke F. Stem cell biology meets systems biology. Development. 2009;136:3525–30. doi: 10.1242/dev.040758. [DOI] [PubMed] [Google Scholar]
- Ronneberger O, Baddeley D, Scheipl F, Verveer PJ, Burkhardt H, Cremer C, Fahrmeir L, Cremer T, Joffe B. Spatial quantitative analysis of fluorescently labeled nuclear structures: problems, methods, pitfalls. Chromosome Res. 2008;16:523–62. doi: 10.1007/s10577-008-1236-4. [DOI] [PubMed] [Google Scholar]
- Sabin FR. On the origin of the cells of the blood. Physiological Reviews. 1922;2:38–69. [Google Scholar]
- Sabin FR. Preliminary note on the differentiation of angioblasts and the method by which they produce blood-vessels, blood-plasma and red blood-cells as seen in the living chick. 1917. J Hematother Stem Cell Res. 2002;11:5–7. doi: 10.1089/152581602753448496. [DOI] [PubMed] [Google Scholar]
- Saitou M, Yamaji M. Germ cell specification in mice: signaling, transcription regulation, and epigenetic consequences. Reproduction. 2010;139:931–42. doi: 10.1530/REP-10-0043. [DOI] [PubMed] [Google Scholar]
- Schermelleh L, Carlton PM, Haase S, Shao L, Winoto L, Kner P, Burke B, Cardoso MC, Agard DA, Gustafsson MG, Leonhardt H, Sedat JW. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 2008;320:1332–6. doi: 10.1126/science.1156947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott MP. Vertebrate homeobox gene nomenclature. Cell. 1992;71:551–3. doi: 10.1016/0092-8674(92)90588-4. [DOI] [PubMed] [Google Scholar]
- Segal E, Widom J. From DNA sequence to transcriptional behaviour: a quantitative approach. Nat Rev Genet. 2009;10:443–56. doi: 10.1038/nrg2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serini G, Ambrosi D, Giraudo E, Gamba A, Preziosi L, Bussolino F. Modeling the early stages of vascular network assembly. EMBO J. 2003;22:1771–9. doi: 10.1093/emboj/cdg176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalaby F, Ho J, Stanford WL, Fischer KD, Schuh AC, Schwartz L, Bernstein A, Rossant J. A requirement for Flk1 in primitive and definitive hematopoiesis and vasculogenesis. Cell. 1997;89:981–90. doi: 10.1016/s0092-8674(00)80283-4. [DOI] [PubMed] [Google Scholar]
- Shen W, Chrobak D, Krishnan K, Lawrence HJ, Largman C. HOXB6 protein is bound to CREB-binding protein and represses globin expression in a DNA binding-dependent, PBX interaction-independent process. J Biol Chem. 2004;279:39895–904. doi: 10.1074/jbc.M404132200. [DOI] [PubMed] [Google Scholar]
- Shen WF, Krishnan K, Lawrence HJ, Largman C. The HOX homeodomain proteins block CBP histone acetyltransferase activity. Mol Cell Biol. 2001;21:7509–22. doi: 10.1128/MCB.21.21.7509-7522.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorenko SP, Clark EA. The dual-function CD150 receptor subfamily: the viral attraction. Nat Immunol. 2003;4:19–24. doi: 10.1038/ni0103-19. [DOI] [PubMed] [Google Scholar]
- Sinclair P, Bian Q, Plutz M, Heard E, Belmont AS. Dynamic plasticity of large-scale chromatin structure revealed by self-assembly of engineered chromosome regions. J Cell Biol. 2010;190:761–76. doi: 10.1083/jcb.200912167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KK. Time's arrow: heterochrony and the evolution of development. Int J Dev Biol. 2003;47:613–21. [PubMed] [Google Scholar]
- Stern CD, Charite J, Deschamps J, Duboule D, Durston AJ, Kmita M, Nicolas JF, Palmeirim I, Smith JC, Wolpert L. Head-tail patterning of the vertebrate embryo: one, two or many unresolved problems? Int J Dev Biol. 2006;50:3–15. doi: 10.1387/ijdb.052095cs. [DOI] [PubMed] [Google Scholar]
- Tam PP, Beddington RS. The formation of mesodermal tissues in the mouse embryo during gastrulation and early organogenesis. Development. 1987;99:109–26. doi: 10.1242/dev.99.1.109. [DOI] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–9. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Till JE, McCulloch EA, Siminovitch L. A Stochastic Model of Stem Cell Proliferation, Based on the Growth of Spleen Colony-Forming Cells. Proc Natl Acad Sci U S A. 1964;51:29–36. doi: 10.1073/pnas.51.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay KD, Dunn NR, Robertson EJ. Mouse embryos lacking Smad1 signals display defects in extra-embryonic tissues and germ cell formation. Development. 2001;128:3609–21. doi: 10.1242/dev.128.18.3609. [DOI] [PubMed] [Google Scholar]
- Tsai SF, Strauss E, Orkin SH. Functional analysis and in vivo footprinting implicate the erythroid transcription factor GATA-1 as a positive regulator of its own promoter. Genes Dev. 1991;5:919–31. doi: 10.1101/gad.5.6.919. [DOI] [PubMed] [Google Scholar]
- Tsien RY. Imagining imaging's future. Nat Rev Mol Cell Biol. 2003;(Suppl):SS16–21. [PubMed] [Google Scholar]
- Ueno H, Weissman IL. Clonal analysis of mouse development reveals a polyclonal origin for yolk sac blood islands. Dev Cell. 2006;11:519–33. doi: 10.1016/j.devcel.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Unger C, Karner E, Treschow A, Stellan B, Felldin U, Concha H, Wendel M, Hovatta O, Aints A, Ahrlund-Richter L, Dilber MS. Lentiviral-mediated HoxB4 expression in human embryonic stem cells initiates early hematopoiesis in a dose-dosedependent manner but does not promote myeloid differentiation. Stem Cells. 2008;26:2455–66. doi: 10.1634/stemcells.2007-0876. [DOI] [PubMed] [Google Scholar]
- Vasiev B, Balter A, Chaplain M, Glazier JA, Weijer CJ. Modeling gastrulation in the chick embryo: formation of the primitive streak. PLoS One. 2010;5:e10571. doi: 10.1371/journal.pone.0010571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voiculescu O, Bertocchini F, Wolpert L, Keller RE, Stern CD. The amniote primitive streak is defined by epithelial cell intercalation before gastrulation. Nature. 2007;449:1049–52. doi: 10.1038/nature06211. [DOI] [PubMed] [Google Scholar]
- Waddington CH. Form and Information. Biological Theory. 2008;3:267–283. [Google Scholar]
- Wadman IA, Osada H, Grutz GG, Agulnick AD, Westphal H, Forster A, Rabbitts TH. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 1997;16:3145–57. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GP. The character concept in evolutionary biology. Academic Press; San Diego, Calif.: 2000. Characters, Units and Natural Kinds: An Introduction; pp. 1–12. [Google Scholar]
- Waldrip WR, Bikoff EK, Hoodless PA, Wrana JL, Robertson EJ. Smad2 signaling in extraembryonic tissues determines anterior-posterior polarity of the early mouse embryo. Cell. 1998;92:797–808. doi: 10.1016/s0092-8674(00)81407-5. [DOI] [PubMed] [Google Scholar]
- Walls JR, Coultas L, Rossant J, Henkelman RM. Three-dimensional analysis of vascular development in the mouse embryo. PLoS One. 2008;3:e2853. doi: 10.1371/journal.pone.0002853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Li L, Shojaei F, Levac K, Cerdan C, Menendez P, Martin T, Rouleau A, Bhatia M. Endothelial and hematopoietic cell fate of human embryonic stem cells originates from primitive endothelium with hemangioblastic properties. Immunity. 2004;21:31–41. doi: 10.1016/j.immuni.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Wang LD, Wagers AJ. Dynamic niches in the origination and differentiation of haematopoietic stem cells. Nat Rev Mol Cell Biol. 2011;12:643–55. doi: 10.1038/nrm3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warr MR, Pietras EM, Passegue E. Mechanisms controlling hematopoietic stem cell functions during normal hematopoiesis and hematological malignancies. Wiley Interdiscip Rev Syst Biol Med. 2011;3:681–701. doi: 10.1002/wsbm.145. [DOI] [PubMed] [Google Scholar]
- Weissman IL, Shizuru JA. The origins of the identification and isolation of hematopoietic stem cells, and their capability to induce donor-specific transplantation tolerance and treat autoimmune diseases. Blood. 2008;112:3543–53. doi: 10.1182/blood-2008-08-078220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilensky U. Netlogo. 1999 http://ccl.northwestern.edu/netlogo/
- Wilensky U, Resnick M. Thinking in Levels: A Dynamic Systems Approach to Making Sense of the World. Journal of Science Education and Technology. 1999;8:3–19. [Google Scholar]
- Winnier G, Blessing M, Labosky PA, Hogan BL. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 1995;9:2105–16. doi: 10.1101/gad.9.17.2105. [DOI] [PubMed] [Google Scholar]
- Woodger JH. The “Concept of Organism” and the Relation Between Embryology and Genetics Part. II. The Quarterly Review of Biology. 1930;5:438–463. [Google Scholar]
- Xu CR, Cole PA, Meyers DJ, Kormish J, Dent S, Zaret KS. Chromatin “prepattern” and histone modifiers in a fate choice for liver and pancreas. Science. 2011;332:963–6. doi: 10.1126/science.1202845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita J, Itoh H, Hirashima M, Ogawa M, Nishikawa S, Yurugi T, Naito M, Nakao K. Flk1-positive cells derived from embryonic stem cells serve as vascular progenitors. Nature. 2000;408:92–6. doi: 10.1038/35040568. [DOI] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–23. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder MC. Generation of HSCs in the embryo and assays to detect them. Oncogene. 2004;23:7161–3. doi: 10.1038/sj.onc.1207931. [DOI] [PubMed] [Google Scholar]
- Yokomizo T, Dzierzak E. Three-dimensional cartography of hematopoietic clusters in the vasculature of whole mouse embryos. Development. 2010;137:3651–61. doi: 10.1242/dev.051094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokomizo T, Takahashi S, Mochizuki N, Kuroha T, Ema M, Wakamatsu A, Shimizu R, Ohneda O, Osato M, Okada H, Komori T, Ogawa M, Nishikawa S, Ito Y, Yamamoto M. Characterization of GATA-1(+) hemangioblastic cells in the mouse embryo. EMBO J. 2007;26:184–96. doi: 10.1038/sj.emboj.7601480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafonte BT, Liu S, Lynch-Kattman M, Torregroza I, Benvenuto L, Kennedy M, Keller G, Evans T. Smad1 expands the hemangioblast population within a limited developmental window. Blood. 2007;109:516–23. doi: 10.1182/blood-2006-02-004564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamir EA, Rongish BJ, Little CD. The ECM moves during primitive streak formation--computation of ECM versus cellular motion. PLoS Biol. 2008;6:e247. doi: 10.1371/journal.pbio.0060247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeb M, Strilic B, Lammert E. Resolving cell-cell junctions: lumen formation in blood vessels. Curr Opin Cell Biol. 2010;22:626–32. doi: 10.1016/j.ceb.2010.07.003. [DOI] [PubMed] [Google Scholar]
- Zeigler BM, Sugiyama D, Chen M, Guo Y, Downs KM, Speck NA. The allantois and chorion, when isolated before circulation or chorio-allantoic fusion, have hematopoietic potential. Development. 2006;133:4183–92. doi: 10.1242/dev.02596. [DOI] [PubMed] [Google Scholar]
- Zhang XB, Schwartz JL, Humphries RK, Kiem HP. Effects of HOXB4 overexpression on ex vivo expansion and immortalization of hematopoietic cells from different species. Stem Cells. 2007;25:2074–81. doi: 10.1634/stemcells.2006-0742. [DOI] [PubMed] [Google Scholar]
- Zovein AC, Turlo KA, Ponec RM, Lynch MR, Chen KC, Hofmann JJ, Cox TC, Gasson JC, Iruela-Arispe ML. Vascular remodeling of the vitelline artery initiates extravascular emergence of hematopoietic clusters. Blood. 2010;116:3435–44. doi: 10.1182/blood-2010-04-279497. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.