Abstract
Cerebellum development involves the coordinated production of multiple neuronal cell types. The cerebellar primordium contains two germinative zones, the rhombic lip (RL) and the ventricular zone (VZ), which generate the different types of glutamatergic and GABAergic neurons, respectively. What regulates the specification and production of glutamatergic and GABAergic neurons as well as the subtypes for each of these two broad classes remains largely unknown. Here we demonstrate with conditional genetic approaches in mice that SMAD4, a major mediator of BMP and TGFβ signaling, is required early in cerebellar development for maintaining the RL and generating subsets of RL-derived glutamatergic neurons, namely neurons of the deep cerebellar nuclei, unipolar brush cells, and the late cohort of granule cell precursors (GCPs). The early cohort of GCPs, despite being deficient for SMAD4, are still generated. In addition, the numbers of GABAergic neurons are reduced in the mutant and the distribution of Purkinje cells becomes abnormal. These studies demonstrate a temporally and spatially restricted requirement for SMAD4 in generating subtypes of cerebellar neurons.
Keywords: SMAD4, rhombic lip, granule cell precursor, deep cerebellar nuclei, unipolar brush cells, mouse
INTRODUCTION
The cerebellum is essential for motor coordination and certain cognitive functions (Schmahmann, 2004). It forms from progenitor cells above the fourth ventricle in dorsal rhombomere 1 (rh1) of the hindbrain. Cerebellar progenitors that give rise to excitatory (glutamatergic) neurons reside in the rhombic lip (RL) whereas those that give rise to inhibitory (GABAergic) neurons reside in the ventricular zone (VZ) (Fig. 1A) (Altman and Bayer, 1977; Hoshino et al., 2005; Machold and Fishell, 2005; Wang et al., 2005). The molecular pathways that generate these two subtypes of neurons remain unclear.
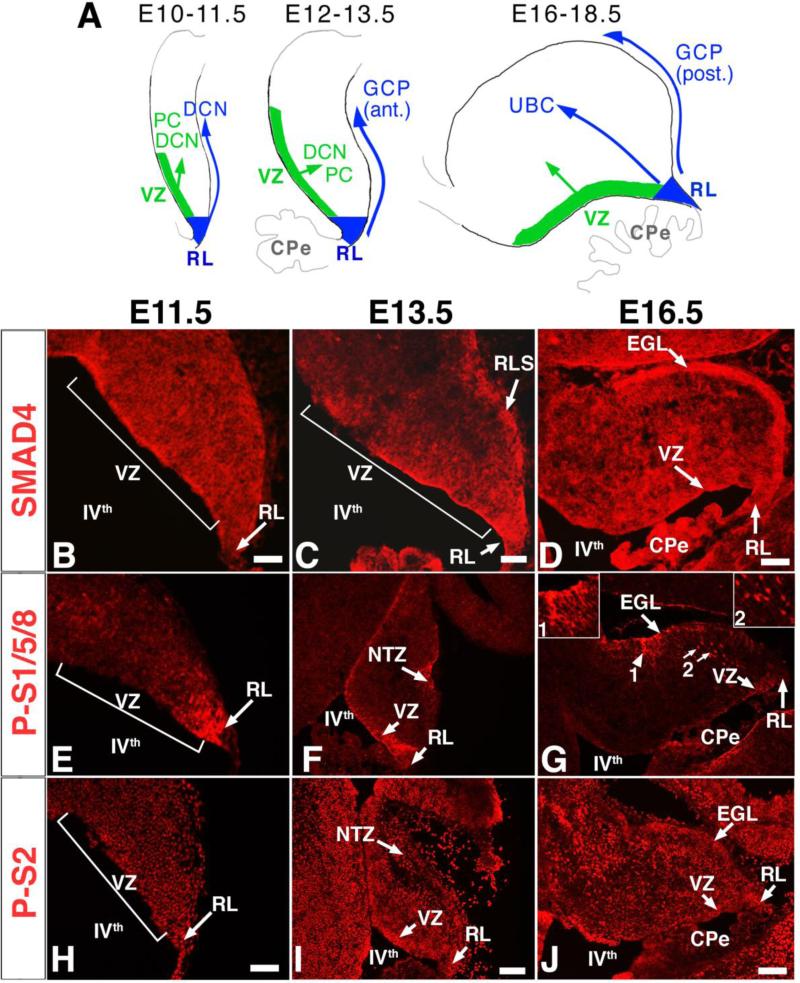
Fig. 1. SMAD4, phospho-SMAD1/5/8, and phospho-SMAD2 are detected in progenitors of the RL and VZ and in differentiated populations during embryonic cerebellar development.
(A) Schema for the development of rhombic lip (RL; blue) and ventricular zone (VZ; green) derived neuronal populations. DCN, deep cerebellar nuclei neurons; GCP, granule cell precursors, PC, Purkinje cell; UBC, unipolar brush cell. Immunostaining was performed on sagittal sections (anterior up, posterior down) for the developmental stages at which different neuronal populations are born, E11.5 (B, E, H), E13.5 (C, F, I) and E16.5 (D, G, J). (G) Besides being expressed at the VZ at E16.5, phospho-SMAD1/5/8 (P-S1/5/8) also appears expressed by processes at the EGL (arrowhead, inset 1) and by a subpopulation of cells in the parenchyma (arrows, inset 2). (J) At E16.5, phospho-SMAD2 (P-S2) is strongly expressed at the VZ, RL, EGL and by cells in the developing cerebelar parenchyma. CPe, choroid plexus; RL, rhombic lip; VZ, ventricular zone; RLS, rhombic lip stream; NTZ, nuclear transitory zone; EGL, external granule layer; IVth, fourth ventricle. Scale bars: (B) 100 μm; (C) 100 μm; (D) 125 μm; (E,H) 125 μm; (F,I) 250 μm; (G,J) 175 μm.
The RL expresses the transcription factor genes Atoh1 (formerly Math1) and Lmx1a. Atoh1 is required for the development of glutamatergic neurons including granule cell precursors (GCPs), large glutamatergic neurons of deep cerebellar nuclei (DCN), and unipolar brush cells (UBCs) (Ben-Arie et al., 1997; Englund et al., 2006; Machold and Fishell, 2005; Wang et al., 2005). Lmx1a is expressed by roof plate cells and RL sub-populations that give rise to posterior GCPs, glutamatergic DCN neurons, and UBCs (Chizhikov et al., 2006; Chizhikov et al., 2010). Interestingly, Lmx1a is required to maintain the RL and form the later developing posterior lobules of the vermis (Chizhikov et al., 2010). Although the role of these transcription factors in early cerebellar development has become clearer, the molecular signals that regulate Atoh1 and Lmx1a expression, that maintain the RL, and that specify subtypes of glutamatergic neurons are still largely unknown.
Most efforts have focused on elucidating which signaling molecules are acting to generate GCPs. In vitro, Bone Morphogenetic Proteins (BMPs) can induce GCPs when added to naïve neural rh1 tissue (Alder et al., 1999). Adding BMP antagonists in mid-hindbrain neural fold explant cultures and in rh1 and roof plate explant co-cultures can significantly reduce, although without completely preventing, Atoh1 and Lmx1a expression (Chizhikov et al., 2006). Conditional double mutants for the type I Bmp receptor genes, Bmpr1a and Bmpr1b, show a dramatic reduction in Atoh1 expression and GCP production (Qin et al., 2006). However, whether BMPs are required for the specification of other RL-derived cell types has not been addressed. As for BMPs, Transforming Growth Factor β ligands, TGFβ1,2,3, are expressed by the roof plate (Allen Brain Atlas) and might also have a role in the specification of cerebellar neurons.
SMAD4 mediates canonical BMP and TGFβ signaling. Upon binding of the ligands to receptors, SMAD4 interacts with the receptor phosphorylated R-SMADs in the cytoplasm (SMAD1, 5, 8 for BMPs and SMAD2, 3 for TGFβ□) (Shi and Massague, 2003). The SMAD4/R-SMAD complexes translocate to the nucleus to activate the transcription of target genes. The BMP/TGFβ pathway can also signal in a SMAD4-independent manner in vivo via p38MAP kinase, mTOR, TAK1, and NRAGE (e.g. Kendall et al., 2005; Rajan et al., 2003; Shim et al., 2009). Which of these pathways transduces BMP and/or TGFβ signals in neural progenitors, in particular in cerebellar progenitors, is unknown. Conditional Smad4 mutants using a Nestin-Cre driver develop a normal cerebellum with only mild decreases in Purkinje cells (PCs) and interneuron numbers in the adult (Zhou et al., 2003). However, in these mutants, loss of Smad4 only occurs starting at ~E11.5 in a subset of cerebellar progenitor cells. Hence it remains unclear to what extent, if any, SMAD4 transmits BMP and/or TGFβ signals during cerebellar development.
Here we show that Smad4 is strongly expressed in the embryonic cerebellum and that R-SMADs for both the BMP and TGFβ pathways are phosphorylated in RL and VZ progenitors. Moreover, loss of Smad4 in the early (~E9), but not later (~E13), cerebellar primordium leads to a failure to maintain the RL and to generate certain RL-derived neuronal cells including DCN neurons, UBCs, and the late cohort of GCPs, resulting in a truncated cerebellum with foliation defects. These studies demonstrate a specific temporal and spatial requirement for Smad4 in generating cerebellar neurons.
MATERIALS AND METHODS
Mice
Mice carrying the floxed Smad4 (Smad4cko), Engrailed1-Cre knockin (En1Cre), and hGFAP-Cre transgene alleles were kindly provided by Chuxia Deng (NIH/NIDDK, MD; Yang et al., 2002), Alex Joyner (Memorial Sloan Kettering Cancer Center, NY; Kimmel et al., 2000), and Albee Messing (University of Wisconsin, Madison; Zhuo et al., 2001), respectively.
RNA in situ hybridization
Embryos and brains were collected in cold PBS, heads were fixed in 4% PFA for 1hr, washed in PBS, soaked in 30% sucrose overnight at 4°C, embedded in OCT, fast frozen in dry ice and stored at -80°C. Postnatal animals were perfused with 4% PFA. RNA in situ hybridization was performed on 10μm thick cryosections using DIG-labeled probes following the protocol described by Hirsch et al. (Hirsch et al., 2007). The plasmid for Atoh1 was provided by Jane Johnson (UT Southwestern Medical Center, TX).
Immunolabeling
Embryos and brains were prepared as above for RNA in situ hybridization. 10-12μm thick cryosections were processed as previously described (Gutin et al., 2006) and incubated with the following primary antibodies: rabbit anti-LMX1A (Millipore AB10533), rabbit anti-phosphoSMAD2 (Millipore AB3849), mouse anti-PAX6 (Hybridoma Bank), rabbit anti-phosphoSMAD1/5/8 (Cell Signaling, 9511), rabbit anti-SMAD4 (Millipore, clone EP618Y, 04-1033), rabbit anti-TBR1 (Millipore, AB9616 and AB10554), rabbit anti-TBR2 (Millipore, AB9618), mouse anti-PAX2 (Babco, PRB276P), rabbit anti-Calretinin (Sigma), mouse anti-NeuN (Millipore, MAB377X), mouse anti-Calbindin (Sigma, C9848), rabbit anti-GABA (Sigma, A2052), rat anti-BrdU (Accurate, OBT0030G), mouse anti-Ki67 (BD Pharmingen, 550609). Sagittal sections were used for BrdU/Ki67 cell counts of the rhombic lip, which was defined as the triangular tip of the posterior-most part of the cerebellum extending ~150 μm on both the ventral (ventricular) and dorsal surfaces from the posterior tip that is connected to the choroid plexus.
Cell death analyses
TUNEL was performed according to the manufacturer's protocol (Roche, 12156792910). Cell counts and data analysis were as previously described (Fernandes et al., 2007; Gutin et al., 2006).
RESULTS
SMADs are expressed and active in the embryonic cerebellum
Fate mapping studies and analysis of mouse mutants have shown that RL-derived glutamatergic DCN neurons and VZ-derived GABAergic DCN neurons and PCs are born with a peak at around E11.5 (Fig. 1A) (Kim et al., 2008; Machold and Fishell, 2005; Wang et al., 2005). At this stage, immunostaining for SMAD4 and phospho-SMAD2, a potential mediator of TGFβ activity, is strong in the entire cerebellar anlage including the RL and the VZ, whereas immunostaining for phospho-SMADs1/5/8, potential mediators of BMP activity, is strong primarily in the RL (Fig. 1B,E,H). Interestingly, the area of phospho-SMAD1/5/8 staining matches that previously described for Math1, which defines region c1 of the cerebellar primordium populations (Chizhikov et al., 2006; Zordan et al., 2008). The specificity of the anti-SMAD4 antibody for SMAD4 protein was demonstrated using the Smad4 mutant (Fig. 2).
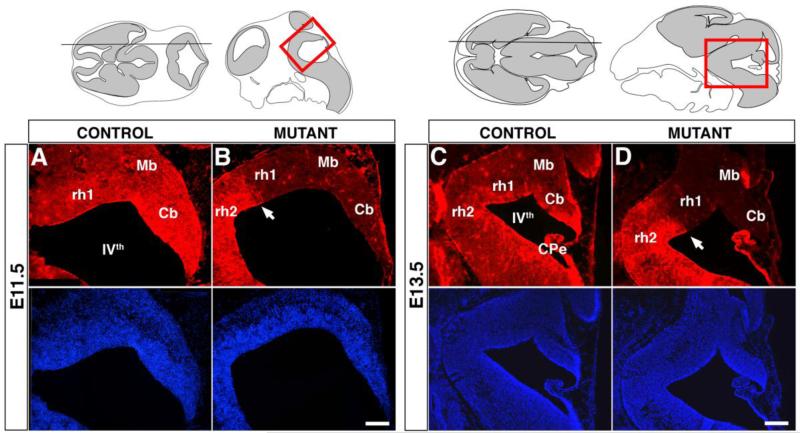
Fig. 2. The expression of SMAD4 is abolished in the cerebellar primordium, posterior midbrain, and rh1 of En1Cre;Smad4cko/cko mutants.
Immunolabelling with SMAD4 antibody (red) and accompanying Hoechst stains (blue) on parasagittal cryosections shows that SMAD4 expression is absent in the entire mutant embryonic cerebellum at E11.5 and E13.5, whereas it is retained in most of the Cpe (except for the anterior most side) and rhombomere 2 where En1Cre is not expressed. Cb, cerebellum; CPe, choroid plexus; Mb, midbrain; rh1, rhombomere 1; rh2, rhombomere 2; IVth, fourth ventricle. Scale bars: (A,B) 175 μm; (C,D) 350 μm.
At E13.5, the first cohort of GCPs is born in the RL and a second wave of GABAergic DCN is generated in the VZ. At this stage, SMAD4 immunoreactivity is strongest in the VZ, RL, and rhombic lip stream and phospho-SMAD1/5/8 is detected in the RL, in the entire VZ with strongest immunoreactivity in the posterior area adjacent to the RL, and in the nuclear transitory zone (NTZ) (Fig. 1C,F). Phospho-SMAD2 immunoreactivity is detected throughout the cerebellar anlage with stronger staining in the areas of the RL, VZ, and NTZ (Fig. 1I).
At E16.5, the RL progenitors are generating UBCs and a late cohort of GCPs. The expression of SMAD4 is strong in the RL, VZ, and external granule layer (EGL), which is composed of the GCPs (Fig. 1D). Phospho-SMAD1/5/8 labels an area of the EGL (arrowhead and inset 1 in Fig. 1G) and the posterior VZ region adjacent to the RL. An additional cell population of unknown identity is labeled in the parenchyma (arrows and inset 2 in Fig. 1G). Phospho-SMAD2 labeling persists in the vast majority of cerebellar cells with the strongest labeling in a subset of RL, EGL, and VZ cells (Fig. 1J). The presence of SMAD4 and phosphorylated receptor SMADs in the germinal zones of the cerebellum throughout neurogenesis is consitent with a function for SMAD4 in progenitors and differentiating neurons.
Loss of Smad4 results in reduced cerebellar size and foliation defects
In order to assess the role of SMAD4 in cerebellum development and in generating specific neuronal subtypes, we used the En1Cre mouse (Kimmel et al., 2000) to delete a floxed allele of Smad4 (Smad4cko; Yang et al., 2002). The En1Cre mouse expresses Cre as early as E8.5 in the neuroepithelium encompassing the posterior mesencephalon and rh1 from which the cerebellum develops (Kimmel et al., 2000; Li et al., 2002). The En1Cre;Smad4cko/cko mutants were obtained at the expected mendelian ratio until P5, but remained smaller than their littermates probably due to difficulties in nursing. They die before P15 displaying severe locomotor and balance defects indicative of cerebellar abnormalities. The En1Cre;Smad4cko/cko mutants lack detectable SMAD4 protein in the posterior mesencephalon and anterior part of rh1 including the entire cerebellar anlage, whereas SMAD4 remains expressed in the posterior part of the choroid plexus and rh2 demonstrating tissue specific inactivation of Smad4 (Fig. 2). Nevertheless, we can not exclude the possibility of residual undetected levels of SMAD4 protein in these mutants.
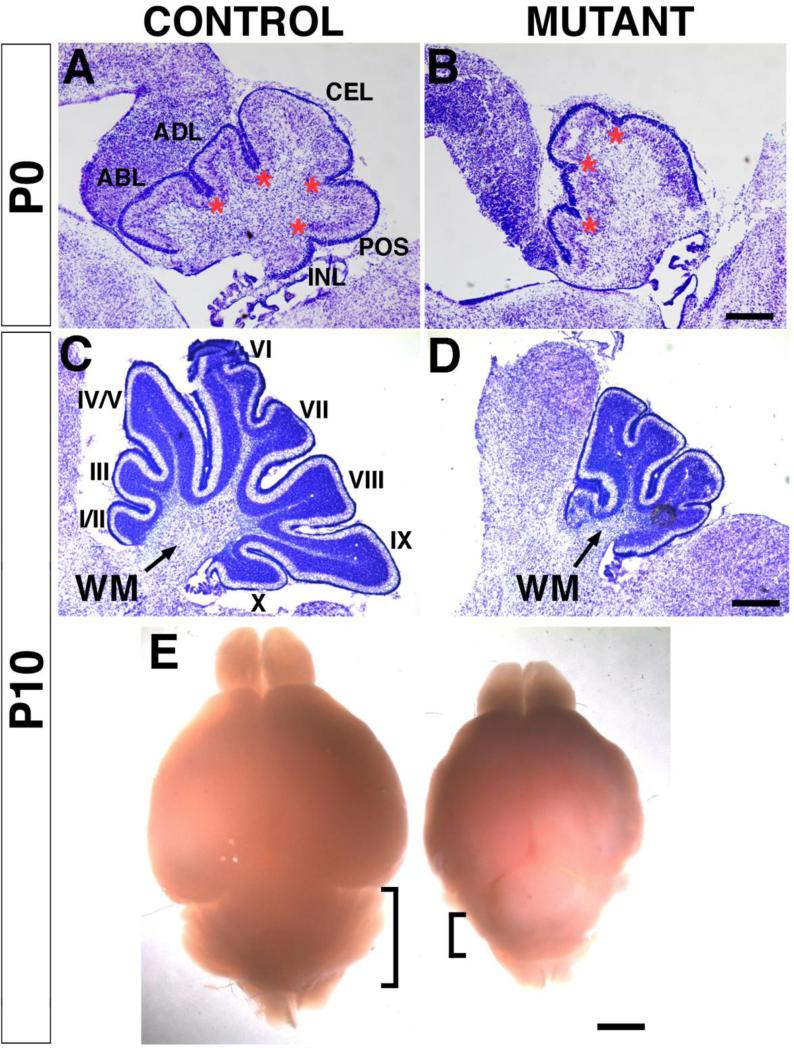
Although the mutants have overall smaller body and brain sizes than their siblings, gross anatomical and histological analysis reveals a disproportionately small cerebellum with abnormal foliation (Fig. 3). In P0 controls, four primary fissures delineate five lobes: anterobasal, anterodorsal, central, posterior, and inferior (Fig. 3A; Altman and Bayer, 1997), whereas in the mutants only three fissures and four lobes are distinguishable (Fig. 3B). At P10, the size difference between mutants and controls becomes more prominent (Fig. 3C,D,E). In the vermis, controls show ten lobules whereas mutants are truncated and only have four small lobules (Fig. 3C, D). The white matter region below the cerebellar cortex where the DCN reside is severely reduced in the mutants (Fig. 3D). Hence the inactivation of Smad4 during early embryogenesis results in a severe reduction in cerebellar size and defects in cerebellar cytoarchitecture.
Fig. 3. The En1Cre;Smad4cko/cko mutant cerebella show reduced size and foliation defects.
Nissl staining of sagittal sections (posterior facing left, anterior right). At P0 (A,B), the mutant cerebellum is slightly smaller compared to controls with one primary fissure and lobule missing. Asterisks denote the base of the fissures. At P10 (C,D), the majority of the ten lobules in the mutant are absent. (E) Whole control (left) and mutant (right) brains. At P10, the whole body and brains of mutant mice are smaller presumably due to an inability to nurse normally, but the cerebellum appears disproportionately small compared to the rest of the brain (bracketed areas marks the cerebella). ABL, anterobasal lobule; ADL, anterodorsal lobule; CEL, central lobule; POS, posterior lobule; INL, inferior lobule; WM, white matter; I-X, lobules. Scale bars: (A,B) 200 μm; (C,D) 400 μm; (E) 1.25 mm.
GABAergic neurons are produced in reduced numbers and PCs are abnormally distributed in Smad4 mutants
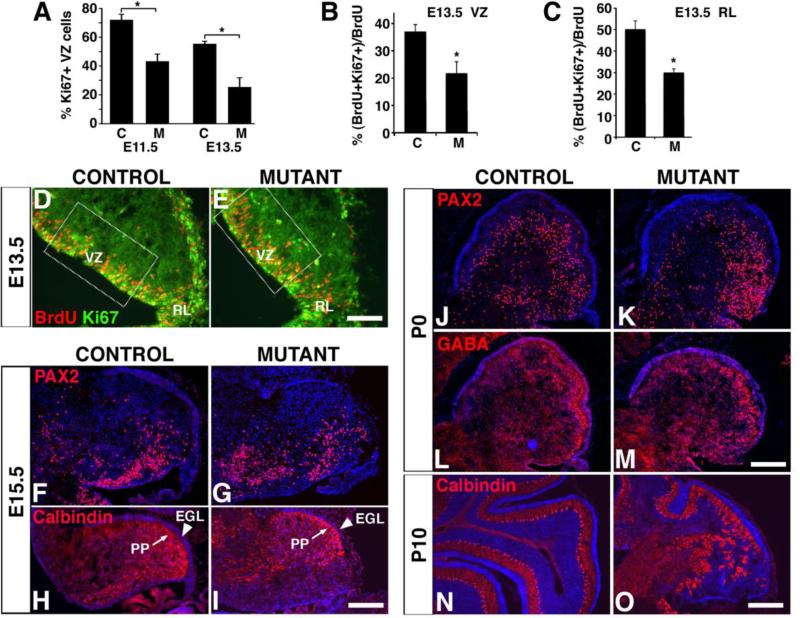
Since SMAD4 and phosphorylated SMAD2 and SMAD1/5/8 are present in the VZ (Fig. 1), a loss of VZ-derived cell types could potentially account at least in part for the reduced cerebellar size and abnormal cytoarchitecture. The VZ generates GABAergic neurons, including PCs, which can be labeled with anti-Calbindin and -GABA antibodies, and the inhibitory interneurons (basket, stellate, and Golgi cells), which can be labeled with anti-PAX2 and -GABA antibodies (Hoshino et al., 2005; Leto et al., 2009; Weisheit et al., 2006). The numbers of PAX2+ interneurons in the En1Cre;Smad4cko/cko mutants are not significantly different compared to controls at E15.5 (346 ± 17 cells/section in controls and 322 ± 38 in mutants; using positionally matched sections; Fig. 4F,G) and only slightly reduced at P0 (692 ± 171 in controls and 522 ± 152 in mutants, p = 0.006; Fig. 4J-M). The total number of PAX2+ cells was also estimated in P10 cerebella by counting all labelled cells in 20 μm sections taken every 400 μm throughout each cerebellum. At this stage, the total number of PAX2+ cells in mutant cerebella is only 24.4 +/- 3.9% that of controls, with no obvious difference in their distribution or density (Supplemental Fig. 1). This decrease in the number of interneurons is at least partially explained by a decrease in the percentage of proliferating (Ki67+) VZ cells at E11.5 and E13.5, which in turn is due to a decrease in the ability of these precursor cells to continue cycling (assessed by administering BrdU 24 hours prior to collecting E13.5 brains and colabelling for BrdU and Ki67; Fig. 4A,B,D,E). Overall, the decrease in interneuron numbers is likely to account only in part for the severe loss in cerebellar size.
Fig. 4. The numbers of VZ-derived GABAergic neurons are reduced in the En1Cre;Smad4cko/cko mutant and PCs become mislocalized.
(A) A significantly smaller percentage of VZ cells are Ki67+ in mutants, ‘M’, compared with controls, ‘C’ at both E11.5 and E13.5. At E13.5, 24 hours after BrdU administration at E12.5, fewer BrdU-labeled cells are also labeled with Ki67 in both the mutant VZ (B) and RL (C). (D,E) Immunostaining for BrdU (red) and Ki67 (green) in E13.5 sagittal sections of the cerebellum. Immunostaining with anti-PAX2 (F,G,J,K), GABA (L,M), and Calbindin (H,I,N,O) antibodies (red) reveals similar numbers and patterns in controls and mutants (see text for quantification). However, by P10 PCs often fail to form a monolayer (N,O). EGL, external granule layer; PP, Purkinje cell plate. Scale bars: (D,E) 100 μm; (F-I) 200 μm; (J-M) 250 μm; (N,O) 175 μm.
PCs are specified starting at around E10.5 and are marked by the expression of Calbindin (Miale and Sidman, 1961). They migrate away from the VZ and, by E14.5, they start to assemble in aggregates forming the Purkinje plate (PP), right below the nascent EGL (Rice and Curran, 2001). In the En1Cre;Smad4cko/cko mutants at E15.5, Calbindin+ PCs are generated and successfully migrate away from the VZ to form the PP below the EGL (Fig. 4H,I). As in controls, PCs at this stage in mutants can be found beginning to form clusters (Supplemental Fig. 2). Their distribution by P10, however, is abnormal. The PCs in the mutant fail to form a normal monolayer in posterior lobules (Fig. 4N,O). The mislocalization of VZ-derived PCs in the posterior area would be consistent with a lack of RL-derived neurons in this area.
RL-derived DCN neurons fail to develop in the Smad4 mutant
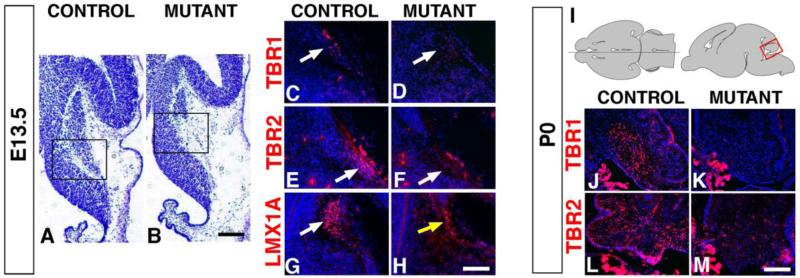
Atoh1+ RL progenitors give rise first to glutamatergic DCN neurons at E10-E11.5, then to an early cohort of GCPs that populate the anterior cerebellum at E12-E13.5, and finally to a late cohort of GCPs that populate the posterior cerebellum and to UBCs at E16-E18.5 (Fig. 1A) (Englund et al., 2006; Machold and Fishell, 2005; Wang et al., 2005). The DCN progenitors accumulate in the NTZ by E13.5 and express the transcription factors TBR1 and TBR2, with only the expression of TBR1 persisting in the neonatal medial cerebellum in the fastigial DCN (Fink et al., 2006). As early as E13.5, the mutant NTZ is severely reduced in size suggesting a defect in the production of DCN progenitors (Fig. 5A,B). The expression of TBR1 and TBR2, which in controls is detected in progenitors entering the NTZ, is missing in the mutants (Fig. 5C-F). Moreover, TBR1+ and TBR2+ progenitors are not detected accumulating in either the rhombic lip stream or regions anterior of the NTZ ruling out possible migration defects for DCN progenitors (Fig. 5 and data not shown). In addition, Lmx1a expression, which also marks DCN progenitors, is severely reduced in the mutant NTZ (Fig. 5G,H). These results strongly suggest that DCN progenitors are not generated.
Fig. 5. RL-derived progenitors for DCN neurons and UBCs are not generated in En1Cre;Smad4cko/cko mutants.
(A,B) Nissl staining of E13.5 sagittal cryosections showing a small mutant NTZ. The black boxed areas indicate the regions enlarged for the sections immunostained for TBR1 (C,D), TBR2 (E,F), and LMX1A (G,H). At E13.5, TBR1+ (C,D) and TBR2+ (E,F) DCN progenitors are absent in the mutant NTZ (arrows). (G,H) The number of LMX1A+ DCN progenitors is severely decreased in mutants (yellow arrow). (I) Schematic showing the location and orientation of the sagittal sections in J-M. (J-M) At P0, TBR1+ and TBR2+ DCN neurons and UBCs are absent in the mutant (note that the image in L is from a slightly more lateral section than for J,K,M and that the bright staining outside the cerebellum marks choroid plexus). Scale bars: (A,B) 300 μm; (C-H) 150 μm; (J-M) 250 μm.
Consistent with a lack of DCN progenitors, P0 mutants lack the glutamatergic neurons of the fastigial DCN as revealed by a severe reduction of TBR1+ neurons in the medial part of the cerebella (Fig. 5J,K). Defective migration is not a cause for the lack of TBR1 staining since no TBR1+ neurons could be detected in more lateral areas (data not shown). Together our results demonstrate that Smad4 function is required to generate glutamatergic DCN progenitors and consequently neurons.
RL-derived UBCs and late-born GCPs fail to develop in the mutant
Once glutamatergic DCN neurons are generated, the RL also produces an early and late population of GCPs, which form the anterior and posterior lobules respectively, and unipolar brush cells (UBCs), which populate the posterior lobules IX and X. In P0 controls, immunolabeling with TBR2 marks UBCs as they migrate from the RL through the developing white matter (Fig. 5L). In the P0 mutants, the number of TBR2+ UBCs is dramatically reduced or absent (Fig. 5M). The absence of accumulating TBR2+ UBCs near the VZ in the mutants suggests that UBCs rather than failing to migrate were not generated. TUNEL analysis also showed no increase in apoptosis in the mutants rendering cell death an unlikely explanation for the lack of UBCs (data not shown).
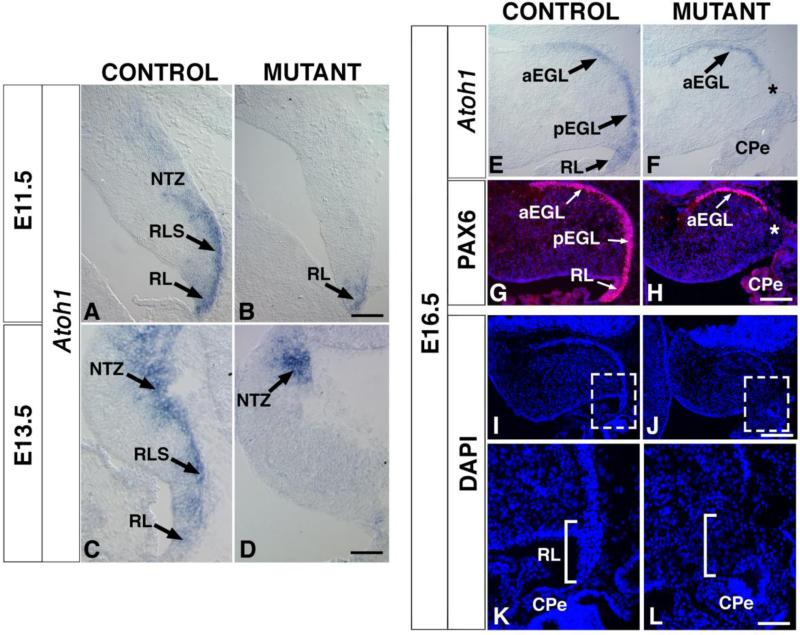
Atoh1 in addition to being expressed in the RL, continues to be expressed in the early cohort of GCPs that forms the anterior EGL and the late cohort that forms the posterior EGL (Fig. 6E). In mutants, however, only the anterior dorsal edge of the cerebellum is labeled suggesting that anterior, but not posterior, GCPs have been generated (Fig. 6F). Consistent with this finding, PAX6, a marker for the entire EGL (Fig. 6G), is only found in the anterior region of mutants in an overlapping manner with Atoh1 expression (Fig. 6H), further demonstrating that only anterior (or early) GCPs have been produced.
Fig. 6. The expression of Atoh1, a marker for the rhombic lip and its derivatives, is severely reduced in En1Cre;Smad4cko/cko mutants.
(A-F) In situ RNA hybridization on sagittal crysosections with Atoh1 probe. (A,B) At E11.5, Atoh1 expression (dark purple) is severely decreased in the mutant RL and absent in RL derivatives (RLS, NTZ). (C,D) At E13.5, Atoh1 expression is absent from the mutant RL, but present in the NTZ area, possibly in the early born GCPs. (E,F) In E16.5 controls, Atoh1 is expressed by GCPs along the entire (anterior and posterior) EGL. In the mutant, Atoh1 expression is only detected in the anterior region, suggesting that only the early born GCPs are produced and form an anterior EGL. (G,H) Immunostaining with anti-PAX6 antibody (red) confirms the absence of posterior GCPs and lack of a posterior EGL (blue: Hoechst). The asterisks in G and I show the zone where the RL should normally be, but is missing in the mutants. (I,J) DAPI stained cerebella. Boxed areas are enlarged in (K) and (L) to show the absence of a rhombic lip in the mutant. CPe, choroid plexus; DCN, deep cerebellar nuclei; aEGL, anterior external granule layer; pEGL, posterior external granule layer; GCP, granule cell progenitors; NTZ, nuclear transitory zone; RL, rhombic lip; RLS, rhombic lip stream; UBC; unipolar brush cells. Scale bars: (A,B) 100 μm; (C,D) 150 μm; (E-H) 150 μm; (I,J) 225 μm; (K,L) 65 μm.
Loss of SMAD4 immunostaining in these anterior GCPs confirms that they have not escaped recombination. While SMAD4 is strongly expressed in E16.5 control cerebella (Supplemental Fig. 3A,A’), in particular the RL and EGL, its expression is absent in the mutant RL and remaining anterior EGL (Supplemental Fig. 3B,B’). SMAD4 staining in the choroid plexus and meninges, where En1Cre is not expressed, serves as an internal positive control. Results were similar for the P0 cerebella (Supplemental Fig. 3C,D). Together our results demonstrate that abolishing Smad4 function leads to a failure to produce all late-born glutamatergic neurons, both UBCs and the late cohort of GCPs.
SMAD4 is required to maintain the rhombic lip
The loss of all late-born glutamatergic neurons could be due to a failure to maintain the RL in the En1Cre;Smad4cko/cko mutants. In E11.5 control animals, Atoh1 is specifically expressed in the RL, the rhombic lip stream, and the prospective NTZ. In the mutants, however, Atoh1 expression is only retained weakly in the RL (Fig. 6A,B). By E13.5, the weak RL expression seen earlier in the mutants disappears and Atoh1 expression can only be detected in the most anterior portion of the remaining NTZ (Fig. 6C,D). By E16.5, Atoh1 expression remains absent in the area of the RL in the mutant (Fig. 6E,F). The progressive loss of Atoh1 expression in the area of the RL is consistent with a role for Smad4 in maintaining RL progenitors. Moreover, morphologically the RL appears completely missing by E16.5 when in controls the RL forms a thin cell-dense extension, whereas in mutants, no such structure is visible by DAPI staining (Fig. 6I-L). A likely mechanism for the loss of the RL is a decrease in the number of RL precursors that remain proliferative at earlier ages. Consistent with this possibility, at E13.5 the percentage of BrdU+ cells that colabel with Ki67 24 hours after a pulse of BrdU is significantly reduced in mutants (Fig. 4C).
Deletion of Smad4 once neurogenesis has started does not disrupt glutamatergic neuron production, but disrupts granule cell layering
The role of Smad4 in cerebellar development and more specifically in generating late-born GCPs and UBCs was further addressed by deleting Smad4 with hGFAP-Cre. hGFAP-Cre recombines cerebellar precursors at ~E13 (Zhuo et al., 2001), five days after En1Cre does, but before UBCs and the late cohort of GCPs that populate the posterior lobules are born. In hGFAP-Cre;Smad4cko/cko mutants, unlike En1Cre;Smad4cko/cko mutants, the RL is maintained throughout development (data not shown). This allows us to further distinguish whether the lack of late-born glutamatergic neurons in the En1Cre;Smad4cko/cko mutants is in fact due to progressive loss of the RL rather than a more direct role for Smad4 in generating specifically late versus early born GCPs and UBCs.
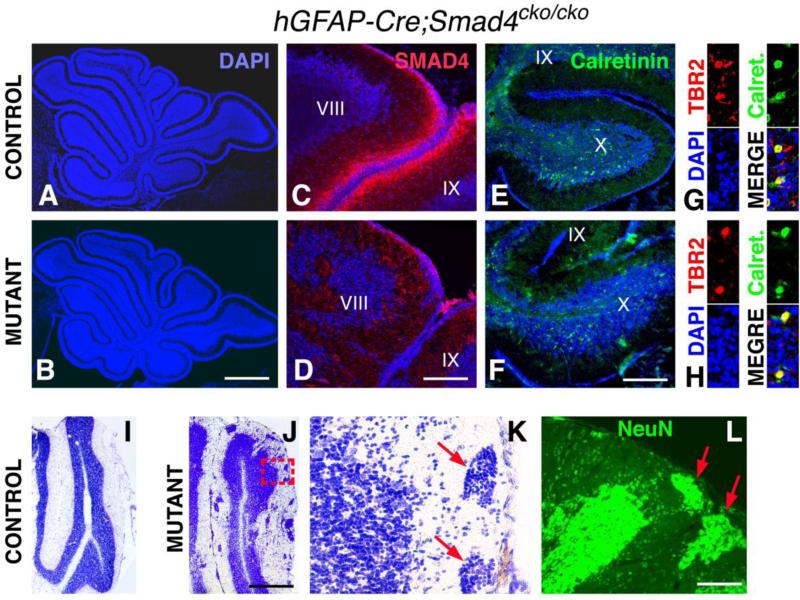
At a gross level, the mutant hGFAP-Cre;Smad4cko/cko cerebellum appears normal in size and lobular organization (Fig. 7A,B) and the animals survive for at least 8 months. It has previously been shown that the early precursors that are recombined using hGFAP-Cre include those that generate early and late GCPs (Zhuo et al., 2001; Yang et al., 2008). Not surprisingly then, at P10 when GCPs are still proliferating and generating granule cells, SMAD4 immunostaining is undetectable in the GCPs of hGFAP-Cre;Smad4cko/cko mutants, whereas staining is detected in the GCP layer of control littermates along the entire periphery of the cerebellum (Fig. 7C,D). For both controls and mutants at this age, SMAD4 immunostaining can not be detected in the differentiated cells that make up the bulk of the lobules. Despite the absence of Smad4 expression in the GCPs, GCPs and the granule cells they produce appear to be generated in roughly normal numbers (compare the sizes of the cerebella and lobules in Fig. 7A,B). Moreover, UBCs, which are generated from the RL at the same time as the late cohort of GCPs, can also be found in roughly normal numbers in their normal location (lobules IX and X) of P10 hGFAP-Cre;Smad4cko/cko mutants by immunolabeling for Calretinin and TBR2 (Fig. 7E-H). These results support the interpretation that Smad4 is required to maintain the RL during cerebellar development rather than directly required for specifying or producing late born glutamatergic neurons.
Fig. 7. hGCPA-Cre;Smad4cko/cko mutants produce UBCs and have disorganized granule cells.
(A,B) DAPI-stained P10 control and mutant cerebella appear normal in size and shape. (C,D) Immunostaining for SMAD4 (red) at P10 is detected throughout the EGL of control but not mutant cerebella (only lobules VIII and IX are shown here). (E,F) Calretinin immunostaining (green) labels UBCs only in the ventral (or posterior) half of lobule IX (partially shown) and throughout lobule X. (G,H) High magnification views of Calretinin+ and TBR2+ co-labeled UBCs in lobule X. (I,J) Cresyl violet stained P20 control and mutant cerebella. (K) High magnification view of boxed area in (J) showing ectopic clumps of cells (red arrows) in the molecular layer. (L) These cells uniformally stain for the mature neuronal marker NeuN (green). Scale bars: (A,B) 800 μm; (C,D) 60 μm; (E,F) 130 μm; (I,J) 600 μm; (K,L) 60 μm.
Unexpectedly, although GCPs and granule cells are present, they appear disorganized at P10 in the hGFAP-Cre;Smad4cko/cko mutants compared to control littermates. Whereas in controls the granule cells are forming a well defined uniform layer, in the mutants the granule cell layer is uneven with ectopic clumps of cells located in the molecular layer (Fig. 7I-L). These clumps are at least superficially reminiscent of tumor initiating cells, which would be consistent with the notion that BMPs can inhibit proliferation of GCPs (Rios et al., 2004) and therefore that the absence of SMAD4 could potentially cause hyperproliferation. However, no cells within these clumps could be labeled with the proliferative marker Ki67 and instead all cells labeled with the mature neuron marker NeuN (Fig. 7L), indicating that these are likely mislocalized granule cells. Moreover, in areas where ectopic clumps of granule cells are observed, the glial processes appear sparser and more discontinous in mutants compared with controls (Supplemental Fig. 4), providing a potential mechanism for the mislocalization of granule cells, namely a migration defect. A radial glia phenotype was not previously observed in Nestin-Cre;Smad4cko/cko mutants, presumably due to highly mosaic recombination (Zhou et al., 2003).
DISCUSSION
In this report we demonstrate an essential role for Smad4 in cerebellar development. Namely, Smad4 is required between E8.5 and E13 for maintaining the RL and for generating RL-derived cell types, some of which are born after E13. Despite the presence of SMAD4 and phosphorylated R-SMADs in the VZ, VZ-derived interneurons are produced in the En1Cre mutants, although with progressively fewer numbers over time compared with controls. In addition, PCs are mislocalized, especially in posterior regions of the developing cerebellum. The lack of glutamatergic DCN neurons and posterior EGL could potentially account, cell non-autonomously, for the mislocalization of VZ-derived neurons. Moreover, because recombination in these mutants occurs in most or all cell types derived from the posterior mesencephalon and rh1, including the mid-hindbrain signaling center, we can not exclude the possibility that any of the phenotypes observed are due to cell non-autonomous effects. Testing cell non-autonomous effects will require lineage-specific deletions of Smad4 in future experiments.
It is not entirely clear what signals are activating SMAD4 in the RL to maintain production of some of the glutamatergic cell types but BMPs are likely involved at least for GCPs. When added to rh1 explant cultures, BMP7 and 6 can induce Atoh1 expression and dissociated rh1 cells treated with BMP7 can differentiate into granule neurons when transplanted into adult cerebella (Alder et al., 1999). Moreover, genetic disruption of the Bmpr1a and Bmpr1b genes showed that the BMP pathway is required for the development of GCPs (Qin et al., 2006).
In the present study, we provide evidence that Smad4 is required to generate late, but not early born GCPs. Based on the results of deleting Smad4 with En1Cre and hGFAP-Cre, the simplest explanation for the early versus late difference is that Smad4 is required to maintain the RL over developmental time. If this is the case, then there could be one population of rather uniform precursors in the RL giving rise to all GCPs. However, over time, these precursors would prematurely be depleted in the mutants. Our data is consistent with this possibility, since we see a loss of RL markers and morphology over time (Fig. 6). Hence Smad4 may be required to maintain RL cells in an early precursor cell state.
However, other possibilities for explaining the lack of late GCPs in the En1Cre;Smad4cko/cko mutants can not be excluded. First, SMAD4, which likely mediates BMP and/or TGFβ signaling, may be required before E13 to induce late but not early GCPs. Consistent with this possibility, BMP inhibitors significantly blocked, but did not completely prevent, the induction of RL cells and formation of the rhombic lip stream (Chizhikov et al., 2006). In addition, the Bmpr1a and Bmpr1b double mutant, although generated using a Cre driver that recombines later (E11.5) than the one used here (E8.5), shows a small number of PAX6+ and ZIC1/2+ GCPs (Derynck and Zhang, 2003; Qin et al., 2006). In this case, GCPs would be a more heterogeneous population of progenitors than previously thought and would be specified using different molecular mechanisms, with the early born ones generated in a SMAD4-independent manner and the late born ones requiring SMAD4. The specification of early GCPs would require signals other than BMPs or TGFβs. WNT and FGF signals are candidates as inducers of early born GCPs (Liu and Joyner, 2001; Sato et al., 2004; Xu et al., 2000). For instance, the Fgf17-/-;Fgf8+/- mutant shows a truncated cerebellum lacking its anterior lobules (Basson et al., 2008; Xu et al., 2000).
Another alternative to explain the specific loss of late born GCPs in the En1Cre;Smad4cko/cko mutants is that BMP and/or TGFβ signaling is required to generate all GCPs from the RL, but compensatory mechanisms allow for the generation of early born GCPs. Perhaps BMP and TGFβ signals do not depend exclusively on SMAD4 in early progenitors. Indeed, BMP and TGFβ ligands can signal through a growing list of potential transducers (e.g. Derynck and Zhang, 2003; Descargues et al., 2008; Xu et al., 2008 ; Shim et al., 2009). Compensatory mechanisms could occur at the cellular level as well: early GCPs in the En1Cre;Smad4cko/cko mutants could be abnormally derived from VZ cells that have been re-specified to become GCPs. Precedence for this possibility exists in that VZ cells can adopt a GCP phenotype in the Ptf1a mutants (Pascual et al., 2007).
Interestingly, although SMAD4 is not required to generate early born GCPs, it is required to generate even earlier born cells, the glutamatergic DCN neurons. Glutamatergic DCN progenitors are the first cells born from Atoh1-expressing RL cells at ~E10.5 and the generation of these DCN progenitors requires Atoh1 (Machold and Fishell, 2005; Wang et al., 2005). The expression of Atoh1 is reduced by E11.5 in our mutants consistent with a role for Smad4 in promoting or maintaining Atoh1 expression in the progenitor pool fated to become DCN neurons.
Likewise, the UBCs are also an RL-derived cerebellar neuronal population that is lacking in the En1Cre;Smad4cko/cko mutants. Little is known about how UBCs, which are born late at ~E16.5, are specified, except that they require at least in part Atoh1 and also derive at least in part from Lmx1a-expressing progenitors (Englund et al., 2006; Chizhikov et al., 2010). As for the late GCPs, in the En1Cre;Smad4cko/cko mutants, UBCs are not specified probably because SMAD4 is necessary for maintaining the Atoh1 and Lmx1a-expressing progenitors that comprise the RL (Fig. 5,6). In summary, we propose that SMAD4-dependent signals act in concert with Lmx1a and Atoh1 to maintain the RL and to generate DCN neurons, UBCs, and a late cohort of GCPs.
Supplementary Material
Highlights.
SMAD4 is required to generate only a subset of cerebellar neurons
Without SMAD4, glutamatergic DCN neurons, UBCs, and late GCPs are not produced
SMAD4 maintains the functional integrity of the rhombic lip
Postnatally, SMAD4 is required to correctly localize granule neurons
AKNOWLEDGEMENTS
We are grateful to Nicholas McKeehan for technical assistance, Celine Zimmer for help with the DIG RNA in situ hybridization protocol, to Chuxia Deng and Alex Joyner for mice and to Jane Johnson for the Atoh1 plasmid; to our laboratory members, to Zaven Kaprielian, Stacey Reeber and Roy Sillitoe for insightful discussions and comments on the manuscript. This work was supported by grants MH083804 and MH070596 from the NIH and from the McDonnell Brain Tumor Foundation (JMH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alder J, Lee KJ, Jessell TM, Hatten ME. Generation of cerebellar granule neurons in vivo by transplantation of BMP-treated neural progenitor cells. Nat Neurosci. 1999;2:535–540. doi: 10.1038/9189. [DOI] [PubMed] [Google Scholar]
- Altman J, Bayer SA. Time of origin and distribution of a new cell type in the rat cerebellar cortex. Exp Brain Res. 1977;29:265–274. doi: 10.1007/BF00237046. [DOI] [PubMed] [Google Scholar]
- Altman J, Bayer SA. Development of the cerebellar system in relation to its evolution, structures and functions. CRC; New York: 1997. [Google Scholar]
- Basson MA, Echevarria D, Ahn CP, Sudarov A, Joyner AL, Mason IJ, Martinez S, Martin GR. Specific regions within the embryonic midbrain and cerebellum require different levels of FGF signaling during development. Development. 2008;135:889–898. doi: 10.1242/dev.011569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Arie N, Bellen HJ, Armstrong DL, McCall AE, Gordadze PR, Guo Q, Matzuk MM, Zoghbi HY. Math1 is essential for genesis of cerebellar granule neurons. Nature. 1997;390:169–172. doi: 10.1038/36579. [DOI] [PubMed] [Google Scholar]
- Chen D, Zhao M, Mundy GR. Bone morphogenetic proteins. Growth Factors. 2004;22:233–241. doi: 10.1080/08977190412331279890. [DOI] [PubMed] [Google Scholar]
- Chizhikov VV, Lindgren AG, Currle DS, Rose MF, Monuki ES, Millen KJ. The roof plate regulates cerebellar cell-type specification and proliferation. Development. 2006;133:2793–2804. doi: 10.1242/dev.02441. [DOI] [PubMed] [Google Scholar]
- Chizhikov VV, Lindgren AG, Mishima Y, Roberts RW, Aldinger KA, Miesegaes GR, Currle DS, Monuki ES, Millen KJ. Lmx1a regulates fates and location of cells originating from the cerebellar rhombic lip and telencephalic cortical hem. Proc Natl Acad Sci U S A. 2010;107:10725–10730. doi: 10.1073/pnas.0910786107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- Descargues P, Sil AK, Karin M. IKKalpha, a critical regulator of epidermal differentiation and a suppressor of skin cancer. EMBO J. 2008;27:2639–2647. doi: 10.1038/emboj.2008.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund C, Kowalczyk T, Daza RA, Dagan A, Lau C, Rose MF, Hevner RF. Unipolar brush cells of the cerebellum are produced in the rhombic lip and migrate through developing white matter. J Neurosci. 2006;26:9184–9195. doi: 10.1523/JNEUROSCI.1610-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes M, Gutin G, Alcorn H, McConnell SK, Hebert JM. Mutations in the BMP pathway in mice support the existence of two molecular classes of holoprosencephaly. Development. 2007;134:3789–94. doi: 10.1242/dev.004325. [DOI] [PubMed] [Google Scholar]
- Fink AJ, Englund C, Daza RA, Pham D, Lau C, Nivison M, Kowalczyk T, Hevner RF. Development of the deep cerebellar nuclei: transcription factors and cell migration from the rhombic lip. J Neurosci. 2006;26:3066–3076. doi: 10.1523/JNEUROSCI.5203-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher E, Howell BW, Soriano P, Cooper JA, Hawkes R. Cerebellar abnormalities in the disabled (mdab1-1) mouse. J Comp Neurol. 1998;402:238–251. [PubMed] [Google Scholar]
- Goffinet AM, So KF, Yamamoto M, Edwards M, Caviness VS., Jr. Architectonic and hodological organization of the cerebellum in reeler mutant mice. Brain Res. 1984;318:263–276. doi: 10.1016/0165-3806(84)90031-2. [DOI] [PubMed] [Google Scholar]
- Gutin G, Fernandes M, Palazzolo L, Paek H, Yu K, Ornitz DM, McConnell SK, Hebert JM. FGF signalling generates ventral telencephalic cells independently of SHH. Development. 2006;133:2937–2946. doi: 10.1242/dev.02465. [DOI] [PubMed] [Google Scholar]
- Hirsch MR, Glover JC, Dufour HD, Brunet JF, Goridis C. Forced expression of Phox2 homeodomain transcription factors induces a branchio-visceromotor axonal phenotype. Dev Biol. 2007;303:687–702. doi: 10.1016/j.ydbio.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Hoshino M, Nakamura S, Mori K, Kawauchi T, Terao M, Nishimura YV, Fukuda A, Fuse T, Matsuo N, Sone M, Watanabe M, Bito H, Terashima T, Wright CV, Kawaguchi Y, Nakao K, Nabeshima Y. Ptf1a, a bHLH transcriptional gene, defines GABAergic neuronal fates in cerebellum. Neuron. 2005;47:201–213. doi: 10.1016/j.neuron.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Jensen P, Smeyne R, Goldowitz D. Analysis of cerebellar development in math1 null embryos and chimeras. J Neurosci. 2004;24:2202–2211. doi: 10.1523/JNEUROSCI.3427-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen P, Zoghbi HY, Goldowitz D. Dissection of the cellular and molecular events that position cerebellar Purkinje cells: a study of the math1 null-mutant mouse. J Neurosci. 2002;22:8110–8116. doi: 10.1523/JNEUROSCI.22-18-08110.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinichenko SG, Okhotin VE. Unipolar brush cells--a new type of excitatory interneuron in the cerebellar cortex and cochlear nuclei of the brainstem. Neurosci Behav Physiol. 2005;35:21–36. doi: 10.1023/b:neab.0000049648.20702.ad. [DOI] [PubMed] [Google Scholar]
- Kendall SE, Battelli C, Irwin S, Mitchell JG, Glackin CA, Verdi JM. NRAGE mediates p38 activation and neural progenitor apoptosis via the bone morphogenetic protein signaling cascade. Mol Cell Biol. 2005;25:7711–7724. doi: 10.1128/MCB.25.17.7711-7724.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EJ, Battiste J, Nakagawa Y, Johnson JE. Ascl1 (Mash1) lineage cells contribute to discrete cell populations in CNS architecture. Mol Cell Neurosci. 2008;38:595–606. doi: 10.1016/j.mcn.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel RA, Turnbull DH, Blanquet V, Wurst W, Loomis CA, Joyner AL. Two lineage boundaries coordinate vertebrate apical ectodermal ridge formation. Genes Dev. 2000;14:1377–1389. [PMC free article] [PubMed] [Google Scholar]
- Leto K, Bartolini A, Yanagawa Y, Obata K, Magrassi L, Schilling K, Rossi F. Laminar fate and phenotype specification of cerebellar GABAergic interneurons. J Neurosci. 2009;29:7079–7091. doi: 10.1523/JNEUROSCI.0957-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JY, Lao Z, Joyner AL. Changing requirements for Gbx2 in development of the cerebellum and maintenance of the mid/hindbrain organizer. Neuron. 2002;36:31–43. doi: 10.1016/s0896-6273(02)00935-2. [DOI] [PubMed] [Google Scholar]
- Liu A, Joyner AL. Early anterior/posterior patterning of the midbrain and cerebellum. Annu Rev Neurosci. 2001;24:869–896. doi: 10.1146/annurev.neuro.24.1.869. [DOI] [PubMed] [Google Scholar]
- Machold R, Fishell G. Math1 is expressed in temporally discrete pools of cerebellar rhombic-lip neural progenitors. Neuron. 2005;48:17–24. doi: 10.1016/j.neuron.2005.08.028. [DOI] [PubMed] [Google Scholar]
- Miale IL, Sidman RL. An autoradiographic analysis of histogenesis in the mouse cerebellum. Exp Neurol. 1961;4:277–296. doi: 10.1016/0014-4886(61)90055-3. [DOI] [PubMed] [Google Scholar]
- Pascual M, Abasolo I, Mingorance-Le Meur A, Martinez A, Del Rio JA, Wright CV, Real FX, Soriano E. Cerebellar GABAergic progenitors adopt an external granule cell-like phenotype in the absence of Ptf1a transcription factor expression. Proc Natl Acad Sci U S A. 2007;104:5193–5198. doi: 10.1073/pnas.0605699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L, Wine-Lee L, Ahn KJ, Crenshaw EB., 3rd Genetic analyses demonstrate that bone morphogenetic protein signaling is required for embryonic cerebellar development. J Neurosci. 2006;26:1896–1905. doi: 10.1523/JNEUROSCI.3202-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan P, Panchision DM, Newell LF, McKay RD. BMPs signal alternately through a SMAD or FRAP-STAT pathway to regulate fate choice in CNS stem cells. J Cell Biol. 2003;161:911–921. doi: 10.1083/jcb.200211021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice DS, Curran T. Role of the reelin signaling pathway in central nervous system development. Annu Rev Neurosci. 2001;24:1005–1039. doi: 10.1146/annurev.neuro.24.1.1005. [DOI] [PubMed] [Google Scholar]
- Rios I, Alvarez-Rodriguez R, Marti E, Pons S. Bmp2 antagonizes sonic hedgehog-mediated proliferation of cerebellar granule neurones through Smad5 signalling. Development. 2004;131:3159–3168. doi: 10.1242/dev.01188. [DOI] [PubMed] [Google Scholar]
- Sato T, Joyner AL, Nakamura H. How does Fgf signaling from the isthmic organizer induce midbrain and cerebellum development? Dev Growth Differ. 2004;46:487–494. doi: 10.1111/j.1440-169x.2004.00769.x. [DOI] [PubMed] [Google Scholar]
- Schmahmann JD. Disorders of the cerebellum: ataxia, dysmetria of thought, and the cerebellar cognitive affective syndrome. J Neuropsychiatry Clin Neurosci. 2004;16:367–378. doi: 10.1176/jnp.16.3.367. [DOI] [PubMed] [Google Scholar]
- Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- Shim JH, Greenblatt MB, Xie M, Schneider MD, Zou W, Zhai B, Gygi S, Glimcher LH. TAK1 is an essential regulator of BMP signalling in cartilage. EMBO J. 2009;28:2028–2041. doi: 10.1038/emboj.2009.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang VY, Rose MF, Zoghbi HY. Math1 expression redefines the rhombic lip derivatives and reveals novel lineages within the brainstem and cerebellum. Neuron. 2005;48:31–43. doi: 10.1016/j.neuron.2005.08.024. [DOI] [PubMed] [Google Scholar]
- Weisheit G, Gliem M, Endl E, Pfeffer PL, Busslinger M, Schilling K. Postnatal development of the murine cerebellar cortex: formation and early dispersal of basket, stellate and Golgi neurons. Eur J Neurosci. 2006;24:466–478. doi: 10.1111/j.1460-9568.2006.04915.x. [DOI] [PubMed] [Google Scholar]
- Xu J, Liu Z, Ornitz DM. Temporal and spatial gradients of Fgf8 and Fgf17 regulate proliferation and differentiation of midline cerebellar structures. Development. 2000;127:1833–1843. doi: 10.1242/dev.127.9.1833. [DOI] [PubMed] [Google Scholar]
- Xu X, Han J, Ito Y, Bringas P, Jr., Deng C, Chai Y. Ectodermal Smad4 and p38 MAPK are functionally redundant in mediating TGF-beta/BMP signaling during tooth and palate development. Dev Cell. 2008;15:322–329. doi: 10.1016/j.devcel.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Li C, Herrera PL, Deng CX. Generation of Smad4/Dpc4 conditional knockout mice. Genesis. 2002;32:80–81. doi: 10.1002/gene.10029. [DOI] [PubMed] [Google Scholar]
- Yang ZJ, Ellis T, Markant SL, Read TA, Kessler JD, Bourboulas M, Schuller U, Machold R, Fishell G, Rowitch DH, Wainwright BJ, Wechsler-Reya RJ. Medulloblastoma can be initiated be deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell. 2008;14:135–145. doi: 10.1016/j.ccr.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou YX, Zhao M, Li D, Shimazu K, Sakata K, Deng CX, Lu B. Cerebellar deficits and hyperactivity in mice lacking Smad4. J Biol Chem. 2003;278:42313–42320. doi: 10.1074/jbc.M308287200. [DOI] [PubMed] [Google Scholar]
- Zhuo L, Theis M, Alvarez-Maya I, Brenner M, Wilecke K,M, Messing A. hGFAP-cre transgenic mice for manipulation of glial and neuronal function in vivo. Genesis. 2001;31:85–94. doi: 10.1002/gene.10008. [DOI] [PubMed] [Google Scholar]
- Zordan P, Croci L, Hawkes R, Consalez GG. Comparative analysis of proneural gene expression in the embryonic cerebellum. Dev. Dyn. 2008;237:1726–1735. doi: 10.1002/dvdy.21571. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.