Abstract
When exposed to the non-metabolized glucose derivative alpha methyl glucoside, both E. coli K-12 (JM109 and MG1655) and E. coli B (BL21) respond by reducing the concentration of the mRNA of the ptsG gene which is responsible for the biosynthesis of the glucose transporter EIICBglu. This occurs through the over-expression of the non-coding small RNA SgrS, which interacts specifically with the mRNA of the ptsG gene and prevents its translation. However, when these bacteria are exposed to a glucose concentration of 40 g/L, over-expression of SgrS is observed only in E. coli B (BL21). Unlike E. coli K-12 (JM109 and MG1655), which are affected by high glucose concentration and produce higher levels of acetate, E. coli B (BL21) is not affected. Based on this information, it was assumed that over-expression of SgrS enables E. coli B (BL21) to reduce its acetate excretion by controlling the glucose transport. When SgrS was over-expressed in both E. coli K-12 strains from a multicopy plasmid, it was possible to reduce their acetate excretion levels to those seen in E. coli B. This observation opens a new approach towards controlling bacterial metabolism through the use of non-coding RNA.
Keywords: small RNA, SgrS, acetate, E. coli, glucose
Introduction
E. coli responds to environmental changes by regulating various functions through complex and interacting networks that affect gene transcription, gene translation and enzyme activity. A class of regulators that currently is of interest is the non-coding small RNAs. More than 70 molecules ranging from 50–250 nucleotides have been identified [1–3]; these molecules can repress or activate the expression of specific genes. A subset of these small RNAs regulates gene translation through base pairing with their target mRNAs and has been investigated intensively. This group of small RNAs interacts with Hfq, an RNA chaperone, to form the base pairing which leads to change in the mRNA translation and stability [2, 3]. The small RNA SgrS, was identified in 2004 [4] as a member of this group. SgrS expression is induced when E. coli K-12 (MG1655) is either exposed to the non-metabolizable glucose derivative αMG [4] or when a mutation is introduced to block the glycolytic pathway [5, 6]; both cases are considered to be stress conditions. It was established that SgrS prevents the translation of the mRNA of the glucose transporter gene ptsG and consequently lowers the concentration of the glucose transporter EIICBglu and the glucose transport into the cell. The proposed trigger for this action is the accumulation of glucose 6 phosphate which causes the induction of sgrS expression [5, 6]. In our previous report [7], we showed that E. coli B (BL21) responded to the presence of αMG in the same way as E. coli K-12 (MG1655 and JM109); both strains over expressed SgrS and lowered the levels of the ptsG transcript and glucose transport. However, when cells were exposed to environmental stress caused by high glucose concentration, only E. coli B (BL21) over-expressed SgrS, lowering ptsG and glucose consumption. Under the same conditions both E. coli K-12 strains did not express SgrS and did not affect glucose transport. We suggested that, together with the different operation of the central carbon metabolism [8], this is an additional mechanism that allows E. coli B (BL21) to respond better to high glucose concentrations and to lower acetate excretion.
Controlling acetate excretion during high density growth of E. coli K-12 is a topic under investigation [9–13] since high concentrations of acetate affect growth and recombinant protein production. Various methodologies were developed to reduce acetate accumulation, among them different growth strategies [9, 10], introducing changes in the central carbon metabolism [11–13] and in the glucose transport process [14, 15] by modifying gene expression and transcription. Based on our finding that SgrS is expressed differently in E. coli K and B, we suggested that perhaps it will be possible to reduce acetate excretion in E. coli K-12 by manipulating the expression of the small RNA SgrS and affecting the translation of the glucose transporter EIICBglu. The effect of over expressing SgrS on acetate excretion in E. coli K-12 is reported in this manuscript.
Material and methods
E.coli strains
The two parental strains used in this study were E. coli K-12 (JM109) (DE3) (endA1, recA1, gyrA96, thi, hsdR17 (rk-, mk+), relA1, supE44, λ-, Δ(lac-proAB), [F’, traD36, proAB, laclqZΔM15], DE3) (Promega Corp, Madison, WI), and E. coli K-12 (MG1655) (F-, λ-, ilvG-, rfb-50, rph-1).
Strains over expressing SgrS were prepared by transforming E. coli K-12 (JM109) and E. coli NM525, an E. coli K-12 (MG1655) containing the lacIq mutation (MG1655 lacIq ) that improved transcriptional repression of the pLCV1 (PLlacO-sgrS) plasmid until induction with IPTG [4].
Transformation procedure: The E. coli K-12 JM109 and E. coli K-12 MG1655 lacIq strains were transformed with the plasmid pLCV1 (PLlacO-sgrS) by electroporation using an electrocell manipulator (BTX Harvard Apparatus, Holliston, MA). The electroporation was performed according to Sambrook and Russell, 2001 [17]. Competent cells were grown at 37°C in LB medium to an OD600~ 0.4. Forty µl cells (8×108) were washed twice with cold water and once with 10% cold glycerol, and suspended in cold GYT medium (10% v/v glycerol, 1.125% w/v yeast extract, 0.25% w/v tryptone). The conditions used in the 2 mm electroporation cuvette, were electrical pulse of 50 µF capacitance, 2.45 kV, and 125 ohm resistance. Cells were recovered in S.O.C. medium (Invitrogen, Carlsbad, CA) and incubated for 1h at 37°C. Cells were plated in LB agar plates containing 100µg/mL ampicillin to select for transformants.
Cell growth
Cells were grown at 37°C in modified LB medium containing 10 g/L tryptone, 15 g/L yeast extract, 5 g/L NaCl and 5 g/L K2HPO4. After sterilization, 10 mM MgSO4 1 ml/L trace elements solution were added, and glucose concentration was adjusted to 40 g/L, 100µg/mL ampicillin were added to the SgrS over-expressing stain. pH was controlled at 7.0 by addition of 50 % (v/v) NH4OH, and dissolved oxygen (DO) was controlled at 30% air saturation. A 5 liter bioreactor (Sartorious) was inoculated with an overnight culture to an OD600= 0.3. Cell density was determined by measuring OD600 with a Pharmacia Biotech Ultrospec 3000 UV/Visible spectrophotometer, when the culture reached an OD600 of 1.0, it was induced with IPTG; 100µM for E. coli K-12 (JM109) and 50µM for E. coli K-12 (MG1655). Samples were collected at specific times and centrifuged at 13,000 g for 5 min; the cell pellet and the supernatant for RNA extraction and metabolites analysis were maintained at −80°C. (Growth experiments were replicated 3 times).
Metabolite analysis
Glucose was determined by YSI 2700 SELECT Biochemistry Analyzer. Acetate was analyzed by HPLC, Hewlett Packard 1100 Series using Aminex resin-based HPX-87H column (Bio-Rad). Separation conditions were as follows: wavelength 210 nm; mobile phase 0.008 N H2SO4, flow rate 0.6 mL/min, temperature 35°C retention time was 14 min.
RNA extraction
The hot phenol method was used: cell pellets were resuspended in 0.5 % SDS, 20 mM NaAc, and 10 mM EDTA and extracted twice with hot acid phenol:chloroform (5:1 pH 4.5) followed by two extractions with phenol:chlorform isoamyl alcohol (25:24:1). Ethanol was added to the extract and the mixture kept at −80°C for 15min. After centrifugation at 14,000 g for 15 min, the pellets were washed in 70% ethanol, air dried and resuspended in ultrapure water (KD medical USA). RNA was quantified using NanoDrop 1000 spectrophotometer (Thermofisher Scientific).
Northern blot analysis
Northern blot analyses to detect sRNA SgrS were performed as described previously [4]. 5 µg of total RNA was separated on a TBE 10 % urea polyacrylamide gel and transferred to a positively charged nylon membrane. A 5’ biotinylated SgrS-specific probe and the Bright-Star Biodetect non-isotopic kit (Ambion, Inc.) were used for probing and detection. The detection of the ptsG mRNA was performed as follows: 5 µg of total RNA was separated on a 1.2 % denaturing agarose gel and transferred to a positively charged nylon membrane [18]. The membranes were probed, washed and conjugated with streptavidin-alkaline phosphatase using the BrightStar Biodetect Kit (Ambion). Chemiluminscent signals were detected using the Fujifilm LAS-4000 imaging system. The image before the saturation point was recorded. The internal controls ssrA and ompA were detected by probing membranes that were stripped with boiling in 0.5 % SDS. The 5’ to 3’ sequences of the probes used were:
sgrS (Bio)-GCAACCAGCACAACTTCGCTGTCGCGGTAAAATAGTG
ptsG (Bio)-CAGCCAGCTGAAATTCGCGGAACCGACGCCCAGCAG
ssrA (Bio)-CGCCACTAACAAACTAGCCTGATTAAGTTTTAACGCTTCA
ompA (Bio)-CCATTGTTGTTGATGAAACCAGTGTCATGGTACTGGGACCAGC
Results
1. Effect of over expression of SgrS in E. coli K-12 (JM 109) on growth, glucose consumption and acetate excretion
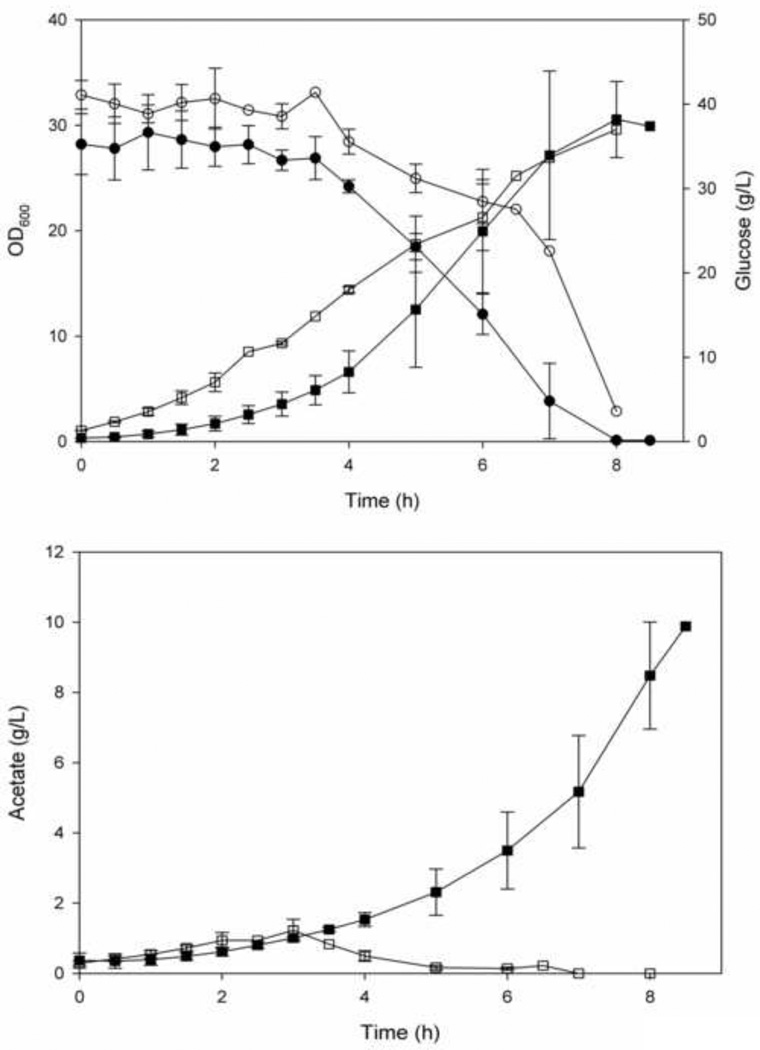
Growth and glucose consumption of E. coli K-12 (JM109) and E. coli K-12 (JM109) over-expressing SgrS are shown in Figure 1A. The patterns of the bacterial growth and the residual glucose concentration of the two strains are similar, but significant difference has been observed in the acetate excretion pattern (Fig 1B); 10 g/L acetate were accumulated in the parent E. coli K-12(JM109) while less than 2 g/L were accumulated in the strain over expressing SgrS.
Figure 1.
Growth, glucose consumption and acetate production in E. coli K-12 (JM109) and E. coli K-12 (JM109) containing plasmid pLCV1 over-expressing SgrS. (a) Growth and glucose consumption (solid and dashed lines, respectively). (b) Acetate excretion. (■, ●) E. coli K-12 (JM109). (□, ○) E. coli K-12 (JM109) over expressing SgrS
2. Transcription of SgrS and ptsG in E. coli K-12 (JM 109) and in E. coli K-12 (JM109) overexpressing SgrS
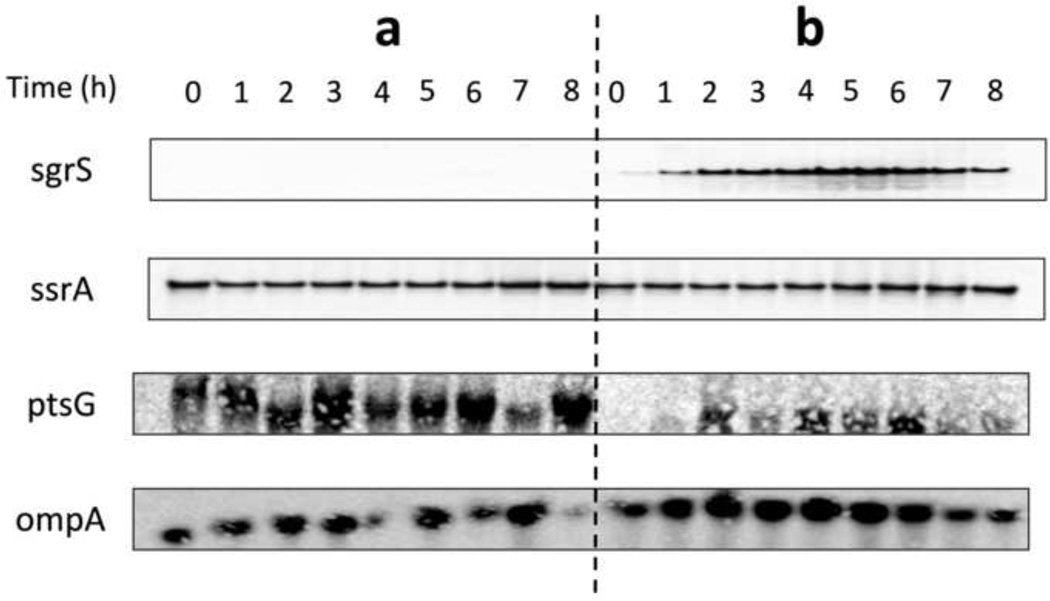
Time course transcription measurements of SgrS and ptsG in E. coli K12 (JM109) and in E. coli K-12 (JM109) over-expressing SgrS are shown in Figure 2. No transcription of SgrS and high transcription of ptsG were detected in the parental E. coli K-12 (JM109) while the opposite was seen in E. coli K-12 (JM109) over-expressing SgrS. The constitutive transcription of ssrA and ompA was used as an internal control.
Figure 2.
Transcription analysis of SgrS and ptsG in E. coli K12 (JM109) and E. coli K-12 (JM109) containing plasmid pLCV1-over expressing SgrS. (a) E. coli K12 (JM109). (b) E. coli K-12 (JM109) over expressing SgrS
3. Effect of over-expression of SgrS in E. coli K-12 (MG1655) on growth, glucose consumption and acetate excretion
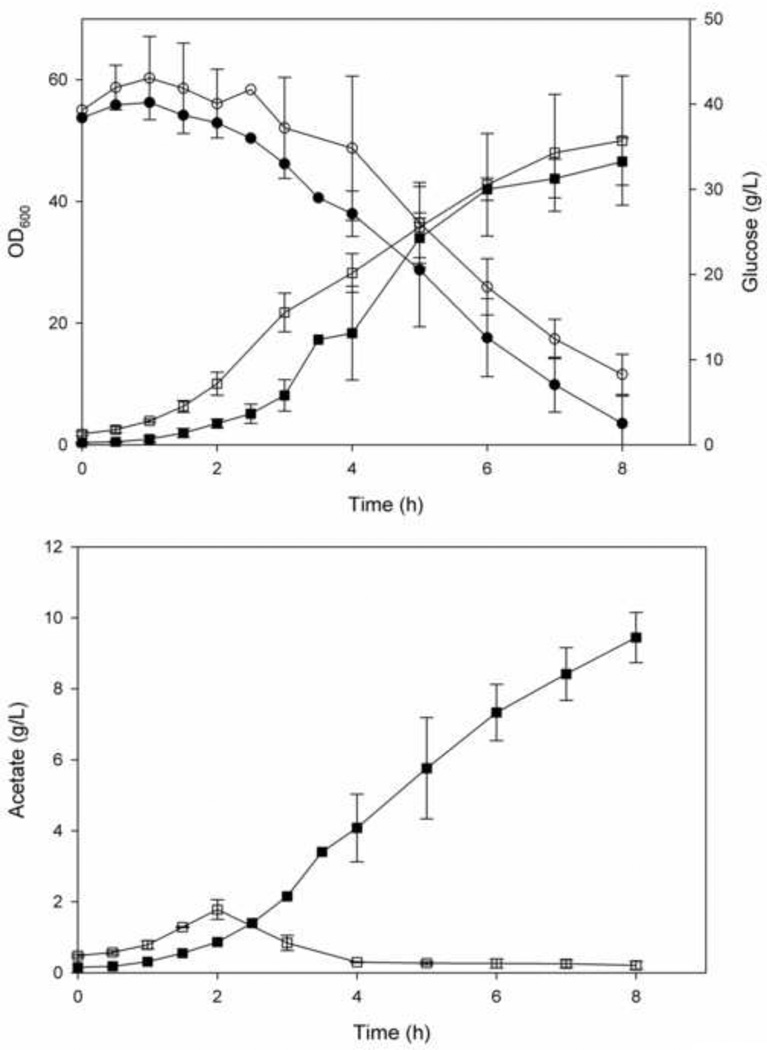
Growth and glucose consumption of parental E. coli K-12 (MG1655) and E. coli K-12 (MG1655 lacIq) over-expressing SgrS are shown in Figure 3A. The growth pattern of the two strains is similar but the glucose consumption of the strain over-expressing SgrS is slightly slower. Significant difference was observed in the acetate excretion pattern (Fig 3B); there was almost no accumulation of acetate in the strain over-expressing SgrS.
Figure 3.
Growth, glucose consumption and acetate production in E. coli K12 (MG1655) and E. coli K-12 (MG1655 lacIq) containing plasmid pLCV1 over expressing SgrS. (a) Growth and glucose consumption (solid and dashed lines, respectively). (b) Acetate excretion. (■, ●) E. coli K12 (MG1655). (□, ○) E. coli K-12 (MG1655 lacIq) over-expressing SgrS
4. Transcription of SgrS and ptsG in E. coli K-12 (MG1655) and in E. coli K-12 (MG1655 lacIq) over-expressing SgrS
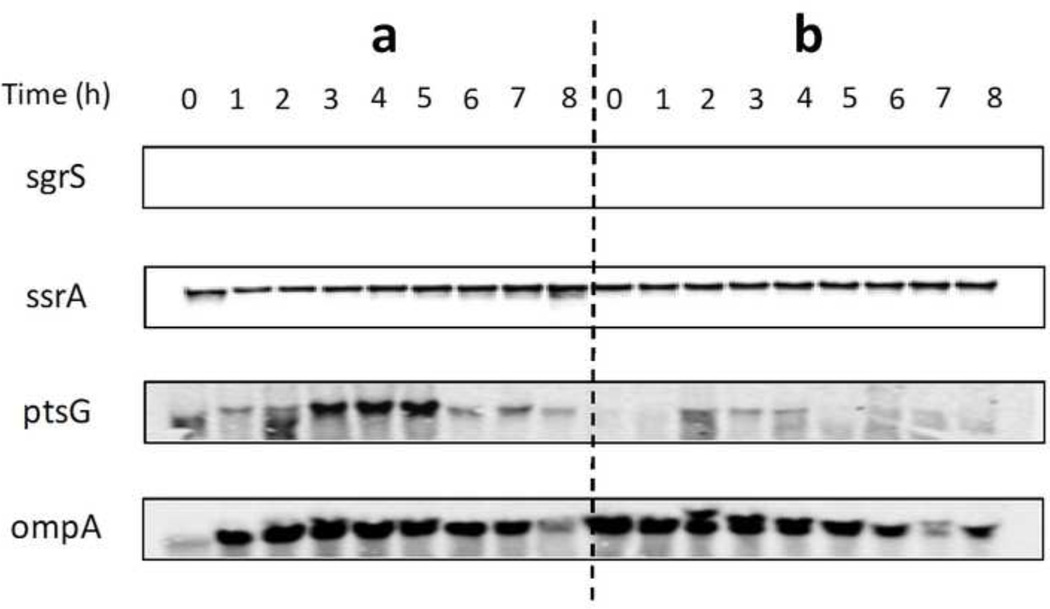
Time course transcription measurements of SgrS and ptsG in E. coli K-12 (MG1655) and E. coli K-12 (MG1655 lacIq) over-expressing SgrS are shown in Figure 4. No transcription of SgrS and high transcription of ptsG were detected in E. coli K-12 (MG1655). However, high transcription of SgrS and low transcription of ptsG were observed in E. coli K-12 (MG1655 lacIq) over-expressing SgrS. The constitutive transcription of ssrA and ompA was used as an internal control.
Figure 4.
Transcription analysis of SgrS and ptsG in E. coli K-12 (MG1655) and E. coli K-12 (MG1655 lacIq) containing plasmid pLCV1 over expressing SgrS. (a) E. coli K12 (MG1655). (b) E. coli K-12 (MG1655 lacIq) over-expressing SgrS
Discussion
Controlling glucose transport is one of the means used by E. coli when responding to stress conditions such as the presence of non-metabolizable glucose derivatives or high glucose concentrations [14, 15]. Glucose transport into cells is mediated by the PTS (phosphoenolpyruvate-dependent sugar phosphotransferase) system that contains several components, of which the EIICBglu is the specific glucose transporter encoded by the ptsG gene [19, 20]. Concentration of EIICBglu is adjusted by the global regulator Mlc that inhibits the ptsG transcription [19–21], and by the small RNA SgrS that inactivates ptsG translation by interacting with the ptsG mRNA [4, 22, 23]. SgrS is expressed in E. coli K-12 (JM109 and MG1655) and E. coli B (BL21) when the cells are exposed to the non-metabolizable glucose derivative αMG; however, when cells are exposed to high glucose concentrations, only E. coli B (BL21) expresses SgrS [7]. The expression of SgrS and the inactivation of the ptsG mRNA is likely responsible for the reduced acetate excretion in the E. coli B (BL21) compared to the E. coli K-12 strain where SgrS is not expressed and there is no reduction in acetate excretion. This difference in response to high glucose concentrations is in addition to the previously identified difference in activity of the central carbon metabolism at high glucose concentrations [8].
Acetate excretion by E. coli K-12 affects both growth and protein production. As a result, efforts are being devoted to lowering acetate excretion from this strain especially when cells are exposed to high glucose [24]. Currently two main strategies are implemented; one involves the development of different growth methods [9, 10] and the other is associated with the manipulation of genes and enzymes related to the central carbon metabolism [11–13] and to glucose transport into cells [14, 15]. In this report, we demonstrate that it is possible to decrease acetate excretion in E. coli K-12 by manipulating the expression of the SgrS sRNA. Over-expression of SgrS in both E. coli K-12 strains (JM109 and MG1655) reduced acetate excretion to a similar level observed in E. coli B (BL21).
Ample information is available on small RNA properties and mechanisms of operation [3], but so far no attempt has been made to implement this information for modifying bacterial growth and metabolism. The lowering of acetate excretion in the E. coli K-12 strains by over-expressing SgrS is a preliminary step in the attempt to regulate bacterial metabolism by using small RNAs. Over-expression of SgrS was achieved by inducing the gene from an inserted plasmid that is not subjected to the internal control mechanism of the bacteria and is not expressed in response to specific growth properties or media composition. It is not known how the bacteria controlled the expression of SgrS and why over-expression by IPTG induction from a plasmid did not eliminate entirely the ptsG transcription as was the case when the cells were exposed to αMG [7]. Clearly, further work is needed to incorporate SgrS expression into the bacterial chromosome and to establish a mechanism to regulate its expression based on the growth properties and media composition.
Conclusions
When growing at high glucose concentrations, E. coli K-12 (JM109 and MG1165) produce high concentrations of acetate. We assumed that this is, in addition to inefficient central carbon metabolism, the result of the inability of this strain to express the small RNA SgrS which inhibits the biosynthesis of the glucose transporter EIICBglu. By over-expressing SgrS in these strains, it was possible to reduce the acetate excretion when the bacteria were exposed to high glucose concentration. This finding suggests that external control of small RNA can potentially be used to regulate bacterial growth and metabolism.
Acknowledgements
Funding was provided by the intramural program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health. The authors would like to thank Dr. S. Gottesman for providing the plasmid and the lacIq strain together with detailed instructions, Ms. W. I. Ng for technical help and Mrs. D. Livnat for critical review of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Babitzke P, Romeo T. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr Opin Microbiol. 2007;10:156–163. doi: 10.1016/j.mib.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 2.Wassarman KM. 6S RNA: a small RNA regulator of transcription. Curr Opin Microbiol. 2007;10:164–168. doi: 10.1016/j.mib.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vanderpool CK, Gottesman S. Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol Microbiol. 2004;54:1076–1089. doi: 10.1111/j.1365-2958.2004.04348.x. [DOI] [PubMed] [Google Scholar]
- 5.Kimata K, Tanaka Y, Inada T, Aiba H. Expression of the glucose transporter gene, ptsG, is regulated at the mRNA degradation step in response to glycolytic flux in Escherichia coli. EMBO J. 2001;20:3587–3595. doi: 10.1093/emboj/20.13.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morita T, El-Kazzaz W, Tanaka Y, Inada T, Aiba H. Accumulation of glucose 6-phosphate or fructose 6-phosphate is responsible for destabilization of glucose transporter mRNA in Escherichia coli. J Biol Chem. 2003;278:15608–15614. doi: 10.1074/jbc.M300177200. [DOI] [PubMed] [Google Scholar]
- 7.Negrete A, Ng WI, Shiloach J. Glucose uptake regulation in E. coli by the small RNA SgrS: comparative analysis of E. coli K-12 (JM109 and MG1655) and E. coli B (BL21) Microb Cell Fact. 2010;9:75–83. doi: 10.1186/1475-2859-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phue JN, Noronha SB, Hattacharyya R, Wolfe AJ, Shiloach J. Glucose metabolism at high density growth of E. coli B and E. coli K: differences in metabolic pathways are responsible for efficient glucose utilization in E. coli B as determined by microarrays and Northern blot analyses. Biotechnol Bioeng. 2005;90:805–820. doi: 10.1002/bit.20478. [DOI] [PubMed] [Google Scholar]
- 9.Lee SY. High cell-density culture of Escherichia coli. Trends Biotechnol. 1996;14:98–105. doi: 10.1016/0167-7799(96)80930-9. [DOI] [PubMed] [Google Scholar]
- 10.Riesenberg D, Guthke R. High-cell-density cultivation of microorganisms. Appl Microbiol Biotechnol. 1999;51:422–430. doi: 10.1007/s002530051412. [DOI] [PubMed] [Google Scholar]
- 11.Contiero J, Beatty C, Kumari S, DeSanti CL, Strohl WR, Wolfe AJ. Effects of mutations in acetate metabolism on high-cell-density growth of Escherichia coli. J Ind Microbiol. 2000;24:421–430. [Google Scholar]
- 12.Phue JN, Lee SJ, Kaufman JB, Negrete A, Shiloach J. Acetate accumulation through alternative metabolic pathways in ackA− pta− poxB− triple mutant in E. coli B (BL21) Biotechnol Lett. 2010;32:1897–1903. doi: 10.1007/s10529-010-0369-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waegeman H, Beauprez J, Moens H, Maertens J, De Mey M, Foulquié-Moreno MR, Heijnen JJ, Charlier D, Soetaert W. Effect of iclR and arcA knockouts on biomass formation and metabolic fluxes in Escherichia coli K12 and its implications on understanding the metabolism of Escherichia coli BL21 (DE3) BMC Microbiol. 2011;11:70–86. doi: 10.1186/1471-2180-11-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Anda R, Lara AR, Hernández V, Hernández-Montalvo V, Gosset G, Bolívar F, Ramírez OT. Replacement of the glucose phosphotransferase transport system by galactose permease reduces acetate accumulation and improves process performance of Escherichia coli for recombinant protein production without impairment of growth rate. Metab Eng. 2006;8:281–290. doi: 10.1016/j.ymben.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 15.Flores N, Leal L, Sigala JC, de Anda R, Escalante A, Martínez A, Ramírez OT, Gosset G, Bolivar F. Growth recovery on glucose under aerobic conditions of an Escherichia coli strain carrying a phosphoenolpyruvate:carbohydrate phosphotransferase system deletion by inactivating arcA and overexpressing the genes coding for glucokinase and galactose permease. J Mol Microbiol Biotechnol. 2007;13:105–116. doi: 10.1159/000103602. [DOI] [PubMed] [Google Scholar]
- 16.Mandin P, Gottesman S. Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. EMBO J. 2010;29:3094–3107. doi: 10.1038/emboj.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sambrook J, Russell DW. Molecular cloning. 3rd edition. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. Plasmids and Their Usefulness in Molecular Cloning; pp. 119–122. [Google Scholar]
- 18.Massé E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci USA. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Decker K, Plumbridge J, Boos W. Negative transcriptional regulation of a positive regulator: the expression of malT, encoding the transcriptional activator of the maltose regulon of Escherichia coli, is negatively controlled by Mlc. Mol Microbiol. 1998;27:381–390. doi: 10.1046/j.1365-2958.1998.00694.x. [DOI] [PubMed] [Google Scholar]
- 20.Kimata K, Inada T, Tagami H, Aiba H. A global repressor (Mlc) is involved in glucose induction of the ptsG gene encoding major glucose transporter in Escherichia coli. Mol Microbiol. 1998;29:1509–1519. doi: 10.1046/j.1365-2958.1998.01035.x. [DOI] [PubMed] [Google Scholar]
- 21.Kim SY, Nam TW, Shin D, Koo BM, Seok YJ, Ryu S. Purification of Mlc and analysis of its effects on the pts expression in Escherichia coli. J Biol Chem. 1999;274:25398–25402. doi: 10.1074/jbc.274.36.25398. [DOI] [PubMed] [Google Scholar]
- 22.Aiba H. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr Opin Microbiol. 2007;10:134–139. doi: 10.1016/j.mib.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 23.Maki K, Morita T, Otaka H, Aiba H. A minimal base-pairing region of a bacterial small RNA SgrS required for translational repression of ptsG mRNA. Mol Microbiol. 2010;76:782–792. doi: 10.1111/j.1365-2958.2010.07141.x. [DOI] [PubMed] [Google Scholar]
- 24.Shiloach J, Fass R. Growing E. coli to high cell density--a historical perspective on method development. Biotechnol Adv. 2005;23:345–357. doi: 10.1016/j.biotechadv.2005.04.004. [DOI] [PubMed] [Google Scholar]