Abstract
The SAGA transcription co-activator plays multiple roles in regulating transcription due to the presence of functionally independent modules of subunits within the complex. We have recently identified a role for the ubiquitin protease activity of SAGA in regulating tissue-specific gene expression in Drosophila. Here, we discuss the modular nature of SAGA and the different mechanisms through which SAGA is recruited to target promoters. We propose that the genes sensitive to loss of the ubiquitin protease activity of SAGA share functional characteristics that require de-ubiquitination of ubH2B for full activation. We hypothesize that de-ubiquitination of ubH2B by SAGA destabilizes promoter nucleosomes, thus enhancing recruitment of Pol II to weak promoters. In addition, SAGA-mediated de-ubiquitination of ubH2B may facilitate binding of factors that are important for the transition of paused Pol II into transcription elongation.
SAGA is essential for metazoan development
The Spt-Ada-Gcn5-acetyltransferase (SAGA) complex is a large, multi-subunit transcription co-activator that is highly conserved from yeast to humans [1, 2]. While SAGA is not essential for viability in the yeast Saccharomyces cerevisiae, mutations in SAGA subunits cause severe developmental defects resulting in lethality in Drosophila melanogaster and in mice [1, 2]. Thus, SAGA appears to have critical roles in regulating gene expression in metazoans that are essential for development. Supporting this notion, mutations in SAGA subunits result in defects in photoreceptor axon targeting in Drosophila [3]. Moreover, Gcn5 mouse mutants show defects in mesoderm development [4]. Thus, metazoan SAGA is associated with specific developmental phenotypes and is essential for viability.
Our recent work suggests that one of the reasons SAGA is critical for metazoan development might be because of the role of its ubiquitin protease activity in regulating tissue-specific developmental gene expression [5]. Notably, the genes that are down-regulated following loss of SAGA ubiquitin protease activity constitute only a small fraction of the total genes bound by SAGA in embryonic muscle [5]. Why are only a fraction of the genes bound by SAGA sensitive to loss of the ubiquitin protease activity of the complex? We believe that the characteristics of this subset of genes might render them particularly sensitive to loss of this specific activity of SAGA, which possesses a variety of activities involved in regulating transcription. In this review we will discuss the modular nature of SAGA that underlies its multiple functions during transcription activation. We will outline various mechanisms that might be used to recruit SAGA to its target genes, and discuss the histone and non-histone substrates of the ubiquitin protease activity of SAGA. We will then present potential mechanisms through which de-ubiquitination of ubH2B by SAGA could potentially activate tissue-specific gene expression.
The modular structure of SAGA supports multiple activities
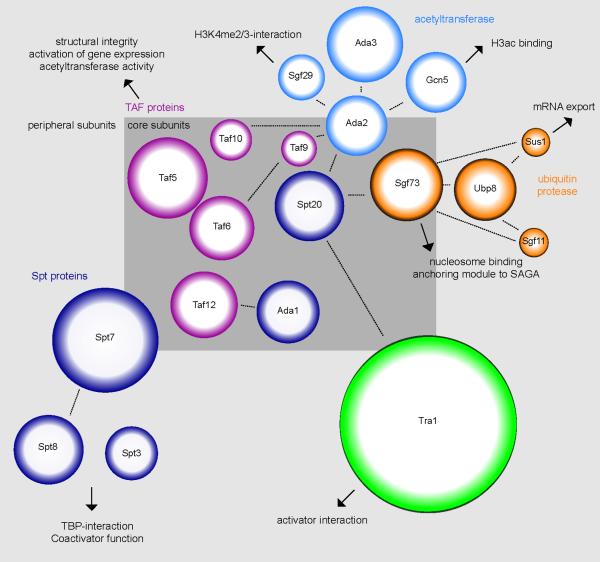
SAGA is composed of 18 to 20 protein subunits that can be separated into four distinct modules based on structural and functional evidence primarily from S. cerevisiae [6–8]. These modules include two distinct enzymatic activities involved in post-translational modification (Figure 1). The first of these SAGA modules contains the acetyltransferase Gcn5 together with Ada2, Ada3 and Sgf29; this association enables Gcn5 to acetylate multiple lysine residues on nucleosomal histone H3 and is important for in vivo HAT activity (glossary) [9–11]. A second module within SAGA consists of the ubiquitin-specific protease Ubp8, which is activated by its association with Sgf11, Sgf73 and Sus1 to catalyze the removal of ubiquitin from substrates that include ubH2B [12–21].
Figure 1. The modular nature of SAGA supports multiple activities.
Subunits within the different modules of S. cerevisiae SAGA are indicated by different colors: acetyltransferase (blue), ubiquitin protease (orange), TAF (pink), Spt (purple) and Tra1 (green). The relative molecular weight of each subunit is indicated by the area of the circle. Subunits that are more central to the complex are shown within the inner box, and peripheral subunits are shown inside the outer box. Different functions of individual subunits are indicated by the arrows and text. Probable physical connections between subunits are indicated by dotted lines.
In addition to these enzymatic modules, SAGA contains a third module consisting of several TAF proteins that are shared with the general transcription factor TFIID [6, 7]. The TAFs might have structural roles within SAGA, because the histone-fold domains of TAF6, TAF9, Ada1 and TAF12 form a complex in vitro that shares remarkable similarities with the histone octamer [22]. However, the TAFs could also regulate transcription directly because the TAF6-like subunit in Drosophila SAGA, SAF6, is required for SAGA-regulated gene expression independent of its histone modifying activities [23]. The fourth module of SAGA contains the transcription activator-binding protein Tra1, together with several Spt proteins [6]. Whereas Tra1 is involved in the recruitment of SAGA to promoters through interactions with transcription activators [24, 25], Spt3 and Spt8 directly interact with components of the general transcription machinery such as TBP [8, 26–28]. Thus, in addition to its histone modifying activities, SAGA is a bona fide transcription co-activator that functions in recruitment of Pol II and PIC formation [29].
Although the general structure and function of these four modules is likely to be conserved, there may be differences in their composition and function in species other than S. cerevisiae. A recent examination of SAGA mutant phenotypes in Schizosaccharomyces pombe suggests that the Tra1/Spt and ubiquitin protease modules could contain sub-modules with distinct functions [25]. Moreover, there may be crosstalk between Gcn5 and the mammalian Ubp8 ortholog, USP22, because deletion of Gcn5 diminishes the ubiquitin protease activity of SAGA toward non-histone substrates [30]. Thus, although this modular composition of SAGA may be subject to further refinement, it can be concluded that SAGA possesses multiple activities that are delineated by distinct groups of subunits.
SAGA recruitment/retention is mediated through multiple interactions with transcriptionfactors and chromatin
The first step in activation of transcription by SAGA requires its recruitment to gene-promoters. In S. cerevisiae, SAGA is recruited to the GAL promoter prior to recruitment of Pol II and PIC formation [26, 29, 31]. Indeed, SAGA binds to promoters of many different genes in a variety of fungal and metazoan species [5, 17, 25, 32–37]. For example, the Ada2 subunit of SAGA localizes to approximately 200 promoters of genes involved in stress-response pathways and metabolic functions in the fungal pathogen Candida albicans [35]. Furthermore, in HeLa cells the SAGA subunits Sgf29, Tra/TRRAP, Spt3, Spt20, ATX7L3 and Ada3 localize to the promoters of genes associated with the endoplasmic reticulum stress response, transcription regulation, replication, development and morphogenesis [17, 32, 33].
How is SAGA recruited to promoters? Multiple studies indicate that Tra1 plays a crucial role in recruiting SAGA to gene-specific promoters (Figure 2A). In S. cerevisiae, Tra1 interacts directly with transcription activators and is important for recruitment of SAGA to individual genes [24, 38–40]. Furthermore, the mammalian ortholog of Tra1, TRRAP, interacts directly with the oncoprotein transcription factors c-Myc and E2F [41]. Is the interaction of Tra1/TRRAP with transcription activators sufficient to recruit SAGA to all its target genes? Recent work in S. pombe suggests that this is not the case [25]. In contrast to S. cerevisiae, in which Tra1 is a component of both SAGA and the essential NuA4 HAT complexes [42, 43], in S. pombe there are two Tra1 paralogs including a SAGA-specific form that is not required for viability [25, 44]. Intriguingly, although Tra1 is required to recruit SAGA to the promoters of some genes in S. pombe, SAGA recruitment to other genes is independent of Tra1 [25].
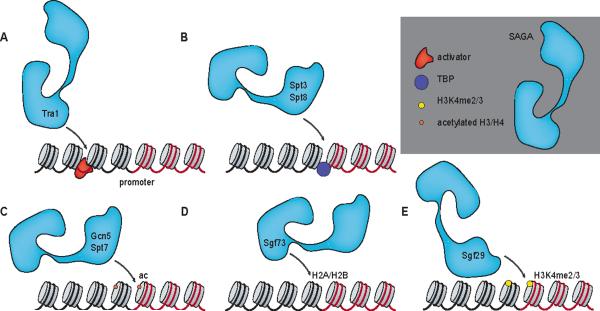
Figure 2. SAGA is recruited through multiple interactions.
Multiple SAGA subunits interact with both protein factors and chromatin marks that are present at gene promoters in S. cerevisiae. (A) Tra1 interacts with transcription activators. (B) Spt3 and Spt8 interact with TBP and could potentially recruit SAGA to genes containing the core transcription machinery. (C) The bromodomains of Gcn5 and Spt7 interact with acetylated nucleosomes. (D) Sgf73 binds H2A-H2B dimers via a novel Zn-binding motif in its SCA7 domain. (E) Sgf29 binds H3K4me2/3. Adapted from [69].
What other SAGA subunits might be involved in recruiting SAGA to promoters? Crosslinking studies suggest that in addition to Tra1, Taf12 might be involved in mediating interactions of transcription activators with SAGA [39, 40]. Furthermore, the binding of Spt3 and Spt8 to TBP could potentially recruit SAGA to promoters via interactions with the core transcription machinery [26–28] (Figure 2B). Moreover, in addition to being recruited to its target genes through interactions with transcription activators or the transcription machinery, SAGA could potentially be recruited or retained at promoters via interactions with chromatin marks [11, 32, 45]. For example, the bromodomains of Spt7 and Gcn5 interact with acetylated nucleosomes and are important for SAGA retention at promoters in S. cerevisiae in the absence of transcription activators (Figure 2C) [45]. In addition, Sgf73/ATXN7 binds H2A-H2B dimers via a novel Zn-binding fold within its SCA7 domain (Figure 2D) [46]. Furthermore, Sgf29 within SAGA binds H3K4me2/3 via its double Tudor-domain (Figure 2E) [11, 32]. Notably, Sgf29 localization correlates with the presence of H3K4me3 in human cells [32]. Moreover, deletion of Sgf29 in S. cerevisiae disrupts SAGA localization genome-wide despite having no effect on the integrity or in vitro HAT activity of the complex [11]. This suggests that SAGA recruitment and/or retention at promoters may be regulated through a combination of interactions with transcription activators, the transcription machinery and chromatin marks.
The finding that there are multiple mechanisms involved in SAGA recruitment has important implications for how SAGA becomes localized to different genes in particular cell types. In Drosophila, SAGA is associated with different genes in embryonic muscle relative to neurons [5]. Are interactions with transcription factors solely responsible for the recruitment of SAGA to different genes in these two distinct cell types? Consistent with the hypothesis that SAGA is recruited to genes through interactions with transcription factors, the number of genes bound by SAGA in different embryonic tissues correlates with the number of transcription factors associated with the complex in these tissues [5]. However, it is not known if any of these individual transcription factors are required for SAGA recruitment in flies. Notably, transcription factor binding sites tend to be clustered within the genome in Drosophila [47, 48]. Thus, combinatorial interactions of transcription factors with SAGA could potentially be required to direct SAGA to its appropriate target genes in metazoans. Furthermore, it is possible that SAGA might be required for Pol II recruitment only at certain genes, while at other genes SAGA could be recruited following binding of Pol II and transcription initiation through interactions with the transcription machinery and chromatin marks.
Saga Modules Play Independent Roles In Regulating Transcription
Saga Contains Multiple Activities That Are Mediated By Distinct Groups Of Subunits Within The Complex, And This Raises The Question As To Whether Different Saga Modules Can Function Independently. Analysis Of Mutants That Lack Individual Saga Subunits Shows That The Different Modules Of Saga Can Indeed Function Independently, And Might Even Play Distinct Roles In Activating Transcription At Particular Genes. For Example, The Hat And Ubiquitin Protease Activities Of The Complex Are Separable [3, 11, 16]. In Addition, Deletion Of Saga Subunits From Different Modules Of The Complex Results In Distinct Phenotypes In Yeast [8, 25]. Notably, Different Modules Of Saga Might Even Have Cooperative Effects On Transcription. For Instance, Deletion Of Ubp8 Exacerbates Transcription Defects Caused By Loss Of Gcn5 In S. Cerevisiae [18]. Furthermore, Deletion Of Gcn5 In Conjunction With Spt3 Or Spt8 Results In More Severe Phenotypes Than The Individual Deletions [8]. Thus, The Different Modules Of Saga Could Play Distinct Roles In Regulating Transcription At Individual Genes, Or Even At Different Sets Of Genes. Could The Different Modules Of Saga Be Independently Recruited To Individual Genes? There Is Some Evidence Supporting Independent Recruitment Of Saga Modules From S. Cerevisiae In Which Ubp8 And Gcn5 Are Observed On The Coding Region Of The Gal1 Gene, While Spt8 Remains Bound Only At The Promoter [34]. However, No Spt8 Ortholog Has Been Identified In Either Drosophila Or Human Saga, Suggesting That Metazoan Saga May Be More Closely Related To The Saga-Related Complex In S. Cerevisiae, Salsa [1]. Thus At Present, It Is Unclear Whether Saga Modules Can Be Recruited Independently Of The Remainder Of The Complex In Metazoans.
Distinct Roles For The Different Modules Of Saga Are Supported By Gene Expression Analysis Studies In Yeast And Flies [3, 25, 49]. In S. Cerevisiae, There Are Overlapping But Also Distinct Sets Of Genes That Show Defects In Expression Following Deletion Of The Saga Subunits Spt3, Spt20 Or Gcn5 [49]. Similarly, Mutations In Components Of The Ubiquitin Protease Module In Drosophila Show Overlapping, But Also Distinct Effects On Gene Expression Relative To Loss Of Ada2B [3]. Moreover, Analysis Of Gene Expression Profiles Of Various Saga Mutants In Both S. Cerevisiae And S. Pombe Show Some Differences That Are Consistent With The Modular Organization Of The Complex [14, 25].
Recent Studies In Drosophila Have Shown That Only A Subset Of Genes Bound By Saga In Embryonic Muscle Are Sensitive To Mutations In Sgf11, Which Eliminates Ubiquitin Protease Activity [5]. Why Are These Genes Particularly Sensitive To Loss Of The Ubiquitin Protease Activity Of Saga? An Examination Of The Subset Of Saga-Bound Genes That Are Down-Regulated In Sgf11 Muscle Showed That These Genes Tend To Be Expressed At Higher Levels In Muscle Relative To Other Tissues In The Embryo [5]. Furthermore, These Genes Were Enriched For Functions Involved In Developmental Processes [5]. The Fact That This Subset Of Saga-Bound Genes Shares Common Characteristics Suggests That These Genes Could Be Transcriptionally Activated Using A Similar Mechanism Requiring the ubiquitin protease activity of SAGA.
De-ubiquitination of ubH2B could activate tissue-specific gene expression
While the non-histone substrates of the SAGA ubiquitin protease activity might play roles in fine-tuning gene activation by SAGA (Box 1), ubH2B itself has the potential to be an important substrate required for activation of sgf11-dependent genes in Drosophila muscle. How could de-ubiquitination of ubH2B activate expression of tissue-specific genes?
H2B ubiquitination requires the phosphorylation of serine 5 within the CTD of Pol II, which occurs during PIC formation at an early stage of the transcription cycle [50, 51]. There are a number of important regulatory events that occur following recruitment of Pol II to promoters and transcription initiation, prior to productive transcription elongation [52]. One of these regulatory events occurs soon after Pol II has begun transcribing, when it pauses around 50 nucleotides into the gene, before continuing to transcribe the remainder of the gene [52]. The decision as to whether Pol II is retained or released from this pause site constitutes an important regulatory step in metazoan transcription [52]. Could the ubiquitin protease activity of SAGA be involved in regulating pausing of Pol II? There are hints that SAGA may indeed be involved in regulating pausing. For instance, the presence of SAGA correlates with high levels of Pol II at the pause site in both Drosophila and human cells [5, 32]. Notably, genes with paused Pol II in early Drosophila embryos tend to be expressed in a tissue-specific manner [53, 54].
ubH2B regulates nucleosome stability and polymerase pausing
How could the ubiquitin protease activity of SAGA regulate the retention or release of paused Pol II? In S. cerevisiae, the ubiquitin protease activity of SAGA facilitates recruitment of the Ctk1 kinase to the inducible GAL gene, where it then phosphorylates serine 2 within the Pol II CTD [34]. This phosphorylation is important for the transition of Pol II from an initiating form to an elongating form, and serves to recruit factors that are important for productive transcription elongation [52]. Although promoter-proximal pausing of Pol II does not appear to play a major regulatory role during transcription activation in S. cerevisiae, in metazoans Pol II pauses, at least transiently, during transcription of most genes [55, 56]. Moreover, the rate of release of paused Pol II is highly regulated at particular subsets of genes such as developmental control genes that are expressed in a tissue-specific manner [53–55]. In metazoans, there are two CTD kinases that phosphorylate serine 2: CDK9 and CDK12/13 [57]. CDK9 associates with a regulatory subunit, Cyclin T, to form the P-TEFb complex, while CDK12 interacts with Cyclin K [57]. In Drosophila, P-TEFb plays an important role in regulating expression of genes, such as the heat shock genes, at which the rate-determining step of transcription occurs at release of paused Pol II [52]. It is possible that SAGA regulates release of Pol II at this type of inducible gene in metazoans because knockdown of Nonstop (Ubp8) in cultured Drosophila cells significantly reduces expression of Hsp70 following heat shock [58]. However, CDK12 rather than CDK9 appears to be the ortholog of S. cerevisiae Ctk1 [57]. Thus, it is unclear whether a mechanism involving the inhibition of kinase binding through persistent ubH2B would hold true for P-TEFb and CDK12, or only CDK12 [34]. Notably, Drosophila CDK12 is associated strongly with developmental puffs on polytene chromosomes indicating that this kinase might play a more important role in the regulation of developmentally paused genes than P-TEFb [57].
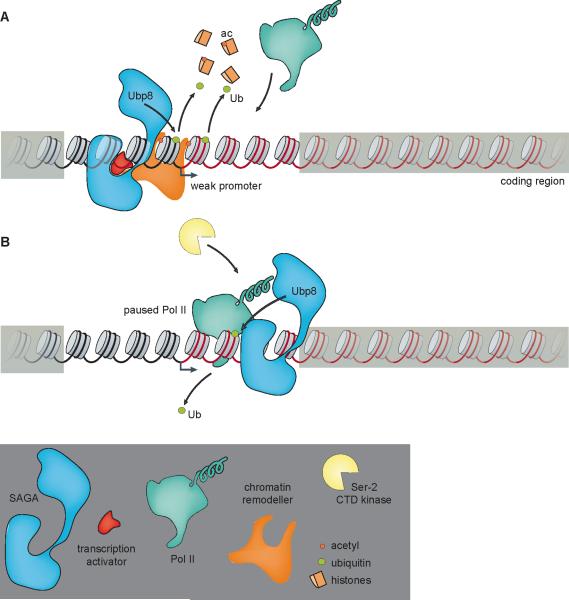
Persistent ubH2B may have other effects in addition to its direct inhibition of Ctk1 binding in S. cerevisiae [34]. Nucleosomes containing ubH2B exhibit decreased sensitivity to micrococcal nuclease digestion and enhanced resistance to salt extraction, both of which are consistent with enhanced stability of the ubH2B-containing nucleosome [59]. Moreover, in vitro-reconstituted ubH2B-containing nucleosomes are more resistant to DNase I digestion than nucleosomes with H2B [60]. In addition, there is a general increase in nucleosome occupancy at promoters in ubp8Δ yeast [59, 61]. Whereas this increase in nucleosome stability and/or occupancy appears to have negative consequences on Pol II recruitment at weakly expressed genes, it has positive effects on highly transcribed genes by promoting nucleosome reassembly in the wake of the elongating polymerase [61]. Interestingly, the increase in the stability and/or occupancy of nucleosomes containing ubH2B is independent of downstream effects on H3K4, H3K79 or H3K36 methylation [61]. Thus, de-ubiquitination of ubH2B by SAGA could enhance Pol II occupancy at weak promoters by destabilizing the nucleosome (Figure 3A). Notably, the HAT activity of SAGA might further destabilize nucleosomes at weak promoters by facilitating recruitment and/or retention of chromatin remodeling complexes (Figure 3A) [62]. Thus, de-ubiquitination of ubH2B may function coordinately with histone acetylation to activate gene expression at some genes. In addition, because de-ubiquitination of ubH2B is important for recruitment of the CTD kinase Ctk1 in S. cerevisiae, it is likely that genes at which transcription is regulated through Pol II pausing might also be particularly sensitive to loss of SAGA ubiquitin protease activity (Figure 3B). Notably, these two classes of genes could overlap significantly because genes with paused Pol II in Drosophila tend also to have promoters that are occluded by nucleosomes [55]. Intriguingly, in addition to its role at promoters, SAGA colocalizes with Pol II on the coding region of genes [5, 34]. Loss of ubH2B correlates with decreases in nucleosome and Pol II occupancy on the coding region of genes in S. cerevisiae [61, 63]. Thus, SAGA-mediated deubiquitination of ubH2B on the coding region could potentially play a repressive role by limiting transcription elongation, especially at highly transcribed genes [61]. Hence, deubiquitination of ubH2B by SAGA might have activating or repressive roles depending on the gene context.
Figure 3. Potential mechanisms of tissue-specific gene activation by de-ubiquitination of ubH2B.
(A) SAGA-mediated de-ubiquitination of ubH2B at weak promoters might destabilize promoter nucleosomes, facilitating recruitment of Pol II. The acetylation of promoter-proximal nucleosomes by SAGA at these same weak promoters could also stimulate chromatin remodeling, thus enhancing the recruitment of Pol II. (B) At genes at which pausing of Pol II constitutes an important rate-determining step in transcription, de-ubiquitination of ubH2B by SAGA might enhance recruitment and/or retention of serine 2 CTD kinases that facilitate release of Pol II into productive transcription elongation. Adapted from [69].
Concluding remarks
SAGA is a multi-functional transcription co-activator that is essential for metazoan development. SAGA is modular in structure, and individual modules have independent roles in regulating transcription. Thus, mutations in different subunits of SAGA exhibit differential effects on transcription. We have recently shown that the ubiquitin protease activity of SAGA is important for expression of tissue-specific, developmental genes in Drosophila embryonic muscle [5]. In addition to ubH2B, there are several alternative substrates of the ubiquitin protease activity of SAGA including transcription regulator proteins (Box #1) [13, 17, 19, 30, 57, 64, 65]. While these alternative substrates might play a role in fine-tuning gene expression, ubH2B has the potential to be the critical substrate of SAGA involved in activation of tissue-specific genes. Deubiquitination of ubH2B could activate expression by destabilizing promoter nucleosomes and enhancing Pol II recruitment to weak promoters, and/or by stimulating release of paused Pol II via recruitment of the serine 2 CTD kinase(s). It will be important to determine whether these different mechanisms are used at distinct or overlapping sets of genes in metazoans. Furthermore, it is likely that additional mechanisms could be involved in activating gene expression through de-ubiquitination of ubH2B. Obtaining a mechanistic understanding of the ways in which different aspects of SAGA activity contribute to regulating gene expression at particular subsets of genes will be crucial to understand the role this important transcription co-activator plays during metazoan development.
Box 1: SAGA de-ubiquitinates both histone and non-histone substrates.
In addition to ubH2B [18, 50, 51], several other substrates have been identified as targets of the ubiquitin protease activity of SAGA [13, 17, 19, 30, 57, 64, 65]. These include the mammalian telomeric-repeat-binding factor 1 (TRF1), the sucrose non-fermenting 1 (Snf1) AMP protein kinase in S. cerevisiae, and the human transcriptional regulator (FUSE)-binding protein 1 (FBP1) [30, 64, 65]. Furthermore, human SAGA de-ubiquitinates another histone, monoubiquitinated H2A (ubH2A) [13, 17, 19, 58]. Could de-ubiquitination of these substrates be involved in activation of tissue-specific gene expression by SAGA? TRF1 is a component of the shelterin complex that protects mammalian telomeres from DNA damage and inhibits access of telomerase to chromosome ends [30]. Thus, stabilization of TRF1 levels via SAGA-mediated deubiquitination seems unlikely to be involved in transcription activation. The transcriptional regulator FBP1 could potentially play a role in regulating transcription activation, but it is best characterized for its regulation of oncogenes and cell-cycle genes such as c-Myc and p21 rather than developmental or tissue-specific genes, and does not have a clearly identifiable ortholog in Drosophila [64]. The third non-histone substrate, Snf1, forms part of a kinase complex that activates genes and is highly conserved in metazoans [65]. Intriguingly, Drosophila mutants that lack the Snf1 ortholog, SNF1A/AMP-activated protein kinase (AMPK), have defects in visceral muscle morphology [66]. UbH2A is associated with transcription repression and is deposited by the Polycomb repressive complex 1 (PRC1) [50, 51]. Thus, it is possible that de-ubiquitination of ubH2A by SAGA could counteract the repressive effects of Polycomb-mediated silencing and activate gene expression. In particular, ubH2A has been suggested to inhibit transcription elongation [67, 68]. This observation raises the possibility that de-ubiquitination of ubH2A by SAGA could stimulate transcription elongation at Polycomb-regulated genes. However, it is currently unclear whether SAGA-mediated de-ubiquitination of ubH2A, rather than ubH2B, is the major target involved in activation of Polycomb repressed genes [19].
Acknowledgements
We thank Jamie Dyer and Kenneth Lee for comments and discussion. This work was supported by grant R37GM047867-18S1 from the NIGMS to Jerry Workman and Susan Abmayr, and funding from the Stowers Institute.
Glossary Box
- HAT
histone acetyltransferases. Catalyze the transfer of an acetyl group from the donor acetyl-CoA to one or more lysine residues, predominantly in the N-terminal tails, of histone proteins.
- ubH2B, ubH2A
mono-ubiquitinated histone H2B, mono-ubiquitinated histone H2A. ubH2B contains a single ubiquitin moiety conjugated to lysine 123 of histone H2B in S. cerevisiae, lysine 119 in S. pombe, or lysine 120 in mammalian cells. In mammalian cells, ubH2A contains a single ubiquitin moiety conjugated to lysine 119 of histone H2A, while this modification is not present in S. cerevisiae.
- TAF, TBP
TBP-associated factors, TATA-binding protein. TAFs interact with TBP within the transcription coactivator TFIID, which is involved in formation of the PIC together with Pol II and the general transcription factors. In addition to being associated with TFIID, some TAFs are also core subunits of other transcription coactivators such as SAGA.
- PIC, Pol II
pre-initiation complex, RNA polymerase II. The PIC consists of Pol II and the general transcription factors TFIIA, TFIIB, TFIID, TFIIE, TFIIF and TFIIH, which bind to promoters during the initial stages of transcription activation.
- TRRAP
Transformation/Transcription domain-Associated Protein. TRRAP is the mammalian ortholog of S. cerevisiae Tra1 and interacts with transcription activators such as c-Myc and E2F.
- H3K4me3
tri-methylated H3 Lys-4. H3K4me3 is catalyzed by the Set1 histone methyltransferase within the COMPASS complex in S. cerevisiae and is associated with active transcription.
- CTD
carboxy-terminal domain. The CTD of the largest subunit of Pol II, Rpb1, consists of multiple repeats of the heptad peptide sequence YSPTSPS, in which all three serine positions are subject to phosphorylation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Koutelou E, et al. Multiple faces of the SAGA complex. Curr Opin Cell Biol. 2010;22:374–382. doi: 10.1016/j.ceb.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rodriguez-Navarro S. Insights into SAGA function during gene expression. EMBO Rep. 2009;10:843–850. doi: 10.1038/embor.2009.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weake VM, et al. SAGA-mediated H2B deubiquitination controls the development of neuronal connectivity in the Drosophila visual system. EMBO J. 2008;27:394–405. doi: 10.1038/sj.emboj.7601966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu W, et al. Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nat Genet. 2000;26:229–232. doi: 10.1038/79973. [DOI] [PubMed] [Google Scholar]
- 5.Weake VM, et al. Post-transcription initiation function of the ubiquitous SAGA complex in tissue-specific gene activation. Genes Dev. 2011;25:1499–1509. doi: 10.1101/gad.2046211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee KK, et al. Combinatorial depletion analysis to assemble the network architecture of the SAGA and ADA chromatin remodeling complexes. Molecular systems biology. 2011;7:503. doi: 10.1038/msb.2011.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu PY, et al. Molecular architecture of the S. cerevisiae SAGA complex. Mol Cell. 2004;15:199–208. doi: 10.1016/j.molcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 8.Sterner DE, et al. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol Cell Biol. 1999;19:86–98. doi: 10.1128/mcb.19.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grant PA, et al. Expanded lysine acetylation specificity of Gcn5 in native complexes. The Journal of biological chemistry. 1999;274:5895–5900. doi: 10.1074/jbc.274.9.5895. [DOI] [PubMed] [Google Scholar]
- 10.Balasubramanian R, et al. Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. The Journal of biological chemistry. 2002;277:7989–7995. doi: 10.1074/jbc.M110849200. [DOI] [PubMed] [Google Scholar]
- 11.Bian C, et al. Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation. EMBO J. 2011;30:2829–2842. doi: 10.1038/emboj.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kohler A, et al. Structural basis for assembly and activation of the heterotetrameric SAGA histone H2B deubiquitinase module. Cell. 2010;141:606–617. doi: 10.1016/j.cell.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samara NL, et al. Structural insights into the assembly and function of the SAGA deubiquitinating module. Science. 2010;328:1025–1029. doi: 10.1126/science.1190049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ingvarsdottir K, et al. H2B ubiquitin protease Ubp8 and Sgf11 constitute a discrete functional module within the Saccharomyces cerevisiae SAGA complex. Mol Cell Biol. 2005;25:1162–1172. doi: 10.1128/MCB.25.3.1162-1172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee KK, et al. Yeast Sgf73/Ataxin-7 serves to anchor the deubiquitination module into both SAGA and Slik(SALSA) HAT complexes. Epigenetics & chromatin. 2009;2:2. doi: 10.1186/1756-8935-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee KK, et al. The deubiquitylation activity of Ubp8 is dependent upon Sgf11 and its association with the SAGA complex. Mol Cell Biol. 2005;25:1173–1182. doi: 10.1128/MCB.25.3.1173-1182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lang G, et al. The Tightly Controlled Deubiquitination Activity of the Human SAGA Complex Differentially Modifies Distinct Gene Regulatory Elements. Mol Cell Biol. 2011;31:3734–3744. doi: 10.1128/MCB.05231-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henry KW, et al. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao Y, et al. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol Cell. 2008;29:92–101. doi: 10.1016/j.molcel.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 20.Zhang XY, et al. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol Cell. 2008;29:102–111. doi: 10.1016/j.molcel.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kohler A, et al. Yeast Ataxin-7 links histone deubiquitination with gene gating and mRNA export. Nature cell biology. 2008;10:707–715. doi: 10.1038/ncb1733. [DOI] [PubMed] [Google Scholar]
- 22.Selleck W, et al. A histone fold TAF octamer within the yeast TFIID transcriptional coactivator. Nature structural biology. 2001;8:695–700. doi: 10.1038/90408. [DOI] [PubMed] [Google Scholar]
- 23.Weake VM, et al. A novel histone fold domain-containing protein that replaces TAF6 in Drosophila SAGA is required for SAGA-dependent gene expression. Genes Dev. 2009;23:2818–2823. doi: 10.1101/gad.1846409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown CE, et al. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science. 2001;292:2333–2337. doi: 10.1126/science.1060214. [DOI] [PubMed] [Google Scholar]
- 25.Helmlinger D, et al. Tra1 has specific regulatory roles, rather than global functions, within the SAGA co-activator complex. EMBO J. 2011;30:2843–2852. doi: 10.1038/emboj.2011.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larschan E, Winston F. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 2001;15:1946–1956. doi: 10.1101/gad.911501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mohibullah N, Hahn S. Site-specific cross-linking of TBP in vivo and in vitro reveals a direct functional interaction with the SAGA subunit Spt3. Genes Dev. 2008;22:2994–3006. doi: 10.1101/gad.1724408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laprade L, et al. Characterization of new Spt3 and TATA-binding protein mutants of Saccharomyces cerevisiae: Spt3 TBP allele-specific interactions and bypass of Spt8. Genetics. 2007;177:2007–2017. doi: 10.1534/genetics.107.081976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhaumik SR, Green MR. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 2001;15:1935–1945. doi: 10.1101/gad.911401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Atanassov BS, et al. Gcn5 and SAGA regulate shelterin protein turnover and telomere maintenance. Mol Cell. 2009;35:352–364. doi: 10.1016/j.molcel.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bryant GO, Ptashne M. Independent recruitment in vivo by Gal4 of two complexes required for transcription. Mol Cell. 2003;11:1301–1309. doi: 10.1016/s1097-2765(03)00144-8. [DOI] [PubMed] [Google Scholar]
- 32.Vermeulen M, et al. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010;142:967–980. doi: 10.1016/j.cell.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 33.Nagy Z, et al. The human SPT20-containing SAGA complex plays a direct role in the regulation of endoplasmic reticulum stress-induced genes. Mol Cell Biol. 2009;29:1649–1660. doi: 10.1128/MCB.01076-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wyce A, et al. H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGA-related complex. Mol Cell. 2007;27:275–288. doi: 10.1016/j.molcel.2007.01.035. [DOI] [PubMed] [Google Scholar]
- 35.Sellam A, et al. Genome-wide mapping of the coactivator Ada2p yields insight into the functional roles of SAGA/ADA complex in Candida albicans. Molecular biology of the cell. 2009;20:2389–2400. doi: 10.1091/mbc.E08-11-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Venters BJ, et al. A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces. Mol Cell. 2011;41:480–492. doi: 10.1016/j.molcel.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zsindely N, et al. The loss of histone H3 lysine 9 acetylation due to dSAGA-specific dAda2b mutation influences the expression of only a small subset of genes. Nucleic Acids Res. 2009;37:6665–6680. doi: 10.1093/nar/gkp722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhaumik SR, et al. In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer. Genes Dev. 2004;18:333–343. doi: 10.1101/gad.1148404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fishburn J, et al. Function of a eukaryotic transcription activator during the transcription cycle. Mol Cell. 2005;18:369–378. doi: 10.1016/j.molcel.2005.03.029. [DOI] [PubMed] [Google Scholar]
- 40.Reeves WM, Hahn S. Targets of the Gal4 transcription activator in functional transcription complexes. Mol Cell Biol. 2005;25:9092–9102. doi: 10.1128/MCB.25.20.9092-9102.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McMahon SB, et al. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell. 1998;94:363–374. doi: 10.1016/s0092-8674(00)81479-8. [DOI] [PubMed] [Google Scholar]
- 42.Grant PA, et al. The ATM-related cofactor Tra1 is a component of the purified SAGA complex. Mol Cell. 1998;2:863–867. doi: 10.1016/s1097-2765(00)80300-7. [DOI] [PubMed] [Google Scholar]
- 43.Allard S, et al. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 1999;18:5108–5119. doi: 10.1093/emboj/18.18.5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Calonge TM, et al. Transformation/transcription domain-associated protein (TRRAP)-mediated regulation of Wee1. Genetics. 2010;185:81–93. doi: 10.1534/genetics.110.114769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hassan AH, et al. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell. 2002;111:369–379. doi: 10.1016/s0092-8674(02)01005-x. [DOI] [PubMed] [Google Scholar]
- 46.Bonnet J, et al. The structural plasticity of SCA7 domains defines their differential nucleosome-binding properties. EMBO Rep. 2010;11:612–618. doi: 10.1038/embor.2010.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moorman C, et al. Hotspots of transcription factor colocalization in the genome of Drosophila melanogaster. Proc Natl Acad Sci U S A. 2006;103:12027–12032. doi: 10.1073/pnas.0605003103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zinzen RP, et al. Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature. 2009;462:65–70. doi: 10.1038/nature08531. [DOI] [PubMed] [Google Scholar]
- 49.Lee TI, et al. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature. 2000;405:701–704. doi: 10.1038/35015104. [DOI] [PubMed] [Google Scholar]
- 50.Atanassov BS, et al. The role of deubiquitinating enzymes in chromatin regulation. FEBS letters. 2011;585:2016–2023. doi: 10.1016/j.febslet.2010.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 52.Fuda NJ, et al. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461:186–192. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zeitlinger J, et al. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muse GW, et al. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gilchrist DA, et al. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mayer A, et al. Uniform transitions of the general RNA polymerase II transcription complex. Nature structural & molecular biology. 2010;17:1272–1278. doi: 10.1038/nsmb.1903. [DOI] [PubMed] [Google Scholar]
- 57.Bartkowiak B, et al. CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev. 2010;24:2303–2316. doi: 10.1101/gad.1968210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ardehali MB, et al. Spt6 enhances the elongation rate of RNA polymerase II in vivo. EMBO J. 2009;28:1067–1077. doi: 10.1038/emboj.2009.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chandrasekharan MB, et al. Ubiquitination of histone H2B regulates chromatin dynamics by enhancing nucleosome stability. Proc Natl Acad Sci U S A. 2009;106:16686–16691. doi: 10.1073/pnas.0907862106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Davies N, Lindsey GG. Histone H2B (and H2A) ubiquitination allows normal histone octamer and core particle reconstitution. Biochimica et biophysica acta. 1994;1218:187–193. doi: 10.1016/0167-4781(94)90009-4. [DOI] [PubMed] [Google Scholar]
- 61.Batta K, et al. Genome-wide function of H2B ubiquitylation in promoter and genic regions. Genes Dev. 2011;25:2254–2265. doi: 10.1101/gad.177238.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weake VM, Workman JL. Inducible gene expression: diverse regulatory mechanisms. Nature reviews. Genetics. 2010;11:426–437. doi: 10.1038/nrg2781. [DOI] [PubMed] [Google Scholar]
- 63.Fleming AB, et al. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol Cell. 2008;31:57–66. doi: 10.1016/j.molcel.2008.04.025. [DOI] [PubMed] [Google Scholar]
- 64.Atanassov BS, Dent SY. USP22 regulates cell proliferation by deubiquitinating the transcriptional regulator FBP1. EMBO Rep. 2011;12:924–930. doi: 10.1038/embor.2011.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wilson MA, et al. Ubp8 and SAGA regulate Snf1 AMP kinase activity. Mol Cell Biol. 2011;31:3126–3135. doi: 10.1128/MCB.01350-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bland ML, et al. AMPK supports growth in Drosophila by regulating muscle activity and nutrient uptake in the gut. Dev Biol. 2010;344:293–303. doi: 10.1016/j.ydbio.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou W, et al. Histone H2A monoubiquitination represses transcription by inhibiting RNA polymerase II transcriptional elongation. Mol Cell. 2008;29:69–80. doi: 10.1016/j.molcel.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Eskeland R, et al. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol Cell. 2010;38:452–464. doi: 10.1016/j.molcel.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weake VM, Workman JL. Chromatin remodelling and the transcription cycle. Nature Reviews Genetics [Poster] 2011:12. [Google Scholar]