Abstract
The glideosome associated protein GAP50 is an essential protein in apicomplexan parasites such as Plasmodium, Toxoplasma and Cryptosporidium, several species of which are important human pathogens. The 44.6 kDa protein is part of a multi-protein complex known as the invasion machinery or glideosome, which is required for cell invasion and substrate gliding motility empowered by an actin-myosin motor. GAP50 is anchored through its C-terminal transmembrane helix into the inner membrane complex and interacts via a short 6 residue C-terminal tail with other proteins of the invasion machinery in the pellicle of the parasite. In this paper we describe the 1.7 Å resolution crystal structure of the soluble GAP50 domain from the malaria parasite P. falciparum. The structure shows an αββα fold with overall similarity to purple acid phosphatases with, however, little homology regarding the nature of the residues in the active site region of the latter enzyme. While purple acid phosphatases contain a phosphate bridged binuclear Fe-site coordinated by seven side chains with the Fe-ions 3.2 Å apart, GAP50 in our crystals contains two cobalt ions each with one protein ligand and a distance between the Co2+ ions of 18 Å.
Keywords: malaria, invasion machinery, glideosome, glideosome associated protein, crystal structure, phosphatase familyfold
1. Introduction
The phylum apicomplexa harbors major pathogenic species such as Cryptosporidumparvum, Toxoplasma gondii and Plasmodium spp., which can cause severe illnesses in humans. Plasmodium parasites cause malaria which is one of the most devastating infectious diseases worldwide with 300 to 500 million cases and one to two million casualties per year (Baird, 2005; Murray et al., 2012; Snow et al., 2005) mostly children and pregnant women in sub Saharan Africa. Of the five Plasmodium species responsible for infecting the human host, P. falciparum and P. vivax are the most important with P. malaria, P. ovale and more recently P. knowlesi causing far fewer cases (Cox-Singh et al., 2008).
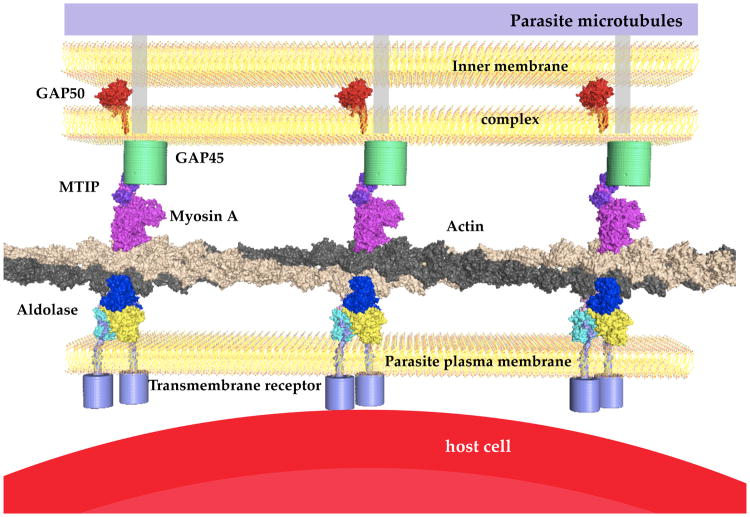
The life cycle of the Plasmodium parasite is very complex and involves various cellular interactions in the mosquito vector as well as in the human host. Sporozoites are injected during a blood meal into the skin by an infected female Anopheles mosquito. After invading hepatocytes, numerous merozoitese merge a few days later into the blood stream, which in their turn invade erythrocytes. After one to three days, depending on species, 10–20 new merozoitesegress from erythrocytes. Throughout the life cycle of the parasite various cell barriers need to be traversed to ensure survival and progeny of the parasite. A specialized multi-protein complex, which fulfills the function of “substrate gliding” and invasion of host cells, is highly conserved throughout the phylum apicomplexa (Kappe et al., 2004; Schmitz et al., 2005; Sibley, 2004; Sibley, 2010). This invasion machinery of the parasite, also called the “glideosome”, is located between the parasite plasma membrane (PPM) and the microtubule-supported inner membrane complex (IMC, Figure 1). The invasion machinery includes an adhesion protein (TRAP, MTRAP or CTRP, depending on the life stage of the parasite) linked via aldolase to short actin filaments (Buscaglia et al., 2003; Jewett and Sibley, 2003). These filaments are part of the actin-myosin motor including the MyosinA-tail interacting protein (MTIP) that connects to the GAP45-GAP50 complex (Baum et al., 2006; Bergman et al., 2003; Gaskins et al., 2004; Green et al., 2006; Herm-Gotz et al., 2002; Meissner et al., 2002; Sahoo et al., 2006).
Figure 1. Schematic overview of the invasion machinery in apicomplexan species.
The schematic drawing represents the known key players of the invasion machinery, space filled models represent known structures either from Plasmodium (Bosch et al., 2007b; Bosch et al., 2006a; Bosch et al., 2006b) or in the case of actin (rabbit) and myosin (chicken) (Holmes et al., 2003). The host cell in this case represents an erythrocyte, therefore the transmembrane receptor is MTRAP. In the liver stage this receptor is replaced by TRAP and in the insect stage with CTRP. The N-terminal sequences of TRAP, MTRAP and CTRP vary and have different length but their C-terminus is very similar and harbors one conserved tryptophan adjacent to charged residues. The transmembrane helix of PfGAP50 was modeled by extending the helical sequence. The exact orientation is unknown but evidence of an interaction with the other proteins of the invasion machinery exists e.g. through pull-down assays (Baum et al., 2006; Bergman et al., 2003; Johnson et al., 2007).
The rationale to study the invasion machinery complex is to obtain insight into this multi-protein assembly, and its mechanism of action, to be able to disrupt the chain of interactions between these proteins and thereby hopefully prevent the invasion of host cells. Previous crystallographic investigations revealed the interaction of P. falciparum aldolase in complex with the C-terminal tail of P. berghei TRAP, where a conformational change enables binding of the important penultimate tryptophan of TRAP into a pocket in the active site region of the enzyme (Bosch et al., 2007b). Additional to this structure, the structure of the complex of MTIP from two species, P. knowlesi and P. falciparum, in complex with the C-terminal tail of P. yoelii MyosinA have been described to atomic resolution (Bosch et al., 2007a; Bosch et al., 2006a). The inhibition of cell invasion by using the wildtype C-terminal MyosinA-tail with an IC50 of 84 μM was demonstrated, confirming the invasion machinery as a valid drug target (Bosch et al., 2006a; Kortagere et al., 2010; Thomas et al., 2010). To reduce the probability for the parasite to become resistant to a particular drug, it is useful to obtain, and use, multiple compounds interfering with different key steps of the parasite’s life cycle. In this connection, studies of multiple proteins of the parasite’s invasion machinery are potentially of great importance and hence we focus here on GAP50, a critical component of the invasion machinery.
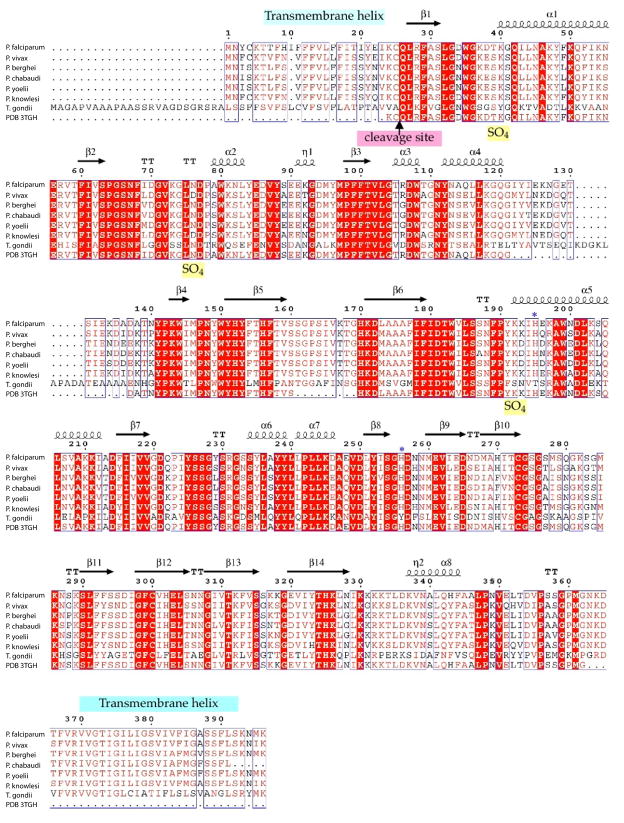
In T. gondii, GAP50 is glycosylated and localized in detergent-resistant membrane regions of the IMC (Johnson et al., 2007). All known GAP50 sequences from apicomplexan species possess a hydrophobic signal sequence and a C-terminal transmembrane helix according to transmembrane helix predictions and annotations in Plasmo DB (Figure 2). The N-terminal signal sequence needs to be cleaved off the protein for maturation and correct localization into the IMC. The fold of the soluble domain of GAP50 belongs to the divergent family of calcineurin-like phosphatases (PFAM PF00149). This soluble domain of GAP50 is conserved within apicomplex a with >80% sequence identity (Anantharaman et al., 2007) (Figure 2). Calcineurin-like phosphatases and purple acid phosphatases belong to the family of metallo-dependent phosphatases. A distinction is that calcineurin-like phosphatases exhibit the motif DIH(X)23GDYVDR(X)27GNHE, while purple acid phosphatases (PAPs) follow the motif DXG(X)nGDXXYXD(X)mGNHD/E, with bold letters indicating metal coordinating residues and conserved nonligand residues in the active site underlined (Rusnak and Mertz, 2000). The tyrosine (bold, underlined) in the active site of the PAPs is responsible for the visible purple color of the PAPs. The 1.7 Å crystal structure of the soluble domain of P. falciparum GAP50 (PfGAP50) described here in intriguingly shows that PfGAP50 also binds divalent metal ions but in a distinctly different manner than in the homologous purple phosphatase. The conservation of residues in a deep hydrophobic pocket leads to the suggestion that GAP50 might employ this conserved region for interactions with as yet unknown partner proteins of the malaria parasite’s invasion machinery.
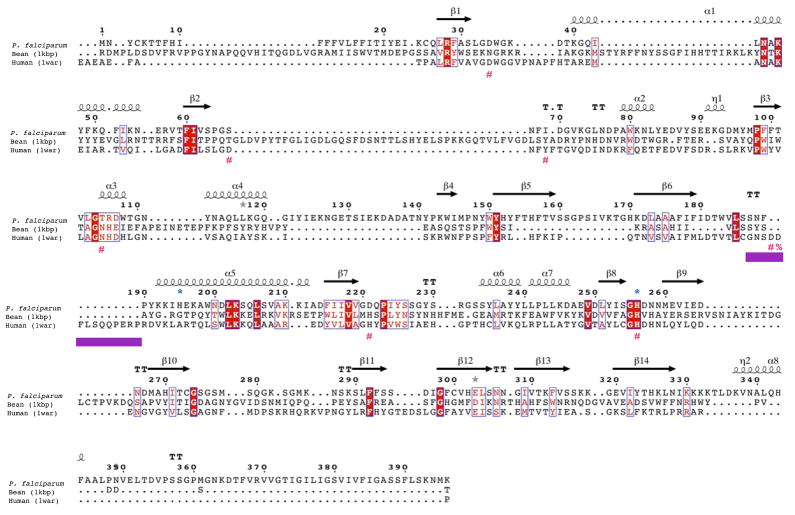
Figure 2. GAP50 sequence alignments.
A) GAP50 sequence alignment of various Plasmodium species and Toxoplasma gondii. (sequence information was obtained through BLAST searches in EuPathDBhttp://eupathdb.org). Predicted transmembrane helices are indicated in light blue bars. The blue *symbols represent residues involved in coordination of the cobalt ions, the Toxoplasma (Daher and Soldati-Favre, 2009) and Plasmodium signal sequence cleavage site (Müller et al., 2010)(Yeoman et al., 2011) is indicated by an arrow.
B) Structure based sequence alignment of Plasmodium falciparum GAP50 Human PAP and kidney bean PAP. The secondary structure elements of the Plasmodium GAP50 structure is shown above the sequence. The repression loop of hPAP is highlighted with a purple box and is inserted between Ser187 and Asn188 of the Plasmodium sequence. The key residue for inhibition of the human enzyme is marked with a red %. The blue *symbols represent residues involved in coordination of the cobalt ions in PfGAP50, red #symbols represent active site residues coordinating the iron atoms in hPAP.
2. Materials and Methods
2.1 Bioinformatic analysis
Psi-Blast (Altschul et al., 1997) searches were performed with the protein sequence from PfGAP50 (PlasmoDB accession code PFI0880c). Transmembrane helix prediction was carried out with the TMHMM server (Krogh et al., 2001) and signal peptide prediction with Signal P-HMM analysis (Emanuelsson et al., 2007).
2.2 Protein Expression and Purification
The gene encoding PfGAP50 was cloned from a P. falciparum cDNA library (Mehlin et al., 2006) into a pRSF vector (Novagene) using the primer combination Lys24 forward (5′-CATCCATGGGCAAATGTCAACTACGCTTTGC-3′) and Asp365 reverse (5′- AGCGGCCGCTTAATCTTTATTTCCCATGGGTCC-3′), spanning the soluble domain of PfGAP50 starting at residue Lys24 to Asp365 with an N-terminal TEV cleavable His6-tag.
Expression was carried out in E. coli BL21 (DE3) at 37°C using baffled flasks in terrific broth (TB) supplemented with 50 μg/ml ampicillin, 34 μg/ml kanamycin, 0.01% Antifoam (Sigma SE-15) and 2 μM CoSO4. Cells were induced with 0.5 mM IPTG at OD600=2 for 4 hours at 37°C. After harvesting, cells were washed once with 50 mM Hepes pH 8.25, 10% glycerol, 300 mM NaCl (buffer A) and pelleted again. The remaining cells were resuspended in a 1:1 ratio in buffer A and either flash frozen or disrupted by using 3 passes of French press at 1000 PSI. Benzonase was added after the first round of French press lysis to a final concentration of 2.5 U/ml and 1 mM MgCl2. The lysate was cleared from cell debris by 45 minutes centrifugation at 50,000×g at 4°C. Batch binding to equilibrated NiNTA in the presence of 10 mM imidazole was carried out for 60 minutes at 4 °C. The protein was eluted by a 300 mM imidazole step gradient and further purified using a hydroxyapatite column equilibrated with buffer A. The flow through containing PfGAP50 was dialysed overnight against 25 mM Hepes pH 8.25, 10% glycerol, 5 mM DTT (buffer B). The dialysed sample was centrifuged for 20 minutes at 50,000×g to pellet occasional precipitated protein impurities and then applied onto a pre-equilibrated Q sepharose column in buffer B. A sodium chloride gradient ranging from 0 to 500 mM was applied and the eluted fractions were subjected to SDS-PAGE analysis. Fractions containing pure PfGAP50 were pooled, concentrated and separated by molecular weight on a Sephadex 75 column equilibrated with 20 mM Hepes pH 8.25, 500 mM NaCl, 10% glycerol. Finally PfGAP50 was concentrated to 8 mg/ml and kept at 4 °C for further use. The protein was stable and crystallizable even after 6 months of storage at 4 °C but could not be stored frozen for crystallization experiments.
2.3 Identification of protein stabilizing agents via thermal shift assay
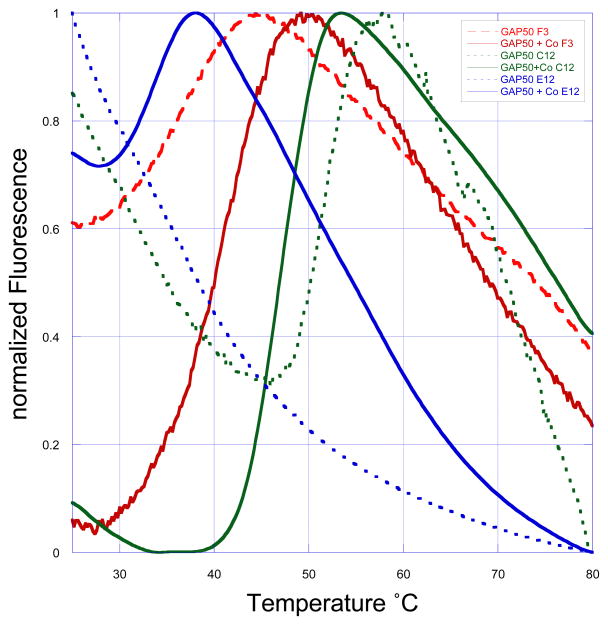
PfGAP50 initially precipitated within 24 hours after breaking the cells. In order to optimize the buffer conditions 96-well thermal shift assays with various buffers, divalent ions, variation in pH and salt concentration were carried out to identify stabilizing agents after the initial NiNTA purification step. The assay was carried out with 10 μM PfGAP50 per reaction in combination with the fluorescent dye SyPro Orange (Molecular Probes, Inc.) using an Opticon-2 (Biorad) RT-PCR machine (Crowther et al., 2010; Ericsson et al., 2006). Four divalent ions (Ca2+, Mg2+, Co2+, Zn2+) were identified to have a stabilizing effect on the protein and a more basic pH was favored versus neutral or acidic pH (Figure 3). Subsequently, small scale expression tests were carried out with 50 ml TB media supplemented with one of the identified ions, then subjected to cell lysis and purification via NiNTA as described earlier. Cells grown in the presence of Co2+ showed the highest soluble protein yield after dialysis, while the amount of precipitated protein was highest in cells grown in the presence of Mg2+. Therefore subsequent expressions were carried out with 2 μM CoSO4 in the growth medium. After the NiNTA capturing step the samples were supplemented with 2 μM CoSO4 to compensate for potential loss or exchange with leaching Ni2+ ions. Co-crystallization attempts with various divalent ions were undertaken, leading to crystals only in the presence of Co2+ or Zn2+. Crystals of PfGAP50 obtained in the presence of Zn2+ diffracted to a maximal resolution of only 6 Å and were therefore not further investigated. The crystals obtained in the presence of Co2+ during the purification steps were of considerable better quality and led to a structure as described in the next sections.
Figure 3. Thermal melt curves.
Examples of normalized thermal melt curves from a 96-well optimization screen using SyPro orange dye. On the y-axis the measured fluorescence from the dye bound to hydrophobic exposed regions is represented, the x-axis indicates the temperature. The temperature is continuously increased in 0.2°C steps over 60 minutes. A high initial fluorescence at room temperature is indicative of an unfolded protein since large hydrophobic regions are exposed allowing the dye to bind. The doted lines represent standard expressed and purified PfGAP50 after the NiNTA capturing step and desalting into buffer A. Thick lines represent PfGAP50 expressed in the presence of 2 μM CoSO4 after the NiNTA capturing step and desalting with buffer A plus 2 μM CoSO4. The decrease of the room temperature fluorescence suggests that the cobalt-supplemented protein is exposing less hydrophobic areas under various conditions (for simplicity only three are shown (F3:50 mM sodium cacodylate pH 6.0, C12:50 mM Tris pH 7.0, E12:50 mM Hepes pH 8.0). Additional to that a distinct melting temperature is observed. An increase of the melting temperature indicates a more stable protein. In the example shown the conditions with the thick green line has a Tm of 48 °C, whereas the red thick line only 39 °C and under the blue conditions 33 °C.
2.4 Initial Crystal Screening and Optimization
PfGAP50 containing 2 μM CoSO4 was shipped at 4°C for high-throughput crystallization screening using the microbatch under oil method at the Hauptman- Woodward Medical Research Institute (Luft et al., 2003). Based on the initial three crystal hits found, further optimization was carried out by the vapor diffusion sitting drop method. The final optimized crystal growth conditions were as follows: 0.2 M (NH4)2SO4, 0.1 M sodium acetate pH 6.0, 3% DMSO or 3% ethanol, 40% PEG 8000. Crystals were setup in hanging drop vapor diffusion experiments using 1 μl of protein solution and 1 μl reservoir solution. Crystals appeared within 24 hours at room temperature. The typical crystal size was approximately 100 × 100 × 20 μm with the shape of hexagonal plates.
2.5 Data Collection and Structure Determination
Initial crystals diffracted to ~6 Å, and could not be flash annealed (Yeh and Hol, 1998). After an extensive search, an optimal cryo protocol was identified using 40% PEG 8000, 18% ethyleneglycol, 0.2 M (NH4)2SO4, 0.1 M sodium acetate pH 6.0. Crystals soaked in this cryo-protectant diffracted to 4 Å and after two rounds of flash annealing reproducibly diffracted beyond 2 Å on a rotating anode generator. The crystals belonged to the trigonal space group P3221 with cell dimensions of a=57.6 Å, b=57.6 Å, c= 210.6 Å. Data reduction and scaling was carried out with XDS/XSCALE (Kabsch, 1993). The asymmetric unit contained one monomer resulting in a solvent content of 56.8 %. A core search model was generated from superposition of ten known PAP structures and structural homologs found through the EBI SSM server (Krissinel and Henrick, 2004). Flexible loops were trimmed off and the remaining 260 residues were mutated according to the PfGAP50 sequence. Only main chain and Cβ-atoms were used for molecular replacement, the remaining atoms were set to an occupancy of zero. Molecular replacement performed with Phaser (McCoy, 2007) using the trimmed coordinates of human purple acid phosphatase (1WAR, 27% sequence identity to PfGAP50) yielded a solution, which was then subjected to manual rebuilding and real space refinement in Coot (Emsley et al., 2010). A higher resolution data set to 1.7 Å was later obtained using synchrotron radiation at SSRL beamline 9-2. This data set was used for TLS refinement with REFMAC (Murshudov et al., 1997) using seven TLS groups determined by the TLSMD Web server (Painter and Merritt, 2005). PfGAP50 was refined to a crystallographic Rwork of 17.2% and an Rfree of 20.2% (Brünger, 1992). Two loops comprising residues 122 to 136 and 161 to 169, were disordered and therefore not included in the structure. The final structure was analyzed with validation tools in Coot (Emsley et al., 2010) as well as MOLPROBITY (Lovell et al., 2003), indicating zero Ramachandran outliers (Ramakrishnan and Ramachandran, 1965). Crystallographic data and refinement statistics are presented in Table 1.
Table 1.
Data collection, phasing and refinement statistics
| Native | |
|---|---|
| Data collection | SSRL 9-2 |
| Space group | P3221 |
| Cell dimensions | |
| a, b, c (Å) | 57.6, 57.6, 210.6 |
| α, β, γ (°) | 90, 90, 120 |
| Wavelength | 0.9794 |
| Resolution (Å) | 20.0 – 1.7 (1.85 – 1.7) |
| Rsym | 0.089 (0.622) |
| I/I | 18.7 (2.3) |
| Completeness (%) | 97.3 (91.6) |
| Redundancy | 5.2 (4.2) |
| Refinement | |
| Resolution (Å) | 20.0 – 1.7 |
| No. reflections | 43472 |
| Rwork/Rfree | 17.2/20.2 |
| No. atoms | |
| Protein | 2545 |
| Ligand/ion | 28 |
| Water | 256 |
| B-factors (Å2) | |
| Protein | 46 |
| Ligand/ion | 79 |
| Water | 64 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 1.092 |
| Ramachandran main chain dihedral analysis** | |
| favored | 295/309 (95.5%) |
| allowed | 14/309 (4.5%) |
| outliers | 0/309 (0.0%) |
| Molprobityside chain rotamer analysis** | |
| bad rotamers | 5/273 (1.83%) |
Highest resolution shell is shown in parentheses.
According to Molprobity (Lovell et al., 2003)
3. Results
3.1 Bioinformatic analysis
Psi-Blast (Altschul et al., 1997) searches with the protein sequence from PfGAP50 PlasmoDB accession code PFI0880c) revealed similarities to purple acid phosphatases with the closest structural homolog being human purple acid phosphatase (hPAP; PDB code 1WAR) (Strater et al., 2005) with 27% sequence identity. The PfGAP50 gene encodes a protein, whose fold belongs to the InterPro superfamily of metallo-dependent phosphatases SSF56300. In this superfamily two iron atoms, Fe(II) and Fe(III), are bridged by a phosphate and coordinated by seven residues, which belong to the purple acid phosphatase (PAP) motif (Strater et al., 2005). In contrast to human PAPs, plant PAPs have replaced the Fe(II) atom by either zinc or manganese, leading to a shift in color compared to Fe(II). Electron transfer between the Fe(II) and Tyr53 lead to a visible shift in color; these differences of metal coordination can be characterized either by UV/Vis or electron paramagnetic resonance (EPR) spectroscopy (Schenk et al., 2001).
Using the TMHMM server (Krogh et al., 2001) two transmembrane helices were predicted in PfGAP50, the first ranging from residue 7 to 23, and the second from residue 370 to 392. The predicted N-terminal transmembrane helix is in fact part of a signal peptide sequence, which is processed and cleaved off (Müller et al., 2010; Yeoman et al., 2011) during maturation of the protein. The C-terminal transmembrane helix remains intact and serves as anchor to the inner membrane complex. We expressed the part of the PfGAP50 gene covering only the soluble part of the protein comprising residues Lys24 to Asp365.
3.2 Overall structure and cobalt sites
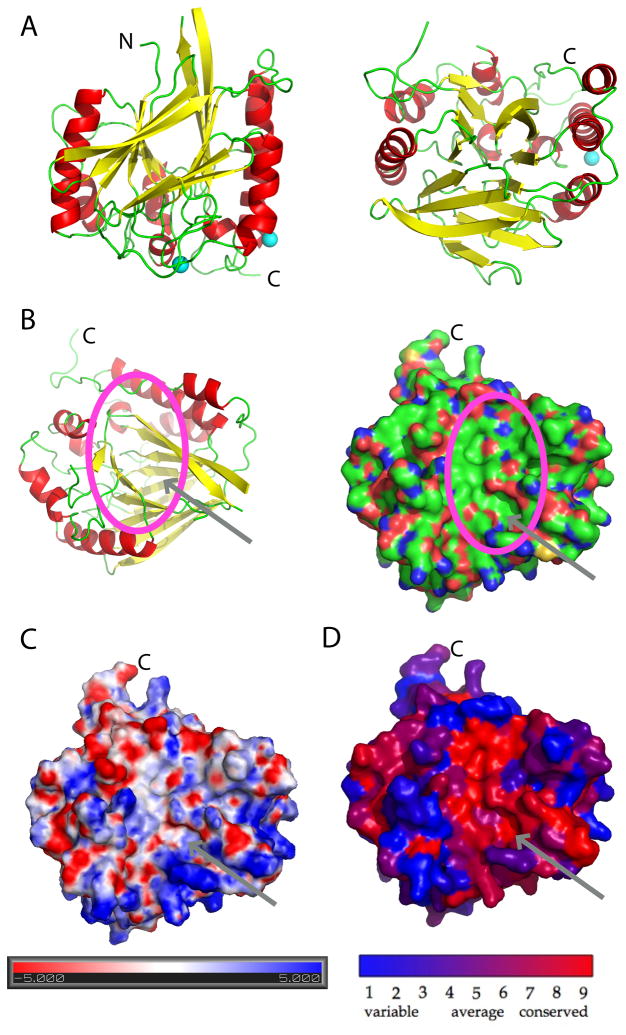
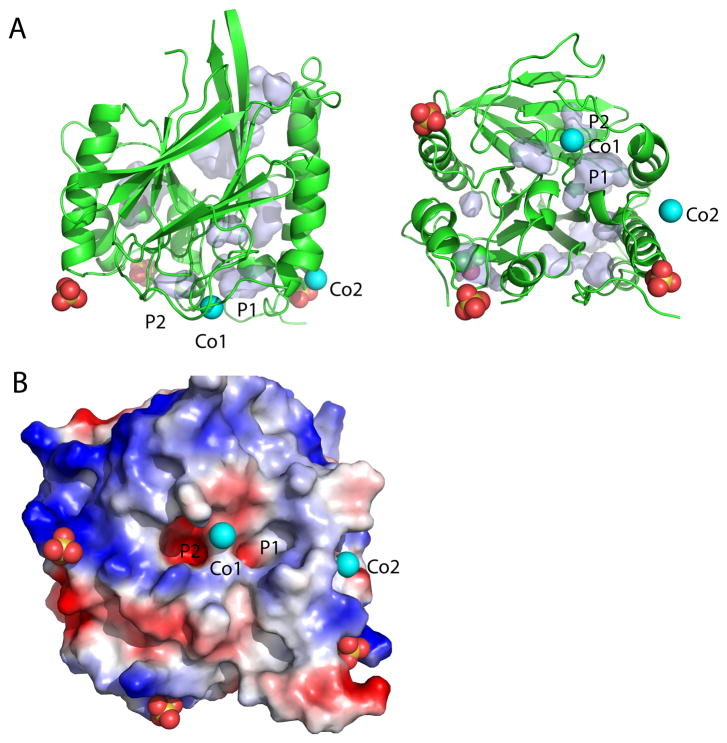
The soluble domain of PfGAP50 displays the αββα fold with 8 helices and a central β-sandwich composed of 14 β-strands with approximate dimensions of 56 Å × 56 Å × 60 Å (Figure 4A). Seven β-strands form one sheet, which is positioned opposite of the second sheet of seven β-strands, with the strands of the two sheets running approximately parallel to each other. The overall approximately conical shape of the protein exhibits one highly conserved concave depression (Figure 4B–D). Three sulfate ions originating from the crystallization buffer were identified in the electron density. Sulfate ion 1 is coordinated by the main chain nitrogen of Lys40 and two water molecules, sulfate ion 2 by main chain nitrogen of Leu75 and Asn76δ and two water molecules, and sulfate ion 3 by the main chain nitrogen of Lys192 and Lys193. Two cobalt ions were suggested by 26 and 21 σ peaks in the difference Fourier combined with anomalous density peaks of 10 σ and 8 σ, respectively, from an in-house data set collected at the copper Kα edge (Figure 5). An excitation scan was performed at the SSRL beamline 9-2 confirming the presence of Cobalt in these crystals (data not shown).
Figure 4. Structural characterization of PfGAP50.
A) The ribbon diagram is represented in a 0 and 90 degree view. Helices are colored red and beta strands yellow, loop regions are kept in green. The two cobalt sites are indicated in cyan.
B) Ribbon representation of PfGAP50 highlighting the concave depression formed between the two β-sheets. This orientation is identical in the following surface representations. A deep hydrophobic patch is visible, indicated by an arrow and circle, atoms are colored by green representing carbon, red oxygen and blue nitrogen atoms.
C) Electrostatic representation of PfGAP50, with negatively charged areas in red and positively charged in blue..
D) View of the concave depression based on conservation across various Apicomplexa including Plasmodium, Toxoplasma, Cryptosporidum, Babesia and Eimeria, where red represents the highest identity of residues and blue the lowest.
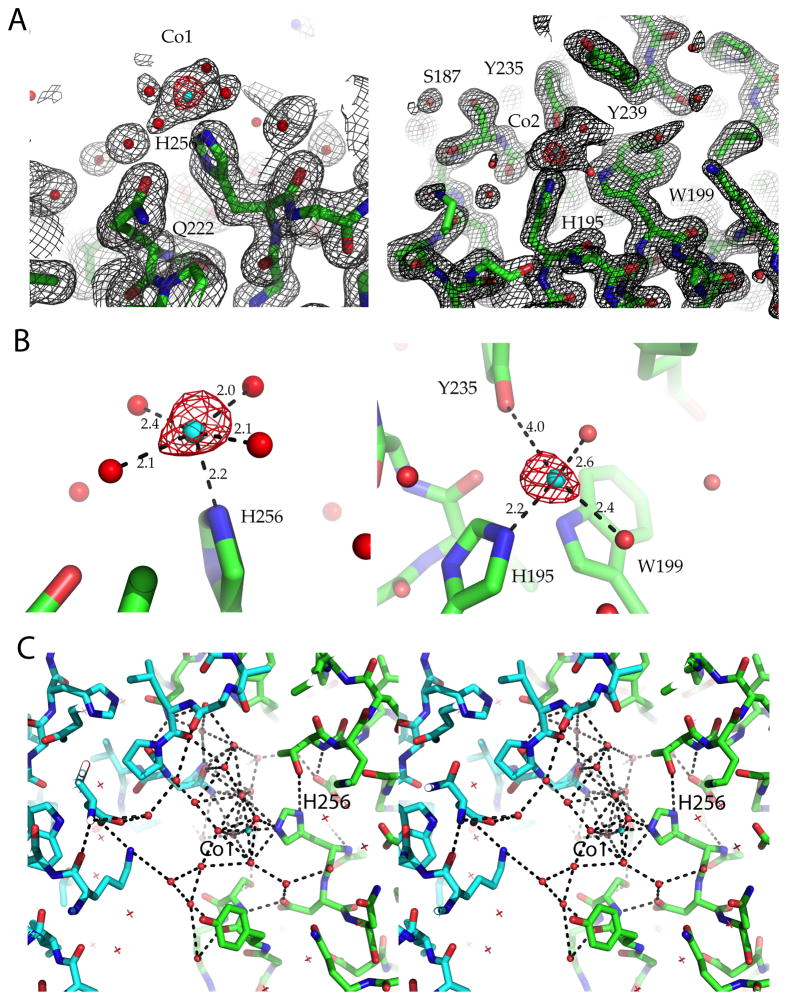
Figure 5. Electron density maps of PfGAP50.
2Fo-Fc electron density map contoured at 1.5σ and the corresponding anomalous difference map in red contoured at 6σ Cobalt ions are shown as cyan spheres and waters as red spheres.
A) Cobalt site 1 (left panel), located near the active site of hPAP after super positioning Cobalt site 2 (right panel). See also Figure 4A for an overview.
B) Distance to binding partners of Cobalt site 1 (left panel) and Cobalt site 2 (right panel), which is approximately 18 Å apart from Cobalt site 1.
C) Stereo figure of the hydrogen bonding network observed in the crystal lattice near the conserved His256 interacting with Cobalt site 1, which in its turn interacts with a crystallographically related GAP50 (cyan backbone) via its water ligands.
Cobalt site 1 is coordinated at a distance of 2.17 Å by the Nε nitrogen of His256, a conserved residue between Plasmodium and the human homolog. The shape and distribution of the FO-FC difference density map suggests 5 additional metal-coordinating water molecules, which surround the metal ion in an approximately octahedral arrangement (Figure 5). The five water molecules are well defined in density, bridging two symmetry related molecules via an extensive hydrogen bonding network. Cobalt site 1 is located close to two pockets, one more of a hydrophobic nature and the other one more charged.
Cobalt site 2 is coordinated by the Nε nitrogen of His195 at a distance of 2.25 Å and four additional water molecules arranged in the plane of an octahedron, with the nitrogen at the apex of the octahedron. Both histidines are conserved throughout the apicomplexa and reside in highly conserved areas of the protein surface (Figure 2). The second Co2+ site is ~18Å apart from the first Co2+ site (Figure 6). No binuclear metal site was observed in PfGAP50, even when using other divalent metals in combination with Co2+ during soaking experiments with crystals at various pH values.
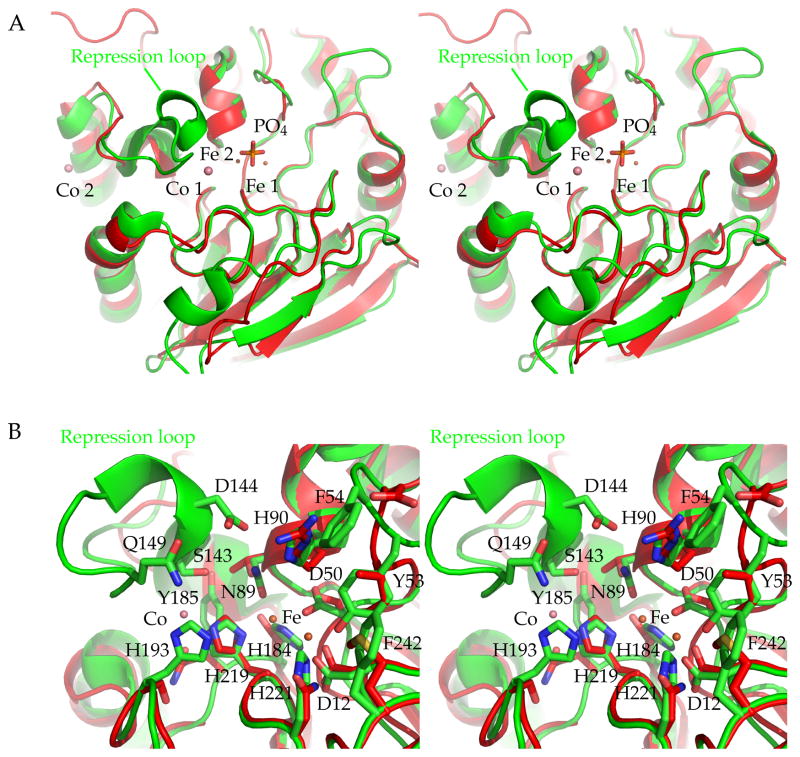
Figure 6. Superposition of hPAP with PfGAP50, active site comparison.
A) Stereo figure overview of the active site region of hPAP active site and PfGAP50, the most prominent feature missing in PfGAP50 is the repression loop. The two cobalt sites of PfGAP50 are indicated with salmon spheres, the iron sites of hPAP are depicted in orange with the bridging phosphate group.
B) Residues involved in coordinating the two iron sites in hPAP. For simplification the phosphate group has been omitted from the figure. Residues and numbering correspond to the hPAP sequence and their side chains are shown in green sticks. The corresponding PfGAP50 residues are shown in red sticks (without numbering).
3.3 Structures of PfGAP50 and hPAP
The overall folds of PfGAP50 and hPAP are quite similar (Figure 7). Ther.m.s deviation is 1.2 Å for 262 structurally equivalent Cα atoms with a sequence identity of 27% for these residues. The central β-sandwich consists in both structures of two antiparallel β-sheets. The surrounding α-helices have slightly different orientations with respect to the β-sandwich in the two proteins. In addition several loops adopt very different conformations (Figure 6). A major difference is the absence in PfGAP50 of the so-called “repression loop” in hPAP, spanning residues Cys140 to Arg153 in the human enzyme, which corresponds in PfGAP50 to an insertion between residues Ser187 and Asn188 (Figure 2).
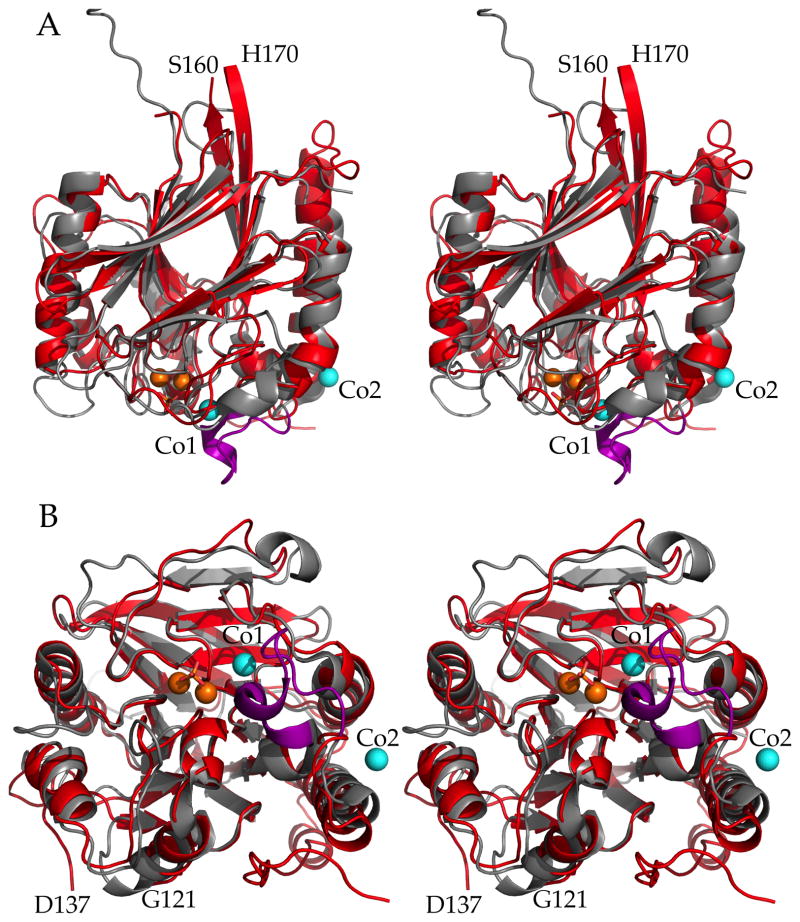
Figure 7. Superposition of PfGAP50 and hPAP.
Stereo figure of PfGAP50 (red) and hPAP (grey) with the latter’s repression loop (purple) superimposed. Cobalt (cyan) and iron (orange) ions are shown as spheres and the bridging phosphate in stick representation.
A) Side view same orientation as Figure 2A.
B) Bottom view (opposite of active site). The loops between Ser160 to His170 and Gly121 to Asp137 are numbered to highlight regions omitted from the PfGAP50 model building due to lack of supportive electron density.
The active site of hPAP is of special interest to compare with its corresponding region in PfGAP50. The key features in hPAP are (i) a binuclear iron metal site with a distance of 3.2 Å between the two iron ions; and (ii) the above-mentioned regulatory repression loop (Strater et al., 2005). By displacing the iron bridging phosphate group through Asp145 of the repression loop, the enzyme is rendered in an inactive state in human and plant purple acid phosphatases (Strater et al., 2005). PfGAP50 has neither of those features: the region corresponding to the repression loop is missing in all apicomplexan species (Figure 2), and no evidence is found in our structure for a binuclear metal site. The active site of hPAP comprises seven residues (Asp12, Asp50, Tyr53, Asn89, His184, His219 and His221) contacting two iron ions, which are bridged by a phosphate group (Strater et al., 2005). Of the residues comprising the hPAP active site only two are maintained in P. falciparum, namely Asp34 (hPAPAsp12) and His256 (hPAPHis219). Moreover only one Co2+ metal site could be identified near the corresponding human active site in PfGAP50 (Figure 6) coordinated by His256. The first Co2+-site of PfGAP50 is 5.6 Å removed from the nearest Fe-site of hPAP when the two protein structures are superimposed (Figure 6). The second Co2+-site of PfGAP50 is ~18Å away from the nearest Fe site in hPAP.
A structure-based sequence alignment of several plasmodial species, hPAP and kidney bean purple acid phosphatase (kbPAP) reveals no conservation of the active site residues of the phosphatases in the GAP50 family of proteins. (Figure 2, Figure 6).
4. Discussion
4.1 The orientation of PfGAP50 in the IMC
Evidence for the localization of GAP50 within the lumen of the IMC arises from various investigations such as fluorescent studies by Johnson et al. (Johnson et al., 2007) with YFP-tagged TgGAP50, which demonstrated the localization of TgGAP50 in the IMC. Two recent publications using GFP-fused PfGAP50 have confirmed the localization of PfGAP50 in the inner membrane complex of P. falciparum (Müller et al., 2010; Yeoman et al., 2011). Furthermore, solubilization experiments and protection from proteolytical cleavage of intact IMC preparations suggest that TgGAP50 is localized within the lumen of the IMC (Johnson et al., 2007). Our model places the bulk of GAP50 in the lumen of the IMC, allowing an interaction of GAP50 (Figure 1), via its C-terminal helix extension with the other components of the invasion machinery (Daher and Soldati-Favre, 2009). Thus far no direct interaction of GAP50 with any member of the invasion machinery has been observed other than in pulldown experiments. An interaction of PfGAP50 with PfGAP45 might be stabilized through the N-terminal palmitoylation and myristoylation of PfGAP45, as demonstrated for the homologous proteins in T. gondii (Daher and Soldati-Favre, 2009; Johnson et al., 2007; Rees-Channer et al., 2006). The conserved surface of GAP50 (>80% identity within apicomplexa) suggests some important transient interaction partners perhaps necessary for regulation of the machinery are missing. The essentiality of GAP50 has been demonstrated in T. gondii (Gaskins et al., 2004) and P. falciparum (Yeoman et al., 2011), where knockouts of the gene were not viable. The importance of the C-terminal tail of TgGAP50 was demonstrated in a C-terminal deletion experiment, where assembly of the members of the invasion machinery failed when examined through pulldown experiments and probed with specific antibodies (Gaskins et al., 2004). Therefore, we assume that GAP50 plays also a critical role in Plasmodium species.
4.2 Active site residues of PAP are poorly conserved in GAP50
The function of GAP50 in the IMC other than serving as an anchor for the invasion machinery remains elusive as the structure does not support a function as a phosphatase. Most of the hPAP active site residues are replaced by non-catalytic residues and enzyme assays performed with pNPP as a general phosphatase substrate showed no activity with recombinant PfGAP50 purified to homogeneity (purity >99% when loading 10 μg recombinant protein on Coomassie stained SDS gels, data not shown). Crystal soaks and co-crystallization experiments with pNPP did not reveal binding of the putative substrate at various pH values, ranging from pH 4 – pH 9 at 2 Å resolution (data not shown). These findings strongly suggest that PfGAP50 and other apicomplexan GAP50 display no enzymatic activity similar to that of the PAPs despite a recent paper describing such activity (Müller et al., 2010). The observed phosphatase activity by the latter authors is not easy to reconcile with our crystal structure and might possibly reflect contaminating phosphatases in the purification procedure used.
Near the region corresponding to the active site of hPAP, we have identified in PfGAP50 a metal binding site close to two surface pockets, one mostly hydrophobic, centered around residue Leu236, and one charged, centered around residue Asp34 (Figure 8). The true nature of the native metal bound to GAP50 in P. falciparum under physiological conditions is unknown at this moment but the stabilizing effect demonstrated through thermal shift assays and the conservation of His195 and His256 of PfGAP50 in all apicomplexa (Figure 2) suggests that the metal might fulfill an important function. The positive influence of the metal is reflected in a decreased initial fluorescence at 25 °C as demonstrated in a thermal shift assay (Figure 3), indicating less exposure of hydrophobic patches accessible to the probe SyPro Orange. Additionally the influence of buffering reagent or pH stabilizes the protein as can be seen in a shift of approximately 15°C towards higher Tm when comparing sodium cacodylate pH 6.0 (blue) to Hepes pH 8.0 (green).
Figure 8. Pockets of GAP50 near the homologous active site region in other proteins.
A) Ribbon diagram of PfGAP50 with identified ligands shown in spheres, the orientation is identical to Figure 4A. Pockets and cavities as calculated by PyMol are as light blue surfaces. Pocket 1 (P1) and pocket 2 (P2) are close to the cobalt atom closest to the active site of the human purple acid phosphatase.
B) Electrostatic potential around pockets 1 and 2 in an orientation as viewed in Figure 4B–D, with P2 being more charged compared to P1.
One possibility is that the metals observed in our PfGAP50 structure form anchor points for bridging with other protein(s) via these metal ions, with the up to now unknown partner proteins completing the coordination sphere. Support for this mechanism might be indicated by the observed interactions of the Co2+ ions in the crystals to neighboring symmetry-related molecules (Figure 5C).
4.3 Conserved GAP50 surface within apicomplexa
The conservation of the exposed surface areas of GAP50, with more than 80% mutual amino acid identity, among various apicomplexan species such as Plasmodium, Toxoplasma, Cryptosporidium, Babesia and Eimeria (Figure 4D, Figure 2) may indicate a yet unknown function of GAP50 in the invasion machinery. The concave depression as seen in Figure 4 might be important for interactions with other so far unknown proteins within the lumen of the IMC. The depression is not only well conserved throughout apicomplexan species but also exhibits a large hydrophobic surface area (Figure 4B). Pulldown assays have so far revealed interactions among the following seven protein components of the invasion machinery: GAP50, GAP45, MTIP, Myosin A, Actin, Aldolase and TRAP (Baum et al., 2006; Bergman et al., 2003; Meissner et al., 2002). Interactions between two pairs of invasion machinery proteins have been confirmed, including crystal structure determinations, between TRAP and aldolase, and between Myosin A and MTIP (Bosch et al., 2007a; Bosch et al., 2007b; Bosch et al., 2006a). All these proteins, except GAP50, are located in the pellicle between the IMC and the parasite plasma membrane. The soluble domain of GAP50 whose structure is presented in this paper is the only domain of the seven identified invasion machinery proteins residing in the lumen of the IMC (Yeoman et al., 2011). The presence of conserved regions on the surface of the soluble domain of GAP50 could be an indication that this particular surface area of GAP50 is possibly involved in transient interactions with other, yet to be discovered, proteins in the lumen of the IMC.
4.4 Pockets for potential future drug development
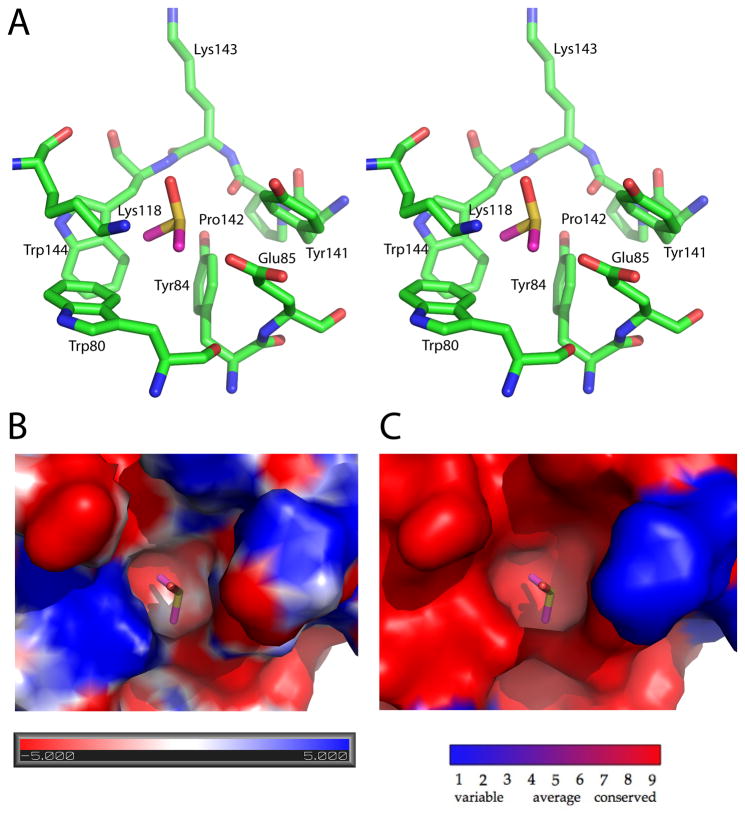
The hydrophobic pocket, filled with a DMSO molecule, centered around residue Tyr 84 is similarly well conserved throughout apicomplexan species and potentially could be exploited for future drug design if interacting partner(s) of GAP50 exist that bind close to this region of the molecule (Figure 9).
Figure 9. DMSO molecule bound to a conserved pocket of PfGAP50.
A) Stereo representation of the pocket forming residues. The DMSO pocket is located between helices α2 and α4, approximately 25 Å away from Cobalt site 1.
B) Surface representation of the electrostatic potential of the DMSO binding pocket with negatively charged regions in red and positively charged in blue.
C) Conservation of the pocket within Plasmodium species, where red represents the highest identity of residues and blue the lowest.
Although we have solved the structure of the soluble domain of GAP50 at high resolution, and discovered unexpected metal binding sites and conserved surface regions, many questions regarding the functioning of this protein and its role in the sophisticated invasion machinery of the malaria parasite remain unanswered and need to be addressed in the future by additional studies. Our structure provides a basis for further investigations in the field.
Acknowledgments
We gratefully acknowledge Stewart Turley for maintaining our in-house data collection system, and Jonathan Kay for maintaining the computer environment at the Biomolecular Structure Center. We particularly thank Pete Dunten, Ana Gonzalez and the staff at SSRL beamline 9-2 for their help and assistance during synchrotron data collection. We wish to acknowledge the contribution of the Hauptmann Woodward High Throughput Crystallization Facility for initial crystal hits. This project was supported by the National Institutes of Health (NIH) Grant 1P50 GM64655-01 “Structural Genomics of Pathogenic Protozoa (SGPP)” (to W.G.J.H.) and the National Institutes of Health Grant AI48226 (to L.W.B.).
Abbreviations
- IMC
inner membrane complex
- GAP
gliding associated protein
- Pf
P. falciparum
- Tg
T. gondii
- hPAP
human purple acid phosphatase
- kbPAP
kidney bean purple acid phosphatase
- TRAP
thrombospondin-related adhesion protein
- MTRAP
merozoite TRAP
- CTRP
circumsporozoite and TRAP-related protein
Footnotes
Protein Data Base accession codes
Coordinates and structure factors have been deposited with the PDB (Accession code 3TGH).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, et al. Gapped blast and psi-blast - a new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman V, Iyer LM, Aravind L. Comparative genomics of protists: New insights into the evolution of eukaryotic signal transduction and gene regulation. Annu Rev Microbiol. 2007;61:453–475. doi: 10.1146/annurev.micro.61.080706.093309. [DOI] [PubMed] [Google Scholar]
- Baird JK. Drug therapy: Effectiveness of antimalarial drugs. N Engl J Med. 2005;352:1565–1577. doi: 10.1056/NEJMra043207. [DOI] [PubMed] [Google Scholar]
- Baum J, Richard D, Healer J, Rug M, Krnajski Z, et al. A conserved molecular motor drives cell invasion and gliding motility across malaria life cycle stages and other apicomplexan parasites. J Biol Chem. 2006;281:5197–5208. doi: 10.1074/jbc.M509807200. [DOI] [PubMed] [Google Scholar]
- Bergman LW, Kaiser K, Fujioka H, Coppens I, Daly TM, et al. Myosin A tail domain interacting protein (MTIP) localizes to the inner membrane complex of Plasmodium sporozoites. J Cell Sci. 2003;116:39–49. doi: 10.1242/jcs.00194. [DOI] [PubMed] [Google Scholar]
- Bosch J, Turley S, Roach CM, Daly TM, Bergman LW, et al. The closed MTIP-Myosin A-tail complex from the malaria parasite invasion machinery. J Mol Biol. 2007a;372:77–88. doi: 10.1016/j.jmb.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch J, Buscaglia CA, Krumm B, Ingason BP, Lucas R, et al. Aldolase provides an unusual binding site for thrombospondin-related anonymous protein in the invasion machinery of the malaria parasite. PNAS. 2007b;104:7015–7020. doi: 10.1073/pnas.0605301104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch J, Turley S, Daly TM, Bogh SM, Villasmil ML, et al. Structure of the MTIP-MyoA complex, a key component of the malaria parasite invasion motor. Proc Natl Acad Sci U S A. 2006a;103:4852–4857. doi: 10.1073/pnas.0510907103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch J, Robien MA, Mehlin C, Boni E, Riechers A, et al. Using fragment cocktail crystallography to assist inhibitor design of Trypanosoma brucei nucleoside 2-deoxyribosyltransferase. J Med Chem. 2006b;49:5939–5946. doi: 10.1021/jm060429m. [DOI] [PubMed] [Google Scholar]
- Brünger AT. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- Buscaglia CA, Coppens I, Hol WGJ, Nussenzweig V. Sites of interaction between aldolase and thrombospondin-related anonymous protein in Plasmodium. Mol Biol Cell. 2003;14:4947–4957. doi: 10.1091/mbc.E03-06-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox-Singh J, Davis TME, Lee KS, Shamsul SSG, Matusop A, et al. Plasmodium knowlesi malaria in humans is widely distributed and potentially life threatening. Clin Infect Dis. 2008;46:165–171. doi: 10.1086/524888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowther GJ, He P, Rodenbough PP, Thomas AP, Kovzun KV, et al. Use of thermal melt curves to assess the quality of enzyme preparations. Anal Biochem. 2010;399:268–275. doi: 10.1016/j.ab.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daher W, Soldati-Favre D. Mechanisms controlling glideosome function in apicomplexans. Current Opinion in Microbiology. 2009;12:408–414. doi: 10.1016/j.mib.2009.06.008. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericsson UB, Hallberg BM, DeTitta GT, Dekker N, Nordlund P. Thermofluor-based high-throughput stability optimization of proteins for structural studies. Anal Biochem. 2006;357:289–298. doi: 10.1016/j.ab.2006.07.027. [DOI] [PubMed] [Google Scholar]
- Gaskins E, Gilk S, DeVore N, Mann T, Ward G, et al. Identification of the membrane receptor of a class XIV myosin in Toxoplasma gondii. J Cell Biol. 2004;165:383–393. doi: 10.1083/jcb.200311137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green JL, Martin SR, Fielden J, Ksagoni A, Grainger M, et al. The MTIP-myosin A complex in blood stage malaria parasites. J Mol Biol. 2006;355:933–941. doi: 10.1016/j.jmb.2005.11.027. [DOI] [PubMed] [Google Scholar]
- Herm-Gotz A, Weiss S, Stratmann R, Fujita-Becker S, Ruff C, et al. Toxoplasma gondii myosin A and its light chain: a fast, single-headed, plus-end-directed motor. Embo J. 2002;21:2149–2158. doi: 10.1093/emboj/21.9.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes KC, Angert I, Kull FJ, Jahn W, Schroder RR. Electron cryomicroscopy shows how strong binding of myosin to actin releases nucleotide. Nature. 2003;425:423–427. doi: 10.1038/nature02005. [DOI] [PubMed] [Google Scholar]
- Jewett TJ, Sibley LD. Aldolase forms a bridge between cell surface adhesins and the actin cytoskeleton in apicomplexan parasites. Mol Cell. 2003;11:885–894. doi: 10.1016/s1097-2765(03)00113-8. [DOI] [PubMed] [Google Scholar]
- Johnson TM, Raifur Z, Jacobson K, Beckers CJ. Immobilization of the type XIV myosin complex in Toxoplasma gondii. Mol Biol Cell. 2007;18:3039–3046. doi: 10.1091/mbc.E07-01-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. Journal of Applied Crystallography. 1993;26:795–800. [Google Scholar]
- Kappe SHI, Buscaglia CA, Bergman LW, Coppens I, Nussenzweig V. Apicomplexan gliding motility and host cell invasion: overhauling the motor model. Trends Parasitol. 2004;20:13–16. doi: 10.1016/j.pt.2003.10.011. [DOI] [PubMed] [Google Scholar]
- Kortagere S, Welsh WJ, Morrisey JM, Daly T, Ejigiri I, et al. Structure-based Design of Novel Small-Molecule Inhibitors of Plasmodium falciparum. Journal of chemical information and modeling. 2010 doi: 10.1021/ci100039k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallographica Section D Biological Crystallography. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Lovell SC, Davis IW, Adrendall WB, de Bakker PIW, Word JM, et al. Structure validation by C alpha geometry: phi, psi and C beta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- Luft JR, Collins RJ, Fehrman NA, Lauricella AM, Veatch CK, et al. A deliberate approach to screening for initial crystallization conditions of biological macromolecules. J Struct Biol. 2003;142:170–179. doi: 10.1016/s1047-8477(03)00048-0. [DOI] [PubMed] [Google Scholar]
- McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallographica Section D-Biological Crystallography. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlin C, Boni E, Buckner FS, Engel L, Feist T, et al. Heterologous expression of proteins from Plasmodium falciparum: results from 1000 genes. Mol Biochem Parasitol. 2006;148:144–160. doi: 10.1016/j.molbiopara.2006.03.011. [DOI] [PubMed] [Google Scholar]
- Meissner M, Schluter D, Soldati D. Role of Toxoplasma gondii myosin A in powering parasite gliding and host cell invasion. Science. 2002;298:837–840. doi: 10.1126/science.1074553. [DOI] [PubMed] [Google Scholar]
- Müller IB, Knöckel J, Eschbach M-L, Bergmann B, Walter RD, et al. Secretion of an acid phosphatase provides a possible mechanism to acquire host nutrients by Plasmodium falciparum. Cell Microbiol. 2010 doi: 10.1111/j.1462-5822.2010.01426.x. [DOI] [PubMed] [Google Scholar]
- Murray CJL, Rosenfeld LC, Lim SS, Andrews KG, Foreman KJ, et al. Global malaria mortality between 1980 and 2010: a systematic analysis. The Lancet. 2012;379:413–431. doi: 10.1016/S0140-6736(12)60034-8. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr Sect D-Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Painter J, Merritt EA. A molecular viewer for the analysis of TLS rigid-body motion in macromolecules. Acta Crystallogr Sect D-Biol Crystallogr. 2005;61:465–471. doi: 10.1107/S0907444905001897. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan C, Ramachandran GN. Stereochemical criteria for polypeptide and protein chain conformations. II. Allowed conformations for a pair of peptide units. Biophysical Journal. 1965;5:909–933. doi: 10.1016/S0006-3495(65)86759-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees-Channer RR, Martin SR, Green JL, Bowyer PW, Grainger M, et al. Dual acylation of the 45 kDa gliding-associated protein (GAP45) in Plasmodium falciparum merozoites. Mol Biochem Parasitol. 2006;149:113–116. doi: 10.1016/j.molbiopara.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Rusnak F, Mertz P. Calcineurin: Form and function. Physiol Rev. 2000;80:1483–1521. doi: 10.1152/physrev.2000.80.4.1483. [DOI] [PubMed] [Google Scholar]
- Sahoo N, Beatty W, Heuser J, Sept D, Sibley LD. Unusual kinetic and structural properties control rapid assembly and turnover of actin in the parasite Toxoplasma gondii. Mol Biol Cell. 2006;17:895–906. doi: 10.1091/mbc.E05-06-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk G, Boutchard CL, Carrington LE, Noble CJ, Moubaraki B, et al. A purple acid phosphatase from sweet potato contains an antiferro magnetically coupled binuclear Fe-Mn center. J Biol Chem. 2001;276:19084–19088. doi: 10.1074/jbc.M009778200. [DOI] [PubMed] [Google Scholar]
- Schmitz S, Grainger M, Howell S, Calder LJ, Gaeb M, et al. Malaria parasite actin filaments are very short. J Mol Biol. 2005;349:113–125. doi: 10.1016/j.jmb.2005.03.056. [DOI] [PubMed] [Google Scholar]
- Sibley LD. Intracellular parasite invasion strategies. Science. 2004;304:248–253. doi: 10.1126/science.1094717. [DOI] [PubMed] [Google Scholar]
- Sibley LD. How apicomplexan parasites move in and out of cells. Current Opinion in Biotechnology. 2010;21:592–598. doi: 10.1016/j.copbio.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow RW, Guerra CA, Noor AM, Myint HY, Hay SI. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature. 2005;434:214–217. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strater N, Jasper B, Scholte M, Krebs B, Duff AP, et al. Crystal structures of recombinant human purple acid phosphatase with and without an inhibitory conformation of the repression loop. J Mol Biol. 2005;351:233–246. doi: 10.1016/j.jmb.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Thomas JC, Green JL, Howson RI, Simpson P, Moss DK, et al. Interaction and dynamics of the Plasmodium falciparum MTIP-MyoA complex, a key component of the invasion motor in the malaria parasite, pp. Mol Biosyst. 2010;6:494–498. doi: 10.1039/b922093c. [DOI] [PubMed] [Google Scholar]
- Yeh JI, Hol WGJ. A flash-annealing technique to improve diffraction limits and lower mosaicity in crystals of glycerol kinase. Acta Crystallographica Section D-Biological Crystallography. 1998;54:479–480. doi: 10.1107/s0907444998004697. [DOI] [PubMed] [Google Scholar]
- Yeoman JA, Hanssen E, Maier AG, Klonis N, Maco B, et al. Tracking Glideosome-associated protein 50 reveals the development and organization of the inner membrane complex of Plasmodium falciparum. Eukaryotic Cell. 2011;10:556–564. doi: 10.1128/EC.00244-10. [DOI] [PMC free article] [PubMed] [Google Scholar]