Abstract
Filoviruses are hemorrhagic fever-causing agents that produce enveloped virions with a filamentous morphology. The viral surface glycoprotein, GP, orchestrates the surprisingly complex process by which filoviruses gain access to the cytoplasm of their host cells. GP mediates viral attachment to cells through multiple, redundant interactions with cell-surface factors. GP then induces virion internalization by a process that resembles cellular macropinocytosis. Within the endo/lysosomal pathway, GP undergoes a series of structural rearrangements, controlled by interactions with host factors, that prime and activate it to bring about fusion between the viral and cellular lipid bilayers. Membrane fusion delivers the viral nucleocapsid core into the cytoplasm, which is the site of filovirus replication. This review summarizes our understanding of the filovirus entry mechanism, with emphasis on recent findings.
Introduction
Filoviruses are negative-sense single-stranded RNA viruses that produce large enveloped infectious particles (virions) with a filamentous morphology. Members of this virus family, most notably Ebola virus (EBOV) and Marburg virus (MARV), have been associated with episodic, but increasingly frequent outbreaks of a highly lethal hemorrhagic fever in sub-Saharan Africa (see [1,2] for recent reviews). Bats are the suspected natural viral reservoirs from which transmission of filoviruses to their accidental human and non-human primate hosts takes place (reviewed in [3]). Infection of dendritic cells (DCs) and macrophages, with attendant coagulopathies due to activation of tissue factor by macrophages, is proposed to be a central feature of filovirus pathogenesis in vivo [1,3]. As the virus continues to multiply relentlessly within its host, more tissue types become infected, ranging from hepatocytes to endothelial cells. Patients eventually succumb to shock and multi-organ failure [1,3].
GP, the virus-encoded surface glycoprotein, is necessary and sufficient to mediate filovirus entry into the cytoplasm of host cells [4,5] (Fig. 1). Its incorporation into surrogate viruses (vesicular stomatitis virus [VSV] and human immunodeficiency virus-1 [HIV-1] pseudotypes) and filamentous filovirus-like particles has allowed this process to be studied in a biosafety level 2 setting [4–6]. Here, we discuss recent findings on filovirus entry, including the identification of new host factors and new steps in this intricate molecular ballet between virus and cell (Fig. 3).
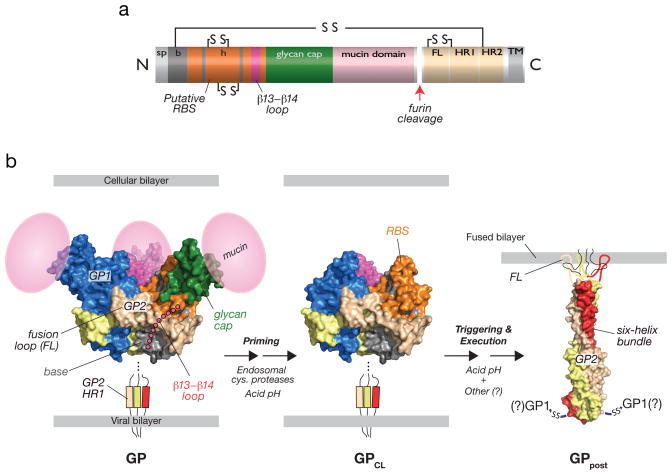
Fig. 1. Structural organization and features of the filovirus glycoprotein, GP, and proposed structural transitions during entry.
(a) Linear diagram of GP indicating sequence features. The cellular Golgi endopeptidase furin cleaves the precursor subunit GP0 to GP1 and GP2. sp, signal peptide. b, GP1 ‘base’ subdomain. h, GP1 ‘head’ subdomain. RBS, putative receptor-binding sequence. FL, GP2 fusion loop. HR1, GP2 N–terminal heptad repeat. HR2, GP2 C–terminal heptad repeat. TM, GP2 transmembrane domain. Lines and ‘SS’ indicate intra- and intersubunit disulfide bonds. (b) Structures of an ectodomain of the EBOV GP pre-fusion trimer (PDB id: 3CSY [9]), GP2 in its putative post-fusion conformation (PDB id: 1EBO [7]), and a structural model of a proteolytically-primed GP intermediate (GPCL). The β13-β14 loop connecting the GP1 base and head subdomains, and the GP1 mucin domain were not visualized in the pre-fusion crystal structures (3CSY and 3S88 [10]) and are shown as cartoons. The GP2 fusion loop was not visualized in the post-fusion crystal structues (1EBO and 2EBO [8]) and is shown as a cartoon. During entry, endosomal cysteine proteases cleave and remove the glycan cap and mucin subdomains of GP1, generating GPCL, in which the putative rbs is fully exposed. In response to an unknown host trigger, GP2 is proposed to undergo release from its pre-fusion conformation and to rearrange to form the six-helix bundle. Formation of this structure is thought to drive fusion of the viral and cellular lipid bilayers. The disposition of GP1 and the intersubunit disulfide bond during membrane fusion and in this post-fusion GP conformer (GPpost) is unknown. Acid pH is required for endosomal cysteine protease activity and is hypothesized to play multiple roles in fusion (see text for details).
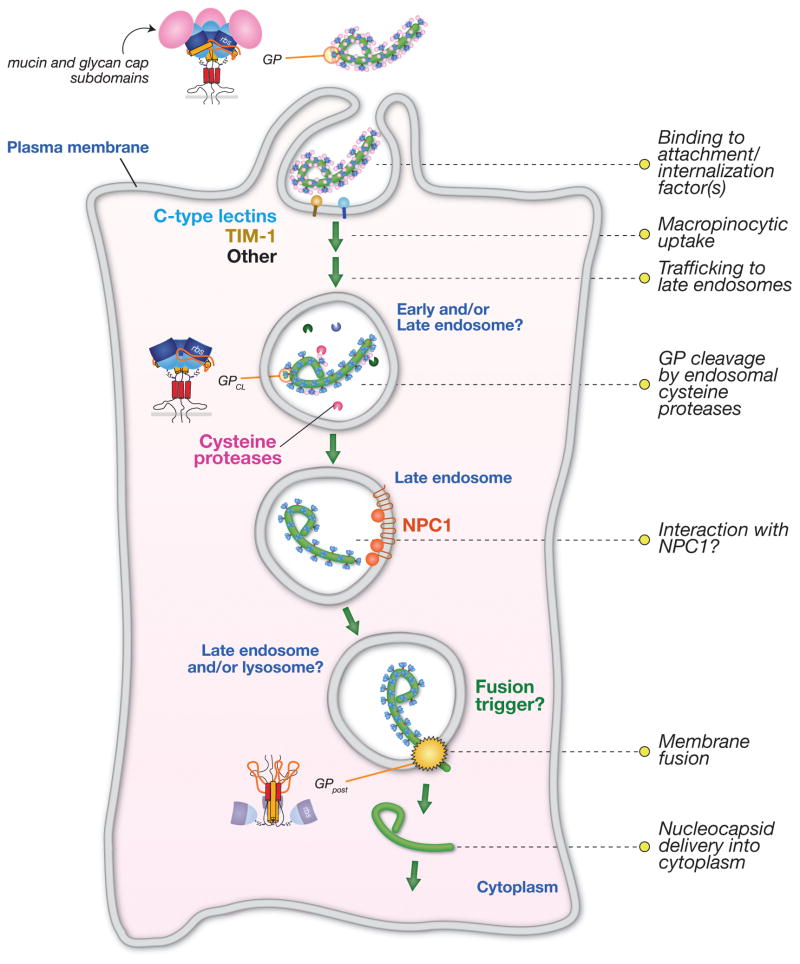
Fig. 3. Schematic model of filovirus entry pathway.
The filovirus glycoprotein, GP, mediates viral attachment to cells via multiple and at least partly redundant interactions with cell-surface molecules. GP then induces viral internalization through a macropinocytosis-like pathway. Within the endocytic pathway, GP is cleaved by endosomal cysteine proteases to generate a primed intermediate that resembles GPCL derived by in vitro proteolysis. Viral particles are delivered to late endosomal/lysosomal compartments, where cleaved GP may interact with NPC1, which plays a critical role at one or more late steps in filovirus entry. It is currently not known if NPC1 is the longsought endosomal receptor or if another host molecule plays this role. An unknown host trigger then induces rearrangements in cleaved GP that drive viral membrane fusion and cytoplasmic escape of the viral nucleocapsid core. The subcellular location of viral membrane fusion has not been defined, but late endosomes and/or late endosome-lysosome hybrids are plausible candidates.
Structural and functional organization of the viral envelope glycoprotein, GP
GP is a ‘class I’ viral membrane fusion glycoprotein that resembles the prototypic HIV-1 Env and influenza virus hemagglutinin (HA) in overall organization ([7–10] reviewed in [11,12]). Mature GP is divided into GP1 and GP2 subunits, generated by cleavage of the GP0 precursor polypeptide by furin during virus assembly (Fig. 1a). GP1 mediates interactions with the viral receptor(s) and regulates the activity of the transmembrane subunit GP2, which carries out membrane fusion. Three GP1–GP2 pairs comprise the mature GP pre-fusion trimer. Crystal structures indicate that it adopts a chalice-like morphology, in which the three GP1 subunits form a bowl that is enwrapped by the three GP2 subunits near its base [9,10] (Fig. 1b).
The membrane-distal subunit, GP1, is organized into three subdomains [9,10] (Fig. 1b). The highly conserved N–terminal half of GP1 forms the ‘base’ (light blue), which interacts extensively with GP2 and clamps it in its pre-fusion conformation. The ‘head’ (green), contains putative receptor-binding sequences (RBS) [13–16]. More variable and heavily glycosylated C–terminal sequences constitute the ‘glycan cap’, which is positioned atop the head, and the mucin domain, whose disposition in the structure is unknown (but is modeled in Fig. 1). The glycan cap is critical for GP folding and plays additional regulatory roles in entry (see below). The mucin domain is dispensable for EBOV GP-dependent entry of pseudotyped viruses in tissue culture [17], but is proposed to mediate viral adhesion to specific cell types within hosts and may contribute to viral immune evasion in natural hosts by shielding key neutralization epitopes [18,19]. The base and glycan cap are connected by a flexible loop that is not visualized in the structure (but is modeled in Fig. 1b).
The transmembrane fusion subunit, GP2, contains a hydrophobic internal fusion peptide (‘fusion loop’) near its N–terminus, and N– and C–terminal α–helical heptad repeat sequences (HR1 and HR2, respectively) [7–9] (Fig. 1a). Comparison of the X-ray crystal structures of GP2 in its pre-fusion and putative post-fusion conformations makes clear that GP2 must undergo large-scale conformational changes during membrane fusion (Fig. 1b). These rearrangements are proposed to facilitate fusion loop insertion into the target cellular membrane and to drive the coalescence of viral and cellular membranes (Figs. 1–2), resulting in cytoplasmic delivery of the viral nucleocapsid core (Fig. 3).
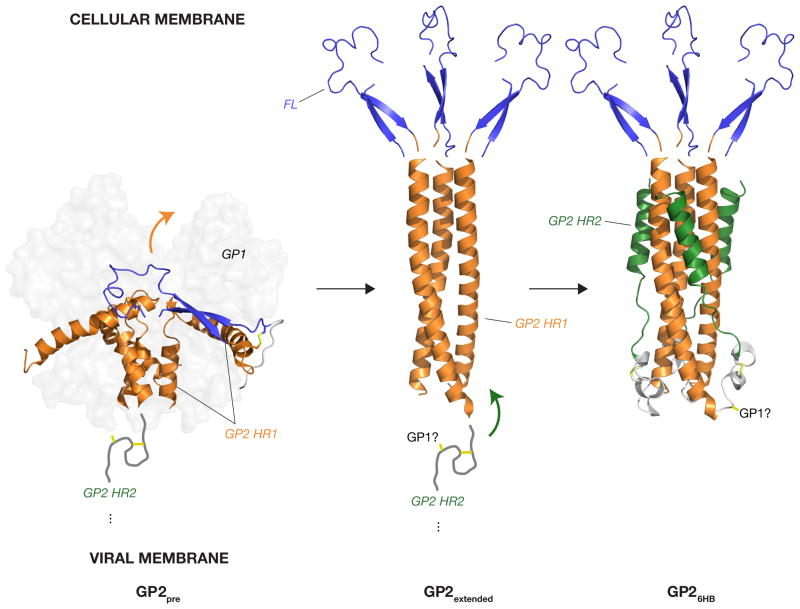
Fig. 2. Proposed structural rearrangements in GP2 during entry.
In response to the fusion trigger, the HR1 sequence in the pre-fusion GP2 conformer (GP2pre) is proposed to undergo release from the GP1 base subdomain and to rearrange to an unbroken α–helix, projecting the GP2 fusion loop (FL) into the target membrane. This rearrangement is mediated by a conformational transition in HR1 . The resulting extended GP2 conformation tipped by fusion loops is then thought to rearrange further, with the sequence betwen HR1 and HR2 reversing direction, and the HR2 sequences packing against grooves in the trimeric HR1 coiled-coil to form the six-helix bundle (6HB) (GP26HB). Orange and green arrows show the direction of motion of HR1 and HR2, respectively. Structures from PDB ids: 3CSY [9] and 1EBO [7]. Fusion loops in GP2extended and GP26HB are derived from the GP pre-fusion structure 3CSY, and their positions are indicated for illustration only. GP2extended is hypothetical. Only one of three GP2 fusion loops in 3CSY is shown for clarity. A cartoon of the GP2 sequence between the HR1 and HR2 with GP1-GP2 and GP2-GP2 disulfide bonds shown (yellow sticks) is based on the structure of the Sudan virus GP pre-fusion trimer (3S88 [10].
Multiple cell-surface molecules mediate viral attachment
Filoviruses can enter and infect many cell types and display a broad mammalian host cell range [4,5,20], suggesting either that they require a single ubiquitous cell-surface receptor, or that multiple cell-surface molecules can redundantly mediate viral attachment. The evidence to date favors the latter scenario.
A variety of C-type lectins (e.g., DC-SIGN, L-SIGN) expressed on key cell types (e.g., macrophages, DCs) can mediate viral attachment and enhance viral infection, as also observed for a number of other enveloped viruses [21–23]. Interactions between GP and C–type lectins are likely mediated by multiple sequences in GP1 and/or GP2 bearing N–linked or O–linked glycans (most notably, the GP1 glycan cap and mucin domain) [24,25]. Not all susceptible cell types express C-type lectins, and their importance as cell attachment factors for filoviruses in vivo remains to be determined. The potential roles of other relatively low-affinity, multivalent interactions between GP and cell surface molecules (e.g., glycosaminoglycans) in promoting viral attachment have not been explored. These initial binding events likely concentrate virions at the plasma membrane, presumably allowing GP to engage additional cell-surface molecules that induce it to change conformation and/or are required for viral internalization [26–28].
Studies in tissue culture with pseudotyped viruses containing EBOV GP indicate that they can only enter and infect cells that are adherent, with even highly susceptible cell lines becoming resistant to infection upon detachment [29]. Cell binding studies with truncated GP1 proteins indicated the existence of a conserved cell-surface protein that contacts GP1 N–terminal sequences (comprising the RBS) in adherent cells, but that is internalized and becomes unavailable to virus following cell detachment [13,14,29]. The identity of this candidate EBOV receptor remains unknown.
TIM-1 is a candidate filovirus receptor
Recently, the T-cell immunoglobulin and mucin domain protein TIM-1, a T-cell costimulatory molecule and phosphatidylserine (PtdSer) receptor [30], was identified as a candidate cell-surface receptor for EBOV and MARV [31]. TIM-1 was shown to interact directly with EBOV GP, to be important for GP-dependent entry in a highly permissive cell line, and to enhance entry when ectopically expressed in weakly permissive cells. Mutations in the proposed GP1 RBS inhibit GP-TIM-1 interaction. Thus, although the the glycan cap appears to sterically obstruct the RBS [9,10], TIM-1 (and other cell-surface factors) may nevertheless be able to access sequences within the RBS prior to proteolytic removal of the glycan cap within endosomes (see below) (Fig. 1b). The functional consequences of GP-TIM1 interaction (beyond viral attachment) are unknown, but it is intriguing to speculate that it induces viral internalization through macropinocytosis (see below).
TIM-1 is expressed on, and may support infection in, many types of human epithelial cells, including cells of the airway [30]. However, important cellular targets of filoviruses within hosts, including macrophages and DCs, do not express it [30]. Whether filoviruses require one or more alternative (non-lectin) receptors for attachment and/or internalization in these cells is unknown at present. The importance of TIM-1 as a filovirus receptor in vivo also remains to be established.
EBOV internalization occurs via a macropinocytosis-like mechanism
Following attachment, filovirus virions must undergo internalization into the endo/lysosomal pathway [4,5]. Recent work indicates that EBOV internalization occurs predominantly via a process resembling macropinocytosis [32,33] (Fig. 3). This process is characterized by the formation of actin-based plasma membrane ruffles and blebs that can enclose large volumes of extracellular fluid and bulky cargoes [34]. A number of host factors and pathways known to regulate and execute membrane ruffling, and subsequently, macropinosome closure, fission, and intracellular trafficking have been implicated in filovirus internalization [32–37].
Dynamin-2 (Dyn2), a GTPase involved in scission of newly formed endocytic vesicles, is dispensable for canonical macropinocytosis [34]; however, its role in filovirus internalization remains unresolved. While EBOV GP-dependent entry was shown to be Dyn2-independent in two studies [32,33], two others observed Dyn2-dependent macropinocytic uptake of virus into several cell types, including macrophages derived from primary human peripheral blood monocytes [36,37].
EBOV, and likely other filoviruses, may exploit cellular macropinocytic pathways for internalization because their large virions (~80 nm diameter; >1 μm length) cannot fit into smaller endocytic vesicles. However, the use of this uptake route is strictly dependent on GP but not virus particle size or morphology, indicating that one or more interactions of GP with the cell surface provide an initiating signal [33,36]. Potential inducers include the candidate receptor TIM-1 and cell-surface lectin attachment factors, both of which have known roles in macropinocytosis [30,38]. Moreover, members of the Tyro3 receptor tyrosine kinase family (e.g., Axl) can enhance GP-induced macropinocytic uptake, apparently without binding directly to GP [37,39]. The Axl ligand Gas6 may mediate indirect Axl-virus interaction by virtue of its capacity to bind to PtdSer on the outer leaflet of the viral membrane, as recently described for several other enveloped viruses [40].
Central role for the endo/lysosomal pathway
Recent work has established that a surprisingly elaborate series of events must occur within host endosomes and/or lysosomes prior to viral membrane fusion and escape into the cytoplasm, but many questions remain. As a case in point, all 13 cellular proteins recently identified in a loss-of-function genetic screen for filovirus entry host factors are involved in the biogenesis, dynamics, and function of the endo/lysosomal system, and 12 of these have never before been implicated in entry by any type of virus [41]. Our current understanding of these intracellular steps in filovirus GP-dependent entry is discussed below.
Virions traffic to late endosomes and/or lysosomes
Internalized pseudotype viruses bearing EBOV GP virions first colocalize with early endosomal antigen-1 (EEA1)-positive compartments (probably sorting endosomes) and are then trafficked to Rab5-positive early endosomes [32]. Viral colocalization with perinuclear Rab7/LAMP-1-positive late endosomes can be observed at later times, and delivery to these compartments appears to be important for entry, since a dominant-negative inhibitor of Rab7 reduces infection [32] (Fig. 3). A variety of other host proteins and signaling networks with known roles in vesicular trafficking are probably involved in the delivery of virions (or entry host factors) to the intracellular sites of filovirus membrane fusion, although details are lacking. These include the phosphatidylinositol-3-kinase and calcium-calmodulin kinase signaling pathways [35,42] and new host factors identified in the genetic screen described above, the PIKFYVE/FIG4 phosphatidyl(3,5)bisphosphate kinase/phosphatase complex and the six-member HOPS (homotypic fusion and vacuole protein sorting complex) [41,43]. It is currently not known if viral particles can escape into the cytoplasm from late endosomes, or if they must instead journey on to lysosomes or another vesicular compartment.
Endosomal cysteine proteases prime EBOV GP for membrane fusion and expose putative receptor-binding sequences
The activity of class I fusion glycoproteins is regulated by ‘priming’ events, which typically involve a single endoproteolytic cleavage of the glycoprotein mediated by a cellular protease within the secretory pathway of the virus-producer cell (e.g., HIV Env --> SU + TM by furin) (reviewed in [11]). This cleavage is essential because it liberates a membrane-interacting fusion peptide (or loop) and/or allows the glycoprotein to rearrange during fusion. The unexpected observation that cleavage of filovirus GP0 to GP1 and GP2 is dispensable both in vitro and in vivo (even though it occurs efficiently) [44,45] led to the insight that GP is primed not by furin in the virus-producer cell, but instead by host endosomal cysteine proteases in the target cell [46,47] (Figs. 1 and 3).
The proteolytic priming of filovirus GP is also unusual in its extent and complexity. For EBOV GP, it requires endosomal cysteine protease cathepsin B (CatB), with cathepsin L (CatL) playing an accessory role [46]. In vitro, CatB or CatL cleave in the disordered loop separating the base and head domains of GP1, releasing the glycan cap and mucin subdomains [9,13,46–48]. CatB, but not CatL, can remove the entire loop to generate an infectious intermediate that no longer requires the activity of CatB within cells [13,47]. It is inferred (but not directly shown) that similar cleaved GP species are generated within cells.
Why are these cleavage events required? Although they do not themselves appear to induce large-scale conformational changes in the GP pre-fusion trimer [49], structural and biochemical evidence argues that they are needed for the triggering and execution of membrane fusion. First, cleavage in the loop connecting the GP1 base and head likely relieves a structural restraint on the underlying GP2 fusion loop, allowing it to emerge from the body of the GP trimer and insert into the target lipid bilayer during membrane fusion [9,10] (Fig. 1b). Second, selection for EBOV GP-containing viruses that no longer require CatB yielded GP mutants with enhanced susceptibility to proteolytic inactivation, suggesting that cleavage by CatB destabilizes the GP pre-fusion trimer and renders it more sensitive to the fusion trigger [50]. Third, heat and denaturant-triggered GP2-membrane association in vitro requires GP1 proteolysis, with each cleavage step progressively destabilizing GP [51].
A second, independent function proposed for GP cleavage is that it unmasks the binding site for an elusive cellular receptor with a presumptive role in viral membrane fusion [9,52]. Support for this idea comes from two observations. First, cleaved virions bind better to cells [52]. Second, while the glycan cap does not block solvent access to RBS residues, it may sterically obstruct a cellular protein’s access to its interaction site within the GP1 head subdomain [9,49]. Because GP cleavage is normally restricted to the endo/lysosomal pathway, it is supposed that virus must engage this putative receptor within endosomes, and not at the plasma membrane, even if the receptor is available at both locations.
Finally, although there is ample evidence from cell culture experiments that endosomal cysteine proteases play a crucial role in EBOV GP priming, the importance of these enzymes for infection remains to be confirmed in vivo. The capacity of EBOV GP to readily adapt to use endosomal cysteine proteases other than CatB and CatL in cultured cells highlights potential redundancies in protease requirements at the priming step [50]. Experiments to determine the roles of additional endosomal cysteine proteases (e.g., cathepsin S, which is highly expressed in macrophages and DCs ) in filovirus infection and pathogenesis are therefore warranted [53].
NPC1 is a critical endo/lysosomal host factor for filovirus entry
The host protein Niemann-Pick C1 (NPC1) was recently identified as a requirement for filovirus entry in independent genetic and chemical screens [41,54]. NPC1 is a large, multipass membrane protein that is expressed in all cells and localizes to late endosomes and lysosomes, where it participates in lysosomal efflux of low density lipoprotein-derived cholesterol . Mutation of NPC1 in humans (and mice) causes Niemann-Pick disease, a rare but fatal neurodegenerative disorder characterized by lysosomal lipid storage at the cellular level [55]. NPC1-mutant fibroblasts derived from Niemann-Pick patients are poorly susceptible to filoviruses, and a small molecule that specifically recognizes NPC1 inhibits EBOV entry in tissue culture [41,54]. The viral requirement for NPC1 appears to be absolute, since fully NPC1-deficient hamster cell lines are completely refractory to infection [41,54]. Mice heterozygous at the NPC1 locus are substantially resistant to authentic virus challenge, strongly suggesting that NPC1 is required for filovirus infection and pathogenesis in vivo [41].
Why is NPC1 required for filovirus entry and infection? Two lines of evidence indicate that this protein plays a specific role in filovirus entry that can be decoupled from its housekeeping function in cholesterol metabolism. First, filovirus entry does not require NPC2, a small, soluble cholesterol-binding protein that cooperates with NPC1, and whose loss phenocopies NPC1 loss in both cells and individuals [41,54,56]. Second, an engineered NPC1 mutant that is defective at cholesterol transport can support viral infection [54]. NPC1 is dispensable for virus-cell attachment, internalization, GP proteolytic priming, and delivery to late endosomal compartments, but it is required for viral escape into the cytoplasm [41] (Fig. 3). Finally, cleaved, but not uncleaved GP could associate with endosome-derived membranes in vitro in an NPC1-dependent manner and co-precipitate NPC1 from membrane extracts, indicating that the two proteins interact directly or indirectlyduring entry [54]. Although more work is needed, these findings taken together suggest a tantalizing scenario in which NPC1 is the long-sought endosomal entry receptor for filoviruses. NPC1 may play a direct role in initiating GP-dependent fusion between viral and cellular membranes. Alternatively or in addition, it may mediate viral trafficking to the intracellular sites of membrane fusion.
The host cell trigger for viral membrane fusion is unknown
In order to rearrange and insert its fusion loop into the target membrane, GP2 must be released from its intimate pre-fusion complex with GP1 (Figs. 1b and 2). The cellular signal that acts on a fully-primed GP intermediate to trigger these conformational changes remains unknown, and investigating its nature has proven challenging due to the lack of robust in vitro or cell-based membrane fusion assays for filoviruses. Endosomal acid pH is necessary for multiple steps in filovirus entry, including cleavage of EBOV GP by endosomal cysteine proteases, but it does not trigger cleaved GP, indicating that additional or alternative host cell factors are involved [4,5,9,46,47,49,51]. Using a new assay that mimics EBOV GP fusion triggering, Brecher and co-workers showed that acid pH is required for fusion loop-dependent binding of cleaved GP to synthetic liposomes, and that reduction of one or more disulfide bonds in GP (or a process mimicking disulfide reduction) may be a part of the fusion trigger [51]. Additionally, viruses containing proteolytically-primed EBOV GP remain sensitive to broad-spectrum inhibitors of endosomal cysteine proteases, raising the possibility that a ‘final cut’ in GP, or instead, the cleavage of an essential host factor, is required for fusion triggering [47,50,57]. Moreover, NPC1 is a strong candidate to be a part of the fusion trigger: it localizes to the limiting membrane of multivesicular late endosomes, where it is ideally placed to interact with primed GP as recently proposed [54], and it is required at a late entry step [41].
Endosomal acid pH plays multiple roles in the execution of viral membrane fusion
While X-ray crystal structures of EBOV GP have given us a good idea of what the first and last frames of the filovirus membrane fusion ‘movie’ look like [7–10] (Figs. 1b and 2), our understanding of the intervening steps is derived largely from analogy to other class I viral fusion machines [11]. In response to the fusion trigger, the pre-fusion HR1 sequence in GP2 is proposed to undergo release from around the GP1 base subdomain and to rearrange to an unbroken α–helix, projecting the GP2 fusion loop into the target membrane (Figs. 1b and 2). NMR structures of a complete fusion loop at neutral and acid pH indicate that it undergoes an acid pH-dependent local conformational change that reorients and compacts a hydrophobic patch at its apex [58]. This may optimize binding of the fusion loop to the target membrane or enhance its bilayer-destabilizing activity [58].
This extended GP2 conformation tipped by fusion loops is then thought to rearrange further, jack-knifing on itself to form a hairpin-like ‘six-helix bundle’ (6HB), in which the HR2s pack against grooves in the trimeric HR1 coiled-coil [7,8] (Figs. 1b and 2). The 6HB, a defining characteristic of class I viral fusion proteins, is proposed to drive viral and cellular bilayer mixing and fusion pore formation [11]. Consistent with the generation of an extended GP2 trimer within cells, a synthetic HR2 peptide directed to endosomes could inhibit filovirus entry, presumably by competing with the authentic HR2s for binding to the HR1 trimer core and preventing 6HB formation [59]. Whether the extended species spontaneously converts to the 6HB or requires additional host factors to do so (as seen with the avian leukosis retrovirus glycoprotein [60]) remains unknown at present. Also unknown is the status of GP1 during these GP2 gymnastics. Finally, studies with a synthetic GP2 6HB construct indicate that it is dramatically stabilized by acid pH [61]. Therefore, endosomal acid pH appears to regulate membrane fusion in at least three distinct ways–it is required for proteolytic priming of GP1 by endosomal cysteine proteases , and it may also be necessary for proper GP2 fusion loop insertion and 6HB formation (Fig. 1b).
Concluding remarks
Recent advances in understanding how filoviruses enter cells have been propelled by new structures of the EBOV glycoprotein, the discovery of new filovirus entry host factors and pathways through educated guesswork, genetic screens, and cell biological experiments, and the study of host-programmed changes in GP structure using in vitro assays. The emerging picture is of a group of viruses that exploit the host endo/lysosomal pathway in an extensive and sophisticated manner to gain access to the cytoplasm (Fig. 3). The findings discussed in this review should stimulate future work along the following fronts: (1) Extending observations made primarily with EBOV to divergent filoviruses, especially MARV; (2) identifying additional GP-host cell interactions that effect viral entry; (3) understanding in greater biochemical and structural detail how proteolytic cleavage of GP prepares it for membrane fusion; (4) determining the specific roles played by TIM-1 and NPC1 in entry and infection; (5) uncovering the GP fusion trigger and fully reconstituting membrane fusion in vitro; (6) defining structural intermediates in membrane fusion; (7) mapping each of these steps in time and space within intact cells; (8) determining their significance for filovirus host range, virulence, and in vivo pathogenesis. It is hoped that studies in this area will ultimately yield filovirus anti-infectives that target the entry step.
Bullet Points.
The filovirus glycoprotein, GP, mediates viral entry into the host cell cytoplasm.
Multiple cell surface molecules play roles in filovirus attachment to cells.
Unknown GP-host cell interactions induce viral internalization into endosomes via a pathway resembling macropinocytosis.
Niemann-Pick C1, an endo/lysosomal cholesterol transport protein, is a critical host factor for filovirus entry that may interact with a cleaved form of GP.
The host trigger for filovirus membrane fusion is unknown, and structural intermediates in the fusion reaction are poorly characterized.
Acknowledgments
We thank John M. Dye and Erica O. Saphire for comments and discussion, and apologize in advance to colleagues whose work could not be cited due to space limitations. The authors are supported by National Institutes of Health grant R01AI088027. E.H.M. is additionally supported by the NIH-funded training program T32 GM007288 at the Albert Einstein College of Medicine.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Feldmann H, Geisbert TW. Ebola haemorrhagic fever. Lancet. 2011;377:849–862. doi: 10.1016/S0140-6736(10)60667-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kortepeter MG, Bausch DG, Bray M. Basic clinical and laboratory features of filoviral hemorrhagic fever. J Infect Dis. 2011;204 (Suppl 3):S810–6. doi: 10.1093/infdis/jir299. [DOI] [PubMed] [Google Scholar]
- 3.Hartman AL, Towner JS, Nichol ST. Ebola and marburg hemorrhagic fever. Clin Lab Med. 2010;30:161–177. doi: 10.1016/j.cll.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 4.Takada A, Robison C, Goto H, Sanchez A, Murti KG, Whitt MA, Kawaoka Y. A system for functional analysis of Ebola virus glycoprotein. Proc Natl Acad Sci USA. 1997;94:14764–14769. doi: 10.1073/pnas.94.26.14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wool-Lewis RJ, Bates P. Characterization of Ebola virus entry by using pseudotyped viruses: identification of receptor-deficient cell lines. Journal of Virology. 1998;72:3155–3160. doi: 10.1128/jvi.72.4.3155-3160.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yonezawa A, Cavrois M, Greene WC. Studies of ebola virus glycoprotein-mediated entry and fusion by using pseudotyped human immunodeficiency virus type 1 virions: involvement of cytoskeletal proteins and enhancement by tumor necrosis factor alpha. Journal of Virology. 2005;79:918–926. doi: 10.1128/JVI.79.2.918-926.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weissenhorn W, Carfí A, Lee KH, Skehel JJ, Wiley DC. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol Cell. 1998;2:605–616. doi: 10.1016/s1097-2765(00)80159-8. [DOI] [PubMed] [Google Scholar]
- 8.Malashkevich VN, Schneider BJ, McNally ML, Milhollen MA, Pang JX, Kim PS. Core structure of the envelope glycoprotein GP2 from Ebola virus at 1. 9-A resolution. Proc Natl Acad Sci USA. 1999;96:2662–2667. doi: 10.1073/pnas.96.6.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *9.Lee JE, Fusco ML, Hessell AJ, Oswald WB, Burton DR, Saphire EO. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454:177–182. doi: 10.1038/nature07082. The first X-ray crystal structures of the Ebola and Sudan virus glycoproteins in their trimeric pre-fusion conformation have illuminated many features of the filovirus entry mechanism and driven new hypotheses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *10.Dias JM, Kuehne AI, Abelson DM, Bale S, Wong AC, Halfmann P, Muhammad MA, Fusco ML, Zak SE, Kang E, et al. A shared structural solution for neutralizing ebolaviruses. Nat Struct Mol Biol. 2011;18:1424–1427. doi: 10.1038/nsmb.2150. The first X-ray crystal structures of the Ebola and Sudan virus glycoproteins in their trimeric pre-fusion conformation have illuminated many features of the filovirus entry mechanism and driven new hypotheses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee JE, Saphire EO. Ebolavirus glycoprotein structure and mechanism of entry. Future virology. 2009;4:621–635. doi: 10.2217/fvl.09.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *13.Dube D, Brecher MB, Delos SE, Rose SC, Park EW, Schornberg KL, Kuhn JH, White JM. The primed ebolavirus glycoprotein (19-kilodalton GP1,2): sequence and residues critical for host cell binding. Journal of Virology. 2009;83:2883–2891. doi: 10.1128/JVI.01956-08. Shows that CatL and CatB prime GP by cleaving within, and removing, the flexible loop that connects the base and head subdomains in GP1. Also defines a presumptive receptor-binding domain in the GP base subdomain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuhn JH, Radoshitzky SR, Guth AC, Warfield KL, Li W, Vincent MJ, Towner JS, Nichol ST, Bavari S, Choe H, et al. Conserved receptor-binding domains of Lake Victoria marburgvirus and Zaire ebolavirus bind a common receptor. J Biol Chem. 2006;281:15951–15958. doi: 10.1074/jbc.M601796200. [DOI] [PubMed] [Google Scholar]
- 15.Manicassamy B, Wang J, Jiang H, Rong L. Comprehensive analysis of ebola virus GP1 in viral entry. Journal of Virology. 2005;79:4793–4805. doi: 10.1128/JVI.79.8.4793-4805.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brindley MA, Hughes L, Ruiz A, McCray PB, Sanchez A, Sanders DA, Maury W. Ebola virus glycoprotein 1: identification of residues important for binding and postbinding events. Journal of Virology. 2007;81:7702–7709. doi: 10.1128/JVI.02433-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeffers SA, Sanders DA, Sanchez A. Covalent modifications of the ebola virus glycoprotein. Journal of Virology. 2002;76:12463–12472. doi: 10.1128/JVI.76.24.12463-12472.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reynard O, Borowiak M, Volchkova VA, Delpeut S, Mateo M, Volchkov VE. Ebolavirus glycoprotein GP masks both its own epitopes and the presence of cellular surface proteins. Journal of Virology. 2009;83:9596–9601. doi: 10.1128/JVI.00784-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Francica JR, Varela-Rohena A, Medvec A, Plesa G, Riley JL, Bates P. Steric shielding of surface epitopes and impaired immune recognition induced by the ebola virus glycoprotein. PLoS Pathog. 2010;6:e1001098. doi: 10.1371/journal.ppat.1001098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kühl A, Hoffmann M, Müller MA, Munster VJ, Gnirss K, Kiene M, Tsegaye TS, Behrens G, Herrler G, Feldmann H, et al. Comparative analysis of Ebola virus glycoprotein interactions with human and bat cells. J Infect Dis. 2011;204 (Suppl 3):S840–9. doi: 10.1093/infdis/jir306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alvarez CP, Lasala F, Carrillo J, Muñiz O, Corbí AL, Delgado R. C-type lectins DC-SIGN and L-SIGN mediate cellular entry by Ebola virus in cis and in trans. Journal of Virology. 2002;76:6841–6844. doi: 10.1128/JVI.76.13.6841-6844.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marzi A, Gramberg T, Simmons G, Möller P, Rennekamp AJ, Krumbiegel M, Geier M, Eisemann J, Turza N, Saunier B, et al. DC-SIGN and DC-SIGNR interact with the glycoprotein of Marburg virus and the S protein of severe acute respiratory syndrome coronavirus. Journal of Virology. 2004;78:12090–12095. doi: 10.1128/JVI.78.21.12090-12095.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsegaye TS, Pöhlmann S. The multiple facets of HIV attachment to dendritic cell lectins. Cell Microbiol. 2010;12:1553–1561. doi: 10.1111/j.1462-5822.2010.01519.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin G, Simmons G, Pöhlmann S, Baribaud F, Ni H, Leslie GJ, Haggarty BS, Bates P, Weissman D, Hoxie JA, et al. Differential N-linked glycosylation of human immunodeficiency virus and Ebola virus envelope glycoproteins modulates interactions with DC-SIGN and DC-SIGNR. Journal of Virology. 2003;77:1337–1346. doi: 10.1128/JVI.77.2.1337-1346.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsuno K, Kishida N, Usami K, Igarashi M, Yoshida R, Nakayama E, Shimojima M, Feldmann H, Irimura T, Kawaoka Y, et al. Different potential of C-type lectin-mediated entry between Marburg virus strains. Journal of Virology. 2010;84:5140–5147. doi: 10.1128/JVI.02021-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marzi A, Wegele A, Pöhlmann S. Modulation of virion incorporation of Ebolavirus glycoprotein: effects on attachment, cellular entry and neutralization. Virology. 2006;352:345–356. doi: 10.1016/j.virol.2006.04.038. [DOI] [PubMed] [Google Scholar]
- 27.Marzi A, Möller P, Hanna SL, Harrer T, Eisemann J, Steinkasserer A, Becker S, Baribaud F, Pöhlmann S. Analysis of the interaction of Ebola virus glycoprotein with DC-SIGN (dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin) and its homologue DC-SIGNR. J Infect Dis. 2007;196 (Suppl 2):S237–46. doi: 10.1086/520607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simmons G, Reeves JD, Grogan CC, Vandenberghe LH, Baribaud F, Whitbeck JC, Burke E, Buchmeier MJ, Soilleux EJ, Riley JL, et al. DC-SIGN and DC-SIGNR bind ebola glycoproteins and enhance infection of macrophages and endothelial cells. Virology. 2003;305:115–123. doi: 10.1006/viro.2002.1730. [DOI] [PubMed] [Google Scholar]
- 29.Dube D, Schornberg KL, Shoemaker CJ, Delos SE, Stantchev TS, Clouse KA, Broder CC, White JM. Cell adhesion-dependent membrane trafficking of a binding partner for the ebolavirus glycoprotein is a determinant of viral entry. Proc Natl Acad Sci USA. 2010 doi: 10.1073/pnas.1008509107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freeman GJ, Casasnovas JM, Umetsu DT, DeKruyff RH. TIM genes: a family of cell surface phosphatidylserine receptors that regulate innate and adaptive immunity. Immunol Rev. 2010;235:172–189. doi: 10.1111/j.0105-2896.2010.00903.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *31.Kondratowicz AS, Lennemann NJ, Sinn PL, Davey RA, Hunt CL, Moller-Tank S, Meyerholz DK, Rennert P, Mullins RF, Brindley M, et al. T-cell immunoglobulin and mucin domain 1 (TIM-1) is a receptor for Zaire Ebolavirus and Lake Victoria Marburgvirus. Proceedings of the National Academy of Sciences. 2011;108:8426–8431. doi: 10.1073/pnas.1019030108. Identified TIM-1 as a candidate attachment receptor for Ebola virus GP in some cell types. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *32.Saeed MF, Kolokoltsov AA, Albrecht T, Davey RA. Cellular entry of ebola virus involves uptake by a macropinocytosis-like mechanism and subsequent trafficking through early and late endosomes. PLoS Pathog. 2010:6. doi: 10.1371/journal.ppat.1001110. These two papers reveal that the Ebola virus GP induces viral internalization via a host pathway resembling macropinocytosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *33.Nanbo A, Imai M, Watanabe S, Noda T, Takahashi K, Neumann G, Halfmann P, Kawaoka Y. Ebolavirus is internalized into host cells via macropinocytosis in a viral glycoprotein-dependent manner. PLoS Pathog. 2010:6. doi: 10.1371/journal.ppat.1001121. These two papers reveal that the Ebola virus GP induces viral internalization via a host pathway resembling macropinocytosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mercer J, Helenius A. Virus entry by macropinocytosis. Nat Cell Biol. 2009;11:510–520. doi: 10.1038/ncb0509-510. [DOI] [PubMed] [Google Scholar]
- 35.Saeed MF, Kolokoltsov AA, Freiberg AN, Holbrook MR, Davey RA. Phosphoinositide-3 kinase-Akt pathway controls cellular entry of Ebola virus. PLoS Pathog. 2008;4:e1000141. doi: 10.1371/journal.ppat.1000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mulherkar N, Raaben M, la Torre de J-C, Whelan SP, Chandran K. The Ebola virus glycoprotein mediates entry via a non-classical dynamin-dependent macropinocytic pathway. Virology. 2011;419:72–83. doi: 10.1016/j.virol.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hunt CL, Kolokoltsov AA, Davey RA, Maury W. The Tyro3 receptor kinase Axl enhances macropinocytosis of Zaire ebolavirus. Journal of Virology. 2011;85:334–347. doi: 10.1128/JVI.01278-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sallusto F, Cella M, Danieli C, Lanzavecchia A. Dendritic cells use macropinocytosis and the mannose receptor to concentrate macromolecules in the major histocompatibility complex class II compartment: downregulation by cytokines and bacterial products. J Exp Med. 1995;182:389–400. doi: 10.1084/jem.182.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shimojima M, Takada A, Ebihara H, Neumann G, Fujioka K, Irimura T, Jones S, Feldmann H, Kawaoka Y. Tyro3 family-mediated cell entry of Ebola and Marburg viruses. Journal of Virology. 2006;80:10109–10116. doi: 10.1128/JVI.01157-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morizono K, Xie Y, Olafsen T, Lee B, Dasgupta A, Wu AM, Chen ISY. The soluble serum protein Gas6 bridges virion envelope phosphatidylserine to the TAM receptor tyrosine kinase Axl to mediate viral entry. Cell Host Microbe. 2011;9:286–298. doi: 10.1016/j.chom.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *41.Carette JE, Raaben M, Wong AC, Herbert AS, Obernosterer G, Mulherkar N, Kuehne AI, Kranzusch PJ, Griffin AM, Ruthel G, et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. Utilizes a genetic approach to identify Niemann-Pick C1 as a critical host factor for filovirus entry. Shows that NPC1 is needed at a late entry step and for filovirus infection and pathogenesis in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kolokoltsov AA, Saeed MF, Freiberg AN, Holbrook MR, Davey RA. Identification of novel cellular targets for therapeutic intervention against Ebola virus infection by siRNA screening. Drug Dev Res. 2009;70:255–265. doi: 10.1002/ddr.20303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huotari J, Helenius A. Endosome maturation. EMBO J. 2011;30:3481–3500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wool-Lewis RJ, Bates P. Endoproteolytic processing of the ebola virus envelope glycoprotein: cleavage is not required for function. Journal of Virology. 1999;73:1419–1426. doi: 10.1128/jvi.73.2.1419-1426.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neumann G, Geisbert TW, Ebihara H, Geisbert JB, Daddario-DiCaprio KM, Feldmann H, Kawaoka Y. Proteolytic processing of the Ebola virus glycoprotein is not critical for Ebola virus replication in nonhuman primates. Journal of Virology. 2007;81:2995–2998. doi: 10.1128/JVI.02486-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 2005;308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schornberg K, Matsuyama S, Kabsch K, Delos S, Bouton A, White J. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. Journal of Virology. 2006;80:4174–4178. doi: 10.1128/JVI.80.8.4174-4178.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *48.Hood CL, Abraham J, Boyington JC, Leung K, Kwong PD, Nabel GJ. Biochemical and structural characterization of cathepsin L-processed Ebola virus glycoprotein: implications for viral entry and immunogenicity. Journal of Virology. 2010;84:2972–2982. doi: 10.1128/JVI.02151-09. Shows that CatL primes GP by cleaving within the flexible loop that connects the base and head subdomains in GP1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bale S, Liu T, Li S, Wang Y, Abelson D, Fusco M, Woods VL, Saphire EO. Ebola virus glycoprotein needs an additional trigger, beyond proteolytic priming for membrane fusion. PLoS Negl Trop Dis. 2011;5:e1395. doi: 10.1371/journal.pntd.0001395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wong AC, Sandesara RG, Mulherkar N, Whelan SP, Chandran K. A forward genetic strategy reveals destabilizing mutations in the Ebolavirus glycoprotein that alter its protease dependence during cell entry. Journal of Virology. 2010;84:163–175. doi: 10.1128/JVI.01832-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *51.Brecher M, Schornberg KL, Delos SE, Fusco ML, Saphire EO, White JM. Cathepsin Cleavage Potentiates the Ebola Virus Glycoprotein to Undergo a Subsequent Fusion Relevant Conformational Change. Journal of Virology. 2011 doi: 10.1128/JVI.05708-11. Describes an in vitro assay to detect and measure deployment of the GP2 fusion peptide, which is presumed to be an initial step in viral membrane fusion. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaletsky RL, Simmons G, Bates P. Proteolysis of the Ebola virus glycoproteins enhances virus binding and infectivity. Journal of Virology. 2007;81:13378–13384. doi: 10.1128/JVI.01170-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Colbert JD, Matthews SP, Miller G, Watts C. Diverse regulatory roles for lysosomal proteases in the immune response. Eur J Immunol. 2009;39:2955–2965. doi: 10.1002/eji.200939650. [DOI] [PubMed] [Google Scholar]
- *54.Côté M, Misasi J, Ren T, Bruchez A, Lee K, Filone CM, Hensley L, Li Q, Ory D, Chandran K, et al. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature. 2011;477:344–348. doi: 10.1038/nature10380. Utilizes a chemical biology approach to identify Niemann-Pick C1 as a critical host factor for filovirus entry. Shows that cleaved GP can directly or indirectly interact with NPC1 derived from endosomal membranes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carstea ED, Morris JA, Coleman KG, Loftus SK, Zhang D, Cummings C, Gu J, Rosenfeld MA, Pavan WJ, Krizman DB, et al. Niemann-Pick C1 disease gene: homology to mediators of cholesterol homeostasis. Science. 1997;277:228–231. doi: 10.1126/science.277.5323.228. [DOI] [PubMed] [Google Scholar]
- 56.Naureckiene S, Sleat DE, Lackland H, Fensom A, Vanier MT, Wattiaux R, Jadot M, Lobel P. Identification of HE1 as the second gene of Niemann-Pick C disease. Science. 2000;290:2298–2301. doi: 10.1126/science.290.5500.2298. [DOI] [PubMed] [Google Scholar]
- 57.Chandran K, Zhang X, Olson NH, Walker SB, Chappell JD, Dermody TS, Baker TS, Nibert ML. Complete in vitro assembly of the reovirus outer capsid produces highly infectious particles suitable for genetic studies of the receptor-binding protein. Journal of Virology. 2001;75:5335–5342. doi: 10.1128/JVI.75.11.5335-5342.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gregory SM, Harada E, Liang B, Delos SE, White JM, Tamm LK. Structure and function of the complete internal fusion loop from Ebolavirus glycoprotein 2. Proceedings of the National Academy of Sciences. 2011;108:11211–11216. doi: 10.1073/pnas.1104760108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miller EH, Harrison JS, Radoshitzky SR, Higgins CD, Chi X, Dong L, Kuhn JH, Bavari S, Lai JR, Chandran K. Inhibition of Ebola virus entry by a C-peptide targeted to endosomes. Journal of Biological Chemistry. 2011;286:15854–15861. doi: 10.1074/jbc.M110.207084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Matsuyama S, Delos SE, White JM. Sequential roles of receptor binding and low pH in forming prehairpin and hairpin conformations of a retroviral envelope glycoprotein. Journal of Virology. 2004;78:8201–8209. doi: 10.1128/JVI.78.15.8201-8209.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Harrison JS, Higgins CD, Chandran K, Lai JR. Designed protein mimics of the Ebola virus glycoprotein GP2 α-helical bundle: stability and pH effects. Protein Sci. 2011;20:1587–1596. doi: 10.1002/pro.688. [DOI] [PMC free article] [PubMed] [Google Scholar]