Abstract
Self-assembling protein nanocapsules can be engineered for various bionanotechnology applications. Using the dodecahedral scaffold of the E2 subunit from pyruvate dehydrogenase, we introduced non-native surface cysteines for site-directed functionalization. The modified nanoparticle’s structural, assembly, and thermostability properties were comparable to the wild-type scaffold (E2-WT), and after conjugation of polyethylene glycol (PEG) to these cysteines, the nanoparticle remained intact and stable up to 79.7 ± 1.8 °C. PEGylation of particles reduced uptake by human monocyte derived macrophages and MDA-MB-231 breast cancer cells, with decreased uptake as PEG chain length is increased. In vitro C4-depletion and C5a-production assays yielded 97.6 ± 10.8 % serum C4 remaining and 40.1 ± 6.0 ng/ml C5a for E2-WT, demonstrating that complement activation is weak for non-PEGylated E2 nanoparticles. Conjugation of PEG to these particles moderately increased complement response to give 79.7 ± 6.0 % C4 remaining and 87.6 ± 10.1 ng/ml C5a. Our results demonstrate that PEGylation of the E2 protein nanocapsules can modulate cellular uptake and induce low levels of complement activation, likely via the classical/lectin pathways.
Keywords: pyruvate dehydrogenase, E2, virus-like particle, polyethylene glycol, cell uptake, complement activation
INTRODUCTION
The use of conventional nanoparticulates such as liposomes, polymers, or gold nanoparticles for bionanotechnology applications, including imaging, therapeutics, biosensing, catalysis, and diagnostics, has been increasingly reported.1,2 Although these particles have been extensively investigated, many issues still exist which raise concern for therapeutic and biomedical applications, including toxicity, physical stability, and heterogeneity.3,4
More recently, recombinant protein-based nanoparticles are being explored as alternatives for such applications.3,5–8 One major advantage of protein nanoparticles over conventional polymeric or inorganic particles is the ability to manipulate precisely their architecture, size, surface functionality, and assembly using recombinant tools and protein engineering. Furthermore, due to their highly organized structure, these systems provide monodisperse capsules that can be engineered at their internal, external, or intersubunit interfaces at specific locations, allowing for high control over surface properties.9 Protein-based particles have included virus-like particles (e.g. Cowpea mosaic virus, adenovirus)10–12 and particles of non-viral origin (e.g. ferritin cages, heat shock protein).3
However, applicability of these protein complex systems for certain purposes may be affected by the biological responses towards them. For example, particles of viral origin may be particularly prone to immunological recognition and clearance. Attachment of polyethylene glycol (PEG) to nanoparticles has been proposed as enabling evasion by phagocytic immune cells and reducing blood plasma protein (e.g. complement proteins) adsorption.13–16 While studies of cellular immune responses to PEGylated virus-like particles are well-documented,12,17 information on humoral innate immune responses to such particles is less prevalent.
In this work, we created a modified protein nanoparticle scaffold with general applicability for site-directed surface attachment of different classes of molecules, and we examined cellular uptake and complement responses to them. The model protein is the dihydrolipoyl acyltransferase enzyme (E2 subunit) of the pyruvate dehydrogenase multienzyme complex. Prior work has shown that this assembly of enzymes can be distilled down to its structural core scaffold (E2-WT) to yield a 24-nm protein nanocapsule composed of 60 identical self-assembling subunits that maintains its structural integrity over a wide range of temperatures and pH values.18–21 In its folded state, the internal hollow cavity of the E2 particle is accessible by small molecules through its openings at the 5-fold axes of symmetry. Redesigning the internal surface enables encapsulation of fluorescent dyes and small molecule drugs, with applicability toward drug delivery.18,22 Inter-subunit interactions can also be re-engineered to introduce environmentally-triggered behavior not observed in the native scaffold.23,24 The E2 scaffold is of non-viral origin, and therefore does not possess any infectious ability in its native wild-type state, which may render it less immunogenic than virus-like particles. Even so, attaching short peptides to the N-terminus via recombinant methods enables surface display of antigens for vaccine development.5,25,26 In this latter case of surface functionalization, the recombinant approach requires extensive DNA manipulation for each different antigen and is not applicable for larger proteins or non-protein based moieties (e.g., synthetic polymers or carbohydrates).
Our investigation examined uptake of this protein scaffold before and after PEGylation by primary human macrophages and by a human breast cancer cell line. We also investigated the capacity of E2-WT and surface-modified E2 nanocapsules to activate human serum complement proteins in vitro, by assaying for C4 consumption (an early marker in the classical/lectin pathway) and generation of the cleavage fragment, C5a. C5a is an anaphylatoxin released following complement activation via the alternative, classical, or lectin pathways, and is therefore a good indicator of complement terminal pathway activation and inflammation. Complement activation can set the course for subsequent adaptive immunological responses, and such responses to PEGylated icosahedral protein nanoparticles of non-viral origin have not yet to our knowledge been reported. Performing these immune assays with the wild-type and PEGylated E2 particle will allow us to determine in vitro whether any changes in uptake and complement activation are provoked. This characterization will help determine general applicability of the E2 particle for future bionanotechnological applications.
MATERIALS AND METHODS
Materials
All buffer reagents were supplied by EMD Chemicals (Gibbstown, NJ), unless otherwise stated. Dithiothreitol (DTT), isopropyl β-D-thiogalactopyranoside (IPTG), dimethylsulfoxide (DMSO), and potassium phosphate were supplied by Fisher Scientific (Pittsburgh, PA). Phenylmethylsulfonyl fluoride (PMSF) and tris(2-carboxyethyl)-phosphine hydrochloride (TCEP) were supplied by Pierce (Rockford, IL), and sodium azide was from Merck KGaA (Gibbstown, NJ). Restriction endonucleases (BamHI, NdeI), T4 DNA ligase, RNase, DNase, and DpnI were from New England Biolabs (Ipswitch, MA) and PfuUltra High-Fidelity DNA polymerase was from Stratagene (Gibbstown, NJ). Dye molecules Alexa Fluor 532 C5-maleimide (AF532) and Alexa Fluor 488 succinimidyl ester (AF488) were purchased from Invitrogen (Carlsbad, CA). Polyethylene glycol (PEG) reagents used were methyl-PEG24-maleimide (PEG1200-maleimide, Pierce), methyl-PEG-maleimide with PEG range of 1800-2200 Da (PEG2000-maleimide, Nanocs, New York, NY), and methyl-PEG-maleimide with PEG range of 4500–5500 Da (PEG5000-maleimide, Nanocs). Host E. coli strains were DH5α (Zymo Research, Orange, CA) and BL21(DE3) (Stratagene, La Jolla, CA). Normal human serum (NHS) was isolated from whole blood and complement activity was confirmed in standard hemolytic assays (data not shown). The MicroVue C5a EIA kit was purchased from Quidel (San Diego, CA).
Design and construction of surface cysteine mutants
To enable chemical attachment of non-native molecules to the surface of the protein nanocapsules, we selected several potential external sites to perform site-directed mutagenesis to cysteine. The thiol side chain of cysteine is advantageous because it enables chemical conjugation at specific sites constructed via protein engineering. Although the E2-WT subunit has one native cysteine, crystallographic structure and our prior investigations18 demonstrated that this native site is buried within the protein and not accessible for chemical conjugation. Selection criteria included external surface accessibility while retaining interactions likely to preserve the protein’s 60-mer structure and stability.20 Other criteria included locations in loop regions to minimize disruption to secondary structure (alpha-helices, beta strands), and avoidance of the N-terminus and inter-subunit interface interactions.20,23,24,27 To retain molecular accessibility into the interior hollow cavity, we also favored positions that did not occlude the native 5-nm entrances to this cavity. Selection was performed using the quaternary protein structure 1b5s (Protein Data Bank) viewed through PyMOL.28 Based on these criteria, a set of five sites were selected for mutagenesis to cysteine: threonine 219 (T219C), aspartic acid 278 (D278C), glutamic acid 279 (E279C), threonine 334 (T334C), and methionine 338 (M338C).
Site-directed mutagenesis was performed following protocols described in Supporting Information. The starting DNA template encodes the wild-type E2 scaffold (E2-WT) in a pGEM vector and includes amino acids 174-427, as described previously.18 Gene sequences of the mutants were confirmed by DNA sequencing and cloned into the expression vector pET-11a. Plasmids were transformed into E. coli strain BL21(DE3) for protein expression.
Protein expression and purification
Protein expression was performed following previously-reported protocol.18 In summary, cells were grown in Luria-Bertani (LB) media containing 100 Mg/ml ampicillin and induced at OD600 0.7–0.9 with 1 mM IPTG at 37 °C for 3.5 hours. For scale-up, purification, and analysis, we selected one of the mutants (E279C) based on criteria described in Results and Discussion. Purification was performed as previously described18 and included using a Q-Sepharose anion exchange column and a Superose 6 size exclusion column (GE Healthcare), with the exception that 1 mM DTT was added to all of the lysis, purification, and storage buffers to prevent cross-linking of protein nanocapsules in solution. Proteins were stored at 4 °C for short-term or −80 °C for long-term storage in 50 mM potassium phosphate (pH 7.4) and 100 mM NaCl (herein referred to as “phosphate buffer”) containing 0.02 % sodium azide, 1 mM DTT, and 5 mM EDTA. Protein concentrations were quantified using a Micro-BCA kit (Pierce).
Confirmation of structural assembly and thermostability
Stability and structural analyses of the protein nanocapsules were carried out as previously described.18 Briefly, secondary protein folding was characterized with far-UV circular dichroism (CD) on a Jasco 810 spectropolarimeter (Jasco, Easton, MD). The thermostability of the five mutants was screened by heating the soluble fractions of freshly lysed cells to elevated temperatures ranging between 37 – 95 °C, followed by centrifugation to remove insoluble proteins. Thermostabilities of the purified E279C mutant were quantified by measuring molar ellipticity at 222 nm from 40 °C to 95 °C. The onset of unfolding (To) and midpoint of unfolding (Tm) temperatures were determined using a nonlinear least squares fitting program in MATLAB according to the method by Greenfield.29 Protein molecular weights were confirmed via electrospray ionization mass spectrometry (ESI-MS, Micromass LCT Premier Mass Spectrometer, Waters) using E2-WT and fluorotriazines as standards for calibration. Particle size was determined using dynamic light scattering (DLS) with a Zetasizer Nano ZS instrument (Malvern, Westborough, MA). Because the E2 assembly comprises 60 identical subunits which are associated by non-covalent interactions, ESI-MS data yields the molecular weight of a single subunit while the milder conditions of DLS measure the size of the particle in solution. Molecular weight (ESI-MS), Tm, To, and particle size are presented as an average ± standard deviation of three separately prepared samples. Transmission electron microscopy (TEM, Philips CM20) was performed on protein samples stained with 2% uranyl acetate on carbon-coated Formvar grids.
Functionalization with polyethylene glycol (PEG) and dye molecules
Stock PEG1200-maleimide, PEG2000-maleimide and PEG5000-maleimide solutions were prepared in DMSO under argon gas, stored dessicated at −20 °C, and brought to room temperature immediately before use. Purified E279C protein was incubated with 8.5 molar excess of TCEP for 40 minutes at room temperature, and PEG-maleimide was added to E279C (1 mg/ml) at a 20-fold molar excess with respect to the E2 subunit and reacted for 2 hours at room temperature with agitation. DTT, TCEP, and excess PEG-maleimide were removed with Zeba desalting columns (40 kDa MWCO, Pierce) using phosphate buffer as the exchange solution. Protocols for attaching dye molecules are described in Supporting Information. AlexaFluor 532 C5-maleimide (AF532M) was conjugated to Cys, and AlexaFluor 488 succinimidyl ester (AF488) reacted with primary amines (lysines) on the protein nanocapsule. The resulting PEG- and dye-conjugated particles were analyzed as described above.
Cellular uptake by macrophages and breast cancer cells
All blood samples were collected in accordance with the guidelines and approval of the University of California Irvine Institutional Review Board. Human monocyte derived macrophages (HMDM) were derived from human peripheral blood monocytes as previously described30 and MDA-MB-231 breast cancer cells (ATCC, Manassas, VA) were cultured as described previously.22 HMDM (harvested using Versene-EDTA) were resuspended at 1×106 cells/ml in phagocytosis buffer (RPMI 1640, 25 mM HEPES, 5 mM MgCl2). MDA-MB-231 cells were plated at 2×105 cells/well in Dulbecco’s Modified Eagle’s Medium supplemented with 10 % fetal bovine serum and 1 % L-glutamine. AF488-labeled protein (E2-WT control and PEGylated E279C) were added to the cell suspensions at a range of concentrations (1, 10, or 30 μg/ml) and incubated for 1 hour (for HMDM, n=4) or 4 hours (for MDA-MB-231, n=3) at 37 °C and 5% CO2. Uptake of AF488-labeled protein was analyzed by flow cytometry using the FACSCalibur (Becton Dickinson, San Jose, CA). Data is presented as an average ± standard deviation of mean fluorescence intensity (MFI) value relative to control (E2-WT) MFI values. Statistical significance was determined using a two-tailed Student’s t-test assuming unequal variances.
Complement activation assays
Activation of the classical/lectin complement pathway by E2-WT and PEGylated E279C was measured using a C4-depletion assay as previously described in detail.31,32 Briefly, proteins were diluted to a final concentration of 1 mg/ml in phosphate buffer and incubated with an equal volume of normal human serum (NHS) for 30 minutes at 30 °C to allow for complement activation, prior to incubation of serial dilutions with antibody-sensitized sheep erythrocytes (EA) in the presence of C4-depleted normal guinea pig serum. Phosphate buffer and aggregated human IgG antibody served as the negative and positive controls, respectively. IgG antibody (10 mg/ml) was incubated at 63°C for 15 minutes, diluted to 2 mg/ml, and centrifuged at 13000 RPM for 5 minutes to remove large aggregates. Samples and controls were then quantified by measuring the amount of EA lysis over the range of dilutions, with lower cell lysis corresponding to greater C4 consumption in the initial NHS/E2 mixture. Dilutions were optimized to ensure that sub-maximal lysis was observed in the most dilute samples. Details of data analysis are described in Supporting Information. Data is reported as % C4 remaining relative to phosphate buffer averaged over 10 independent experiments and statistical significance was determined using a two-tailed Student’s t-test assuming unequal variances.
Since the C4-assay measures early activation, it was import to evaluate whether activation progressed to terminal complement pathway activation. Therefore, production of C5a was measured to determine the activation extent of the entire complement pathway and whether the alternative pathway may be involved. Human serum was incubated with both PEGylated and non-functionalized E2 protein using the same procedures as described for the C4 depletion assays with the exception that EDTA was added to final concentration of 20 mM following incubation to arrest further complement activation, and C5a production (and its more stable cleavage product, C5a-des-Arg) was measured using the MicroVue C5a EIA kit following the manufacturer’s instructions. Data is reported as absolute C5a concentration averaged over 3 separate experiments, and statistical significance was determined using a two-tailed Student’s t-test assuming unequal variances.
Levels of background endotoxin in the samples and solutions were determined using a Pyrogent Plus Gel Clot LAL Assay (Lonza, Walkersville, MD). Endotoxin concentrations in protein nanoparticle samples were nearly 80 times lower than the minimum values which activate complement in vitro.33 This indicates that any complement activity observed is not due the presence of endotoxins.
RESULTS AND DISCUSSION
All surface cysteine mutants were expressed, soluble, and thermostable
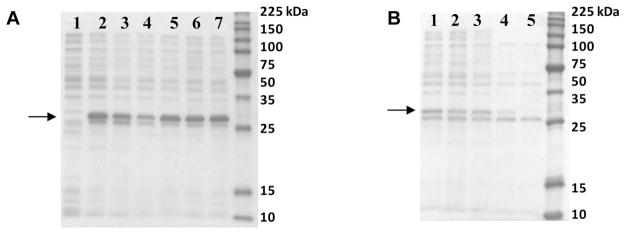
All five mutants with external cysteines (Cys) yielded soluble proteins and were overexpressed at the relatively high levels observed for the wild-type E2 scaffold (E2-WT) (Figure 1A). Our thermostability screen showed that all mutants remain soluble up to approximately 80 °C, which is remarkably stable and similar to the thermostability of E2-WT (Figure 1B). Our previous investigations found that protein scaffold solubility and correct 60-mer assembly is closely coupled;18 therefore, these thermostability results suggest correct quaternary complex assembly of all five Cys mutants.
Figure 1.
SDS-PAGE of mutants with introduced external cysteine residues. A: The soluble fraction of BL21(DE3) cell lysates show that all Cys mutants are expressed and soluble. Lanes: (1) pET-11a (no gene), (2) E2-WT, (3) T219C, (4) E279C, (5) D278C, (6) T334C, and (7) M338C. B: Temperature screens show that all Cys mutants remain soluble and assembled until approximately 80 °C. Shown is one representative data set, for the E279C soluble fraction after heating for 20 min at (1) 37, (2) 60, (3) 70, (4) 80, and (5) 95 °C. The arrows highlight the bands with the E2 subunit.
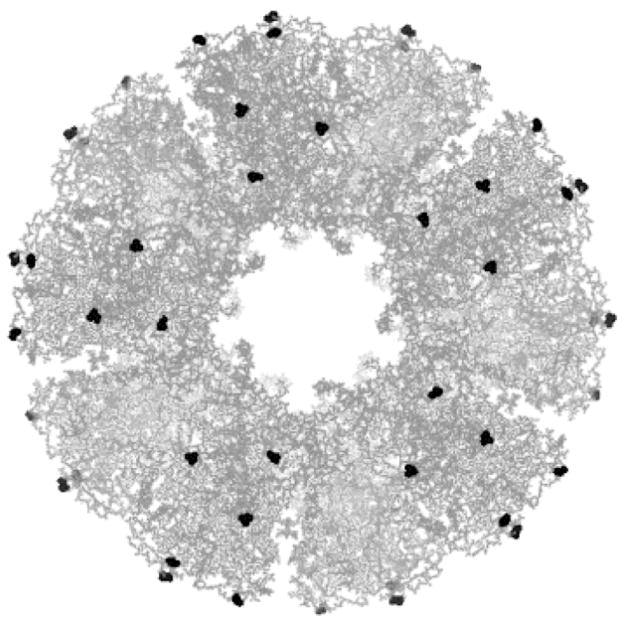
Because protein nanocapsule structure, expression, and thermostability appeared comparable between all mutants, we chose one mutant to investigate further based on examining the PDB crystallographic structure of the E2 scaffold. Position 279C (Figure 2) is surface accessible, does not obstruct the entrance leading to the internal hollow cavity, and gives relatively uniform spacing over the external surface to minimize steric hindrance for chemical coupling. Therefore, E279C was chosen for subsequent studies.
Figure 2.
Fully assembled E2 scaffold with position 279 highlighted in black. This protein assembly consists of 60 identical subunits, each with one cysteine-279. Structure is displayed at the five-fold axis of symmetry and generated by PyMOL (DeLano 2002).
E279C exhibit characteristics similar to E2-WT
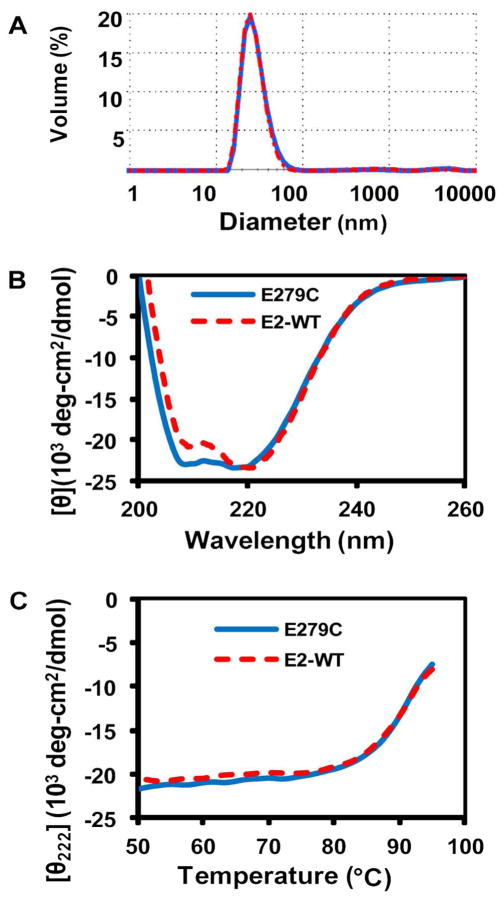
Following purification of E279C, ESI-MS results revealed a single, distinct peak with a molecular weight of 28091 ± 1 Da corresponding to a single subunit of the nanocapsule, (theoretical is 28091 Da), confirming high purity and incorporation of Cys into E279C (Figure S-1). Protein folding, quaternary structure and assembly, and thermostability of the E279C mutant were similar to those of the E2-WT scaffold. DLS analysis of intact 60-mer particles showed a single peak with an average hydrodynamic diameter of 28.0 ± 0.8 nm, compared to the previously-reported value of 26.6 ± 0.6 nm for E2-WT (Figure 3A).18 DLS also revealed that the DTT reducing agent added to the phosphate buffer prevented cross-linking between Cys and subsequent aggregation of the protein; removal of DTT yielded large aggregates > 100 nm.
Figure 3.
Representative characterization data of purified E279C. A: DLS measurement of E2-WT (26.6 ± 0.6 nm, n=3, red dashed line) and E279C (28.0 ± 0.8 nm, n=3, blue solid line). B: Molar ellipticity versus wavelength shows characteristic minima at 208 and 222 nm for E2-WT and E279C, indicative of high alpha-helical structure. C: Thermostability profile (50–95 °C) measuring ellipticity at 222 nm. The calculated average To and Tm for E279C are 80.0 ± 1.5 °C and 89.5 ± 1.9 °C, respectively (n=3).
The CD wavelength scans revealed molar ellipticity minima at 208 and 222 nm, indicative of the E2 scaffold’s characteristic high alpha-helical content (Figure 3B). Additionally, a thermostability scan at 222 nm showed that the E279C protein is highly stable, with an onset of unfolding temperature (To) of 80.0 ± 1.5 °C and a midpoint of unfolding temperature (Tm) of 89.5 ± 1.9 °C, both of which are close to E2-WT values (Figure 3C). Protein solubility (Figure 1) and correct 60-mer assembly have been found to be closely coupled in the E2 complex.18 This demonstrates that introduction of a non-native cysteine residue into the external surface of the E2-WT subunit does not alter its overall conformation, assembly, or thermostability, rendering it a good candidate for further applications and studies.
Nanocapsule surface can be functionalized with both small molecule dyes and polymeric macromolecules
The E2 scaffold is a self-assembled complex of 60 subunits; therefore, a single cysteine mutation theoretically yields 60 accessible thiol sites on the external surface of the protein nanocapsule. To evaluate the chemical accessibility of these thiols, we reacted AlexaFluor 532 C5-maleimide (AF532) with protein and obtained conjugation ratios of 15.2 ± 1.7 and 58.3 ± 12.5 (dye molecules per protein nanocapsule) for E2-WT and E279C, respectively. These values are comparable to those obtained for Cys engineered into the internal hollow cavity of E2.18 Although the E2-WT control indicates a low level of nonspecific interactions or conjugation to AF532,34 the significantly higher conjugation ratio of AF532 to E279C mutant shows that the engineered surface cysteines are indeed accessible for conjugation to near-saturation.
Conjugation of PEG with the E279C particle was confirmed through ESI-MS (Supporting Information). Since PEG1200-maleimide from our source is monodisperse, we expected a distinct peak for PEG1200-E279C at 29330 Da for a single PEG conjugated subunit. The presence of this single peak at 29330 ± 1 Da (Figure S-2) and the absence of the non-functionalized E279C peak at 28091 Da show that Cys-279 had reacted with PEG to near-completion. The specificity of the PEG reagents for E279C thiols was further confirmed by performing the PEGylation procedures with E2-WT; ESI-MS yielded a peak for non-functionalized E2-WT only, demonstrating no detectable reaction with the E2-WT control.
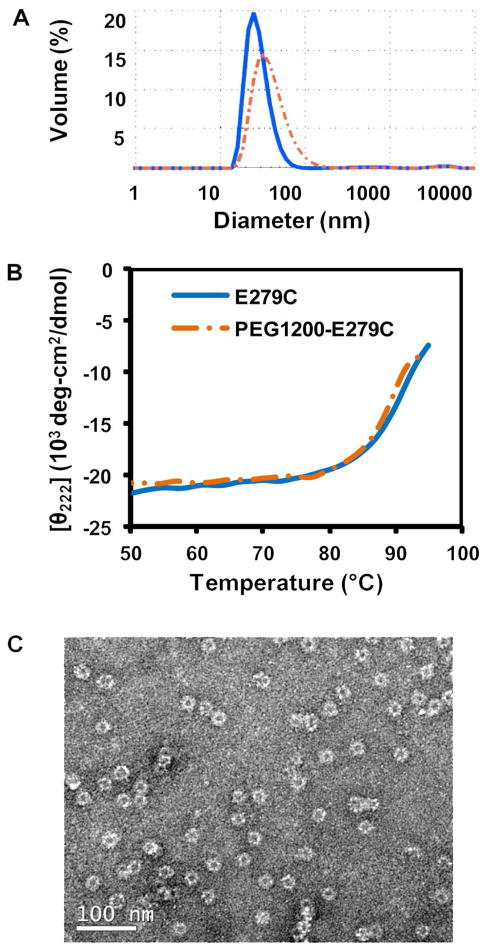
Characterization of PEGylated particles confirmed that intact structure and thermostability had been retained after functionalization (Figure 4). DLS measured the hydrodynamic particle diameters to be 37.3 ± 3.6, 41.6 ± 6.4, and 41.4 ± 4.6 nm for PEG1200-E279C, PEG2000-E279C, and PEG5000-E279C, respectively (Figures 4A and S-3). The average particle diameters are within the expected range, and secondary aggregation peaks are absent. CD data showed that PEGylation does not affect the structural integrity or the high thermostability of the nanocapsules, with the onset of unfolding (To) at 79.7 ± 1.8 °C, and the midpoint of unfolding transition temperature (Tm) at 89.2 ± 1.3 °C for PEG1200-E279C (Figures S-4 and 4B). TEM micrographs of PEG1200-E279C further confirm the presence of intact particles following the conjugation reaction (Figure 4C). A summary of the physicochemical characterization for the E279C and PEG1200-E279C particles can be found in Table S-2 (Supporting Information).
Figure 4.
Representative characterization data of PEGylated E279C particles. A: DLS measurement of particle sizes of E279C (28.0 ± 0.8 nm, n=3, solid blue line) and PEG1200-E279C (37.3 ± 3.6 nm, n=3, orange dashed line). B: Thermostability profile of PEG1200-E279C at 222 nm. The calculated average To and Tm for PEG1200-E279C are 79.7 ± 1.8 °C and 89.2 ± 1.3 °C, respectively (n=3). C: Transmission electron micrograph of PEG1200-E279C particles.
Our strategy leaves solvent-exposed lysines on the PEGylated nanoparticles available for secondary functionalization. These reactive sites can be used to couple a range of molecules to the particle, including fluorescent dyes or therapeutic compounds. We demonstrated the availability of these sites by labeling with AF488 (Supporting Information). These nanocapsules, functionalized with two different chemical entities, were used to investigate in vitro cellular uptake. The ability to introduce multiple functionality to the protein particles is important toward applicability. Our prior work demonstrates that by functionalizing the E2 particles with doxorubicin, we can obtain drug loading, pH-dependent release, and cytotoxicity toward breast cancer cells.22
PEGylation inhibits cellular uptake of nanoparticles
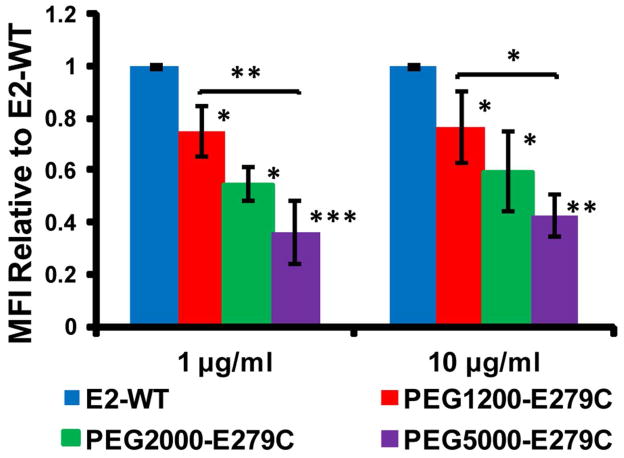
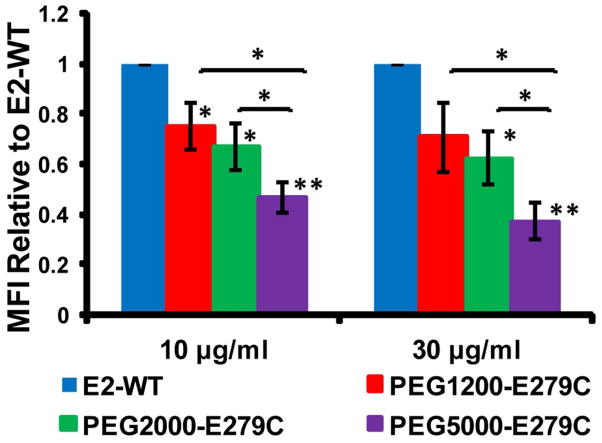
Uptake by HMDM (from multiple donors) and the MDA-MB-231 human breast cancer cell line were significantly lower for PEGylated E279C nanoparticles relative to non-PEGylated (E2-WT control) in all protein concentrations tested (Figures 5, 6, and S-5). For HMDM cells, uptake levels, relative to E2-WT, decreased to 74.8 ± 13.8, 54.6 ± 15.7, and 36.3 ± 8.3 % MFI for PEG1200-E279C, PEG2000-E279C, and PEG5000-E279C, respectively, at a protein concentration of 1 μg/ml. A similar trend was seen at 10 μg/ml (76.3 ± 12.4, 59.4 ± 21.0, and 32.6 ± 8.3 % MFI, respectively) (Figure 5). Observing uptake properties in the absence of serum allows us to determine the intrinsic recognizability of our particle without other contributing factors (e.g. opsonins or antibodies). For MDA-MB-231 cells, relative uptake levels decreased to 70.8 ± 13.9, 62.4 ± 10.5, and 37.4 ± 7.2 % MFI, respectively, for 30 Mg/ml protein, with a similar trend also seen at 10 μg/ml (75.3 ± 9.1, 67.3 ± 9.3, 46.8 ± 5.8 % MFI, respectively) (Figure 6).
Figure 5.
Uptake of E2-WT and PEGylated E279C by human monocyte derived macrophages. Mean fluorescence intensity (MFI) values were normalized to E2-WT MFI. Significant differences in uptake between E2-WT and PEGylated proteins are indicated (*p < 0.05, **p < 0.01, ***p < 0.001; n=4).
Figure 6.
Uptake of E2-WT and PEGylated E279C by MDA-MB-231 cells. Mean fluorescence intensity (MFI) values were normalized to E2-WT MFI. Significant differences in uptake between E2-WT and PEGylated proteins are indicated (*p < 0.05, **p < 0.01; n=3).
PEGylation of biomaterials and nanoparticles has been utilized as a strategy to evade immune recognition, improve solubility, and increase in vivo half-life.15,16,35–38 For nanoparticles similar in size to ours, reduced in vitro cellular uptake has been observed following PEG functionalization.16,39,40 Our data is consistent with this and also shows the trend of reduced uptake with increasing PEG chain length, with PEG5000-E279C particles showing significantly less uptake compared to PEG1200-E279C over the particle concentrations examined. This dependence on PEG content was also observed in previous studies with gold nanoshells41 and virus-like particles,16 and showed that reduced uptake was greatly diminished as the PEG molecular weight was further increased from 5 kDa to 20 kDa.41
Our observation of reduced cellular uptake with increasing PEG content may have implications for modulating the cellular recognition of our particle in vivo. This effect is possibly due to inhibition of specific receptor-mediated interactions, which has been proposed for other protein particle systems.16,40 Explanations in the studies mentioned have suggested shifting of polymer architecture from mushroom-like to brush-like or increased effective shielded area.16,41 Vonarbourg et al. showed that the ability of PEG to mask particles was increased as particle diameter decreased from 100 to 20 nm, which is similar to the size of our protein nanocapsule.42 Decreased cellular uptake is a property that could decrease immunogenicity and potentially allow longer in vivo circulation times of our particle. Conversely, decreased cellular recognition may also reduce efficacy for applications such as drug delivery, where it may be beneficial to evade the phagocytic immune cells while remaining recognizable by target cells. Previous studies have overcome this barrier by affinity ligand functionalization of the PEG free ends.36,40,43
PEGylation of protein nanocapsule activates complement
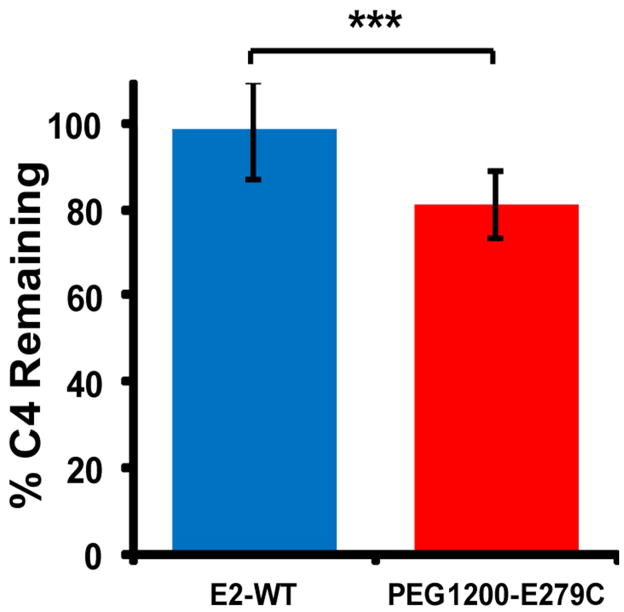
We characterized the capacity of our protein nanocapsules to activate complement, an innate immune mechanism that initiates inflammation and recognizes and clears foreign pathogenic material. Following protein nanoparticle incubation with serum (as a source of complement proteins), a higher percentage of C4 remaining in the sample corresponds to a lower degree of classical/lectin complement pathway activation, with no activation when 100 % of C4 remains. Unmodified E2-WT promotes only a small degree of classical/lectin complement activation, with 97.6 ± 10.8 % C4 remaining after incubation with human serum, relative to phosphate buffer alone (Figure 7). In comparison, PEG1200-E279C consistently exhibited an enhanced activation of complement relative to non-PEGylated E2-WT within individual assays, with 79.7 ± 6.0 % C4 remaining. Heat-aggregated IgG (positive control) gave near-complete consumption of C4 over all the dilutions tested, showing full activation. Observed inter-assay variability may be due to donor variability in sheep erythrocytes and guinea pig serum. Our data indicate that PEGylation of the E279C nanocapsules moderately increases classical/lectin complement activation.
Figure 7.
In vitro classical/lectin complement activation by E2-WT and PEG1200-E279C, as measured by C4 depletion in human serum. Higher percent C4 remaining corresponds to lower activation and is relative to phosphate buffer (negative control). Heat-aggregated human IgG (positive control) showed near-complete C4 consumption. Significant differences in activation are indicated (***p < 0.001).
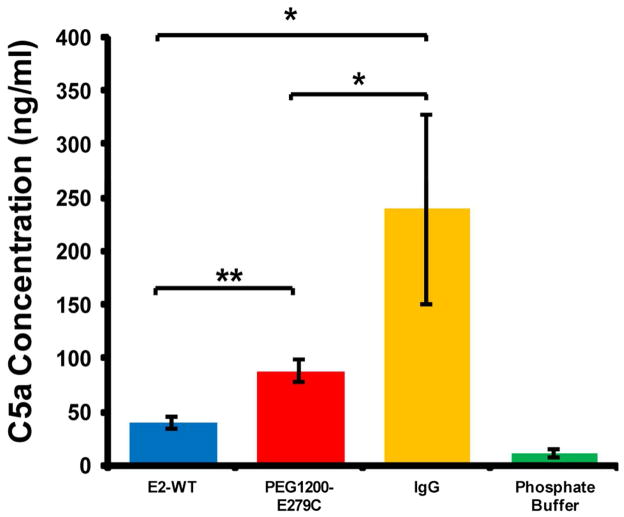
Since C4 is a protein involved early in the classical/lectin pathway, it is not a definitive marker of full pathway activation, nor does it provide information about possible involvement of the alternative pathway. We therefore additionally measured C5a production in human serum following incubation with the E2 protein nanoparticles, with a higher amount of C5a produced in vitro corresponding to a greater degree of complement activation. C5a serum concentrations resulting from incubation with E2-WT and PEG1200-E279C averaged 40.1 ± 6.0 ng/ml and 87.6 ± 10.1 ng/ml, respectively (Figure 8). Both E2-WT and PEG1200-E279C produced greater amounts of C5a in human serum relative to the phosphate buffer background (11.2 ± 3.3 ng/ml) and significantly less than the heat-aggregated IgG positive control (239.0 ± 89.2 ng/ml).
Figure 8.
Measurement of C5a concentration in human serum following incubation with E2-WT, PEGylated E279C, phosphate buffer, and aggregated human IgG. A higher amount of C5a indicates greater complement activation. Significant differences in concentration are indicated (*p < 0.05, **p < 0.01). E2-WT, PEG1200-E279C, and IgG all showed significantly greater C5a concentrations relative to phosphate buffer (p < 0.001).
In both the C4 depletion and C5a production assay results, the PEGylated E279C mutant consistently induced more complement activity compared to E2-WT. Our data indicates that the unmodified protein nanoparticle is a weak activator of complement relative to a known strong classical pathway activator (aggregated IgG). Measurements of background endotoxin levels in our serum/protein solutions was determined to be approximately 80 times lower than the minimum amount reported to activate complement, indicating that any observed complement activation is due to the properties of the sample itself and not contaminating levels of endotoxin.33 Furthermore, free PEG1200-maleimide at levels comparable to that of residual PEG1200-maleimide after PEG1200-E279C purification exhibited no complement activation, further indicating that the activation observed is due to the attachment of PEG on the protein nanoparticle. PEGylation of these particles moderately activate the classical/lectin pathway, and given that relative amounts of C5a produced are similar to relative consumption amounts observed in the C4 assays, this suggests that the alternative pathway does not substantially contribute to or amplify complement activation by these particles.
The adsorption of proteins on nanomaterial surfaces can have many adverse in vivo effects such as opsonization of the nanomaterial or triggering inflammation, and it has been suggested that PEGylation can minimize such effects.13,14,37,42,44 While nanoparticle conjugation with PEG and reduction of PEGylated nanoparticle size has been observed to reduce complement activation in vitro, it does not eliminate it completely.37,45–47 In contrast, the rapid clearance of PEGylated nanoparticles in vivo and PEG-mediated complement activation in vitro and in vivo have also been reported previously,37,44,47–50 although the mechanism of activation is still unclear. One explanation for the apparent contradictory observations in these prior investigations may be that PEG is masking the complement activation properties of the particle itself, but is introducing a lower, yet non-negligible, amount of polymer-mediated complement activation. This would in part explain why we have observed complement activation after PEGylation of a particle that intrinsically is not a strong activator, an observation also made with liposomal systems.50 However, since only modest activation due to PEG relative to the positive control was observed, and also because reported concentrations of protein particles used for in vivo applications are typically much lower than those which activate complement in our in vitro assay, 25,35,38,40 we expect that the complement activation is likely to be less significant when in vivo conditions are examined.
CONCLUSIONS
We have engineered a protein nanocapsule (E279C), based on the E2 scaffold, which can be functionalized on the surface with various molecules. This scaffold properly assembles and remains highly thermostable, even following conjugation with synthetic polymers. We found that introducing PEG on the surface of this scaffold decreased cellular uptake by both human monocyte derived macrophages and a human breast cancer cell line. PEGylation of the protein nanocapsule also moderately increased complement activation capacity relative to the unfunctionalized E2-WT scaffold, which itself yields a low level of activation. PEG activates weakly the classical/lectin pathways with little, if any, alternative pathway amplification. Our work demonstrates a generally applicable strategy of generating surface modifications to this protein nanocapsule platform, and that these modifications can alter cellular uptake and complement activation properties.
Supplementary Material
Acknowledgments
We gratefully acknowledge assistance from Dr. Matthew Shindel for TEM images and MATLAB programming to determine thermostability parameters, Prof. Andrea Tenner for helpful discussions, suggestions, and use of laboratory facilities for complement and uptake assays, Dr. Merce Dalmau for initial discussions regarding mutagenesis, Dr. Wytze Van der Veer in the UCI Laser Spectroscopy Facility, and Dr. John Greaves in the UCI Mass Spectrometry Facility. TEM was performed at the UCI Materials Characterization Center. This work was supported by NIH (R21 EB010161-01) and the Department of Defense Breast Cancer Research Program (BC085958).
Footnotes
Supporting Information Available. Details of recombinant nanoparticle construction, dye functionalization, complement analysis, and additional results from mass spectrometry, dynamic light scattering, and flow cytometry. This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.De M, Ghosh PS, Rotello VM. Advanced Materials. 2008;20(22):4225–4241. [Google Scholar]
- 2.Nie S, Xing Y, Kim GJ, Simons JW. Annu Rev Biomed Eng. 2007;9:257–88. doi: 10.1146/annurev.bioeng.9.060906.152025. [DOI] [PubMed] [Google Scholar]
- 3.Lin YH, MaHam A, Tang ZW, Wu H, Wang J. Small. 2009;5(15):1706–1721. doi: 10.1002/smll.200801602. [DOI] [PubMed] [Google Scholar]
- 4.Forrest ML, Aillon KL, Xie YM, El-Gendy N, Berkland CJ. Advanced Drug Delivery Reviews. 2009;61(6):457–466. doi: 10.1016/j.addr.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Domingo GJ, Orru S, Perham RN. Journal of Molecular Biology. 2001;305(2):259–267. doi: 10.1006/jmbi.2000.4311. [DOI] [PubMed] [Google Scholar]
- 6.Koudelka KJ, Manchester M. Curr Opin Chem Biol. 2010;14(6):810–7. doi: 10.1016/j.cbpa.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 7.Steinmetz NF. Nanomedicine-Nanotechnology Biology and Medicine. 2010;6(5):634–641. doi: 10.1016/j.nano.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strable E, Finn MG. Curr Top Microbiol Immunol. 2009;327:1–21. doi: 10.1007/978-3-540-69379-6_1. [DOI] [PubMed] [Google Scholar]
- 9.Uchida M, Klem MT, Allen M, Suci P, Flenniken M, Gillitzer E, Varpness Z, Liepold LO, Young M, Douglas T. Advanced Materials. 2007;19(8):1025–1042. [Google Scholar]
- 10.Douglas T, Young M. Science. 2006;312(5775):873–875. doi: 10.1126/science.1123223. [DOI] [PubMed] [Google Scholar]
- 11.Lee LA, Niu ZW, Wang Q. Nano Research. 2009;2(5):349–364. [Google Scholar]
- 12.Manchester M, Destito G, Schneemann A. Viruses and Nanotechnology. 2009;327:95–122. doi: 10.1007/978-3-540-69379-6_5. [DOI] [PubMed] [Google Scholar]
- 13.Shan XQ, Yuan Y, Liu CS, Tao XY, Sheng Y, Xu F. Biomedical Microdevices. 2009;11(6):1187–1194. doi: 10.1007/s10544-009-9336-2. [DOI] [PubMed] [Google Scholar]
- 14.Sant S, Poulin S, Hildgen P. J Biomed Mater Res A. 2008;87(4):885–95. doi: 10.1002/jbm.a.31800. [DOI] [PubMed] [Google Scholar]
- 15.Dobrovolskaia MA, McNeil SE. Nat Nanotechnol. 2007;2(8):469–78. doi: 10.1038/nnano.2007.223. [DOI] [PubMed] [Google Scholar]
- 16.Steinmetz NF, Manchester M. Biomacromolecules. 2009;10(4):784–92. doi: 10.1021/bm8012742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ludwig C, Wagner R. Current Opinion in Biotechnology. 2007;18(6):537–545. doi: 10.1016/j.copbio.2007.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dalmau M, Lim S, Chen HC, Ruiz C, Wang SW. Biotechnol Bioeng. 2008;101(4):654–64. doi: 10.1002/bit.21988. [DOI] [PubMed] [Google Scholar]
- 19.Domingo GJ, Chauhan HJ, Lessard IAD, Fuller C, Perham RN. European Journal of Biochemistry. 1999;266(3):1136–1146. doi: 10.1046/j.1432-1327.1999.00966.x. [DOI] [PubMed] [Google Scholar]
- 20.Izard T, Aevarsson A, Allen MD, Westphal AH, Perham RN, de Kok A, Hol WG. Proc Natl Acad Sci U S A. 1999;96(4):1240–5. doi: 10.1073/pnas.96.4.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Milne JLS, Shi D, Rosenthal PB, Sunshine JS, Domingo GJ, Wu XW, Brooks BR, Perham RN, Henderson R, Subramaniam S. Embo Journal. 2002;21(21):5587–5598. doi: 10.1093/emboj/cdf574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ren DM, Kratz F, Wang SW. Small. 2011;7(8):1051–1060. doi: 10.1002/smll.201002242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dalmau M, Lim S, Wang SW. Nano Lett. 2009;9(1):160–6. doi: 10.1021/nl8027069. [DOI] [PubMed] [Google Scholar]
- 24.Dalmau M, Lim S, Wang SW. Biomacromolecules. 2009;10(12):3199–206. doi: 10.1021/bm900674v. [DOI] [PubMed] [Google Scholar]
- 25.Caivano A, Doria-Rose NA, Buelow B, Sartorius R, Trovato M, D’Apice L, Domingo GJ, Sutton WF, Haigwood NL, De Berardinis P. Virology. 2010;407(2):296–305. doi: 10.1016/j.virol.2010.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Domingo GJ, Caivano A, Sartorius R, Barba P, Backstrom M, Piatier-Tonneau D, Guardiola J, De Berardinis P, Perham RN. Vaccine. 2003;21(13–14):1502–9. doi: 10.1016/s0264-410x(02)00664-3. [DOI] [PubMed] [Google Scholar]
- 27.Zhou ZH, Liao WC, Cheng RH, Lawson JE, McCarthy DB, Reed LJ, Stoops JK. Journal of Biological Chemistry. 2001;276(24):21704–21713. doi: 10.1074/jbc.M101765200. [DOI] [PubMed] [Google Scholar]
- 28.DeLano W. DeLano Scientific. Palo Alto, USA: 2002. [Google Scholar]
- 29.Greenfield N. Nat Protocols. 2006;1(6):2527–35. doi: 10.1038/nprot.2006.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fraser DA, Tenner AJ. J Immunol. 2010;185(7):3932–9. doi: 10.4049/jimmunol.1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cribbs DH, Velazquez P, Soreghan B, Glabe CG, Tenner AJ. NeuroReport. 1997;8(16):3457–3462. doi: 10.1097/00001756-199711100-00009. [DOI] [PubMed] [Google Scholar]
- 32.Webster SD, Tenner AJ, Poulos TL, Cribbs DH. Neurobiol Aging. 1999;20(3):297–304. doi: 10.1016/s0197-4580(99)00020-2. [DOI] [PubMed] [Google Scholar]
- 33.Kaca W, Roth R. Biochimica Et Biophysica Acta-General Subjects. 1995;1245(1):49–56. doi: 10.1016/0304-4165(95)00077-o. [DOI] [PubMed] [Google Scholar]
- 34.Smyth DG, Nagamatsu A, Fruton JS. Journal of the American Chemical Society. 1960;82(17):4600–4604. [Google Scholar]
- 35.De Geest B, Snoeys J, Van Linthout S, Lievens J, Collen D. Hum Gene Ther. 2005;16(12):1439–51. doi: 10.1089/hum.2005.16.1439. [DOI] [PubMed] [Google Scholar]
- 36.Eto Y, Gao JQ, Sekiguchi F, Kurachi S, Katayama K, Maeda M, Kawasaki K, Mizuguchi H, Hayakawa T, Tsutsumi Y, Mayumi T, Nakagawa S. Journal of Gene Medicine. 2005;7(5):604–612. doi: 10.1002/jgm.699. [DOI] [PubMed] [Google Scholar]
- 37.Mosqueira VCF, Legrand P, Gulik A, Bourdon O, Gref R, Labarre D, Barratt G. Biomaterials. 2001;22:2967–79. doi: 10.1016/s0142-9612(01)00043-6. [DOI] [PubMed] [Google Scholar]
- 38.Raja KS, Wang Q, Gonzalez MJ, Manchester M, Johnson JE, Finn MG. Biomacromolecules. 2003;4(3):472–6. doi: 10.1021/bm025740+. [DOI] [PubMed] [Google Scholar]
- 39.Kelf TA, Sreenivasan VK, Sun J, Kim EJ, Goldys EM, Zvyagin AV. Nanotechnology. 2010;21(28):285105. doi: 10.1088/0957-4484/21/28/285105. [DOI] [PubMed] [Google Scholar]
- 40.Ogawara KI, Rots MG, Kok RJ, Moorlag HE, van Loenen AM, Meijer DKF, Haisma HJ, Molema G. Human Gene Therapy. 2004;15(5):433–443. doi: 10.1089/10430340460745766. [DOI] [PubMed] [Google Scholar]
- 41.Kah JC, Wong KY, Neoh KG, Song JH, Fu JW, Mhaisalkar S, Olivo M, Sheppard CJ. J Drug Target. 2009;17(3):181–93. doi: 10.1080/10611860802582442. [DOI] [PubMed] [Google Scholar]
- 42.Vonarbourg A, Passirani C, Saulnier P, Simard P, Leroux JC, Benoit JP. J Biomed Mater Res A. 2006;78(3):620–8. doi: 10.1002/jbm.a.30711. [DOI] [PubMed] [Google Scholar]
- 43.Jung Y, Park HJ, Kim PH, Lee J, Hyung W, Yang J, Ko H, Sohn JH, Kim JH, Huh YM, Yun CO, Haam S. Journal of Controlled Release. 2007;123(2):164–171. doi: 10.1016/j.jconrel.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 44.Moghimi SM, Szebeni J. Prog Lipid Res. 2003;42(6):463–78. doi: 10.1016/s0163-7827(03)00033-x. [DOI] [PubMed] [Google Scholar]
- 45.Danielsson A, Elgue G, Nilsson BM, Nilsson B, Lambris JD, Totterman TH, Kochanek S, Kreppel F, Essand M. Gene Ther. 2010;17(6):752–62. doi: 10.1038/gt.2010.18. [DOI] [PubMed] [Google Scholar]
- 46.Gbadamosi JK, Hunter AC, Moghimi SM. FEBS Lett. 2002;532(3):338–44. doi: 10.1016/s0014-5793(02)03710-9. [DOI] [PubMed] [Google Scholar]
- 47.Hamad I, Al-Hanbali O, Hunter AC, Rutt KJ, Andresen TL, Moghimi SM. ACS Nano. 2010;4(11):6629–38. doi: 10.1021/nn101990a. [DOI] [PubMed] [Google Scholar]
- 48.Arima Y, Toda M, Iwata H. Biomaterials. 2008;29(5):551–60. doi: 10.1016/j.biomaterials.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 49.Moghimi SM, Andersen AJ, Hashemi SH, Lettiero B, Ahmadvand D, Hunter AC, Andresen TL, Hamad I, Szebeni J. J Control Release. 2010;146(2):175–81. doi: 10.1016/j.jconrel.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 50.Szebeni J, Baranyi L, Savay S, Milosevits J, Bunger R, Laverman P, Metselaar JM, Storm G, Chanan-Khan A, Liebes L, Muggia FM, Cohen R, Barenholz Y, Alving CR. Journal of Liposome Research. 2002;12(1–2):165–172. doi: 10.1081/lpr-120004790. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.