Abstract
Background
Congenital malformations involving the Müllerian ducts are observed in around 5% of infertile women. Complete aplasia of the uterus, cervix, and upper vagina, also termed Müllerian aplasia or Mayer–Rokitansky–Kuster–Hauser (MRKH) syndrome, occurs with an incidence of around 1 in 4500 female births, and occurs in both isolated and syndromic forms. Previous reports have suggested that a proportion of cases, especially syndromic cases, are caused by variation in copy number at different genomic loci.
Methods
In order to obtain an overview of the contribution of copy number variation to both isolated and syndromic forms of Müllerian aplasia, copy number assays were performed in a series of 63 cases, of which 25 were syndromic and 38 isolated.
Results
A high incidence (9/63, 14%) of recurrent copy number variants in this cohort is reported here. These comprised four cases of microdeletion at 16p11.2, an autism susceptibility locus not previously associated with Müllerian aplasia, four cases of microdeletion at 17q12, and one case of a distal 22q11.2 microdeletion. Microdeletions at 16p11.2 and 17q12 were found in 4/38 (10.5%) cases with isolated Müllerian aplasia, and at 16p11.2, 17q12 and 22q11.2 (distal) in 5/25 cases (20%) with syndromic Müllerian aplasia.
Conclusion
The finding of microdeletion at 16p11.2 in 2/38 (5%) of isolated and 2/25 (8%) of syndromic cases suggests a significant contribution of this copy number variant alone to the pathogenesis of Müllerian aplasia. Overall, the high incidence of recurrent copy number variants in all forms of Müllerian aplasia has implications for the understanding of the aetiopathogenesis of the condition, and for genetic counselling in families affected by it.
INTRODUCTION
The incidence of congenital malformations involving the Müllerian ducts in the general population is ~5 per 1000, and for infertile women it is more frequent at 35–63 per 1000.1,2 Among the most common uterine anomalies are uterine duplications, uterine indentations, and partial uterine aplasias, which occur as a result of incomplete Müllerian and Wollfian duct fusions during development. The Mayer–Rokitanski–Küster–Hauser (MRKH) syndrome (OMIM 277000), occurring in 1 of 4500 live female births, is the most severe type where complete aplasia of the uterus, cervix, and upper vagina is found, leading to failure to menstruate and infertility, despite normal secondary sexual characteristics.3–5
The different MRKH subtypes are clinically classified into type I (typical or isolated) patients with normally developed fallopian tubes, ovaries, and urinary tract, and type II (atypical) patients, with Fallopian or ovarian abnormalities, and additional malformations, which typically involve the urinary tract and spine.6,7 The acronym MURCS (Müllerian–renal–cervicothoracic somite abnormalities, OMIM 601076) applies to some of these cases. Craniofacial (dysmorphism, microtia) and cardiovascular malformations, and learning difficulties/mental retardation, may also be associated.8 The reproductive and psychosocial consequences of the disorder are severe; despite this, from the aetiological perspective, it has been relatively poorly studied. Nonetheless, over the last decade, evidence has accumulated to suggest that genetic factors may be important.
Notwithstanding that patients with Müllerian aplasia are usually precluded from reproducing, familial clustering of the disorder has been reported, with apparently autosomal dominant inheritance.9,10 For example, in the article by Shokeir,10 figures 6 and 9 show Müllerian aplasia segregating in apparently autosomal dominant fashion in two or more generations. In a small proportion of cases, a specific genetic aetiology has been identified. Müllerian aplasia with hyper- and rogenism and renal malformations (OMIM 158330) is due to mutations in WNT4.11 Beside single gene involvement, there is evidence implicating copy number variation in the pathogenesis of Müllerian aplasia. Case reports in the literature have linked MURCS to microdeletions at 17q1212 and 22q11.213; in one small series of 14 cases, both of these loci were identified and two more suggested, a microduplication at 1q21.1 and microdeletion at Xq21.31.14 Müllerian aplasia has been reported in association with thrombocytopenia–absent radius syndrome, due to microdeletion at chromosome 1q21.1.15
Typically, syndromes due to recurrent copy number variants exhibit wide phenotypic variability, a fact which has been recognised since the early descriptions of the 22q11.2 microdeletion syndrome16 and which continues to be recognised in the new syndromes which have been described since the advent of high resolution array based studies (reviewed in Mefford and Eichler17). A further example of this variability is given by the 16p11.2 microdeletion syndrome. This was initially described in cohorts of patients with autism.18,19 Later, the recognition was made that patients with 16p11.2 microdeletions have a more complex phenotype than those patients with presumed multifactorial forms of autism spectrum disorder, including dysmorphism, congenital anomalies, growth disturbance, motor delay, and epilepsy.20,21 Most recently, a strong association between 16p11.2 microdeletions and obesity was reported, especially where cognitive disability was also present.22 A detailed explanation of how such pronounced phenotypic variability can arise from copy number variation at a single locus is lacking; environmental factors, epigenetic changes, and ‘second hits’23 may all play a part. For the present, it is likely that the full range of phenotypic variability of these disorders has yet to be explored, and that more associations may be identified through the study of different patient cohorts.
We report copy number analysis of DNA samples from a cohort of 63 individuals with Müllerian aplasia, of which 38 were classified as isolated or typical Müllerian aplasia (60 3%) and 25 were classified as syndromic or atypical Müllerian aplasia (39 7%), from which 11 had a diagnosis of MURCS association.
METHODS
Patient samples
Patient samples and phenotypic data were collected by the Department of Obstetrics and Gynecology, Erlangen, Germany. The project was approved by the Ethics Review Board of Friedrich-Alexander-University, Erlangen-Nuremberg, Germany. Informed consent for genetic studies was obtained in each case.
Copy number assay
Copy number analysis was performed on one of two platforms: Agilent 244K oligonucleotide array, as described,24 and the Affymetrix SNP 6.0 genotyping platform, as described.25 An initial pilot study was performed using the Agilent 244K array; subsequently, the Affymetrix SNP 6.0 array was used in order to facilitate comparison with a cohort of 7366 population controls, which had been assayed using the same platform. These control individuals were of European ancestry and were recruited from the WTCCC2 and GAIN (Genetic Association Information Network) consortia as described.25
Affymetrix SNP 6.0 data were analysed using Affymetrix powertools and Birdsuite software. Copy number variant calls made using Agilent software were converted to genotyping calls in order to enable comparison with the control cohort. We used a cut-off for calling copy number variants of 200 Kb.
Validation of results
Quantitative PCR (qPCR) on patient DNA was used to confirm deletions or amplifications at specific genomic loci. Results were normalised to ACTB DNA levels, which served as a control. Primer and probe sequences are shown in table 1. Experiments were performed as described earlier26 on the ABI PRISM 7900HT Sequence Detection System (Applied Biosystems, Foster City, California, USA). Parental samples were not available for analysis; thus, information about whether a given copy number variant arose de novo was not available.
Table 1.
Primer and probe sequences used for quantitative PCR*
| Region | Primer/probe | Sequence |
|---|---|---|
| 1q21.1 | Pex11b fw | CACGCTGATGTGCTTGTGATG |
| Pex11b rev | TTTGACAATGATGAGGCCTGAA | |
| Pex11b probe | TGCGGCCTGCACTGGCCC | |
| 1q21.1 | CD160 fw | TTGAACCCAGGAGTCCACAAG |
| CD160 rev | ACAGACGGCGGGAAACTCTT | |
| CD160 probe | CCAGGACCGCCTCCGAAGGTG | |
| 2q11.2 | NCAPH fw | GGGCGGCCTCCTCCTT |
| NCAPH rev | TTCCAGGAAAACCACCATTTTAA | |
| NCAPH probe | CTCCTAAAGCGTGCTCGGTGTCTCTCC | |
| 2q11.2 | Kiaa1310 fw | GGTGCTGGCCAAGCAAGT |
| Kiaa1310 rev | GCTCACCGACCCCAAGGT | |
| Kiaa1310 probe | TACTGTCTGTCCACGCGAGGTCTTTCTG | |
| 16p11.2 | MAZ fw | GGCTCAAAGGGCCCAATAG |
| MAZ rev | CCTCCCTGTGCCCAGAAGT | |
| MAZ probe | AGGGATGCCCATGTACCACTCAGGC | |
| 16p11.2 | Tmem219 fw | GCACCCCACTTGGAAGCA |
| Tmem219 rev | TGAGGCTCGCGGACTTTAA | |
| Tmem219 probe | TCAGATCTTGGCCCTACCCCTCCTGT | |
| 17q12 | LHX1 fw | CATGTGCCTGGGAAGAAAGG |
| LHX1 rev | TGCCCTGTCTCTTCCAAGCT | |
| LHX1 probe | AGCCTGACTCGGCCCAGAAGCC | |
| 17q12 | Dusp14 fw | TCTGGTGCATGGATAGAAGCAA |
| Dusp14 rev | CCACCGCAGAGAAAGACTCAA | |
| Dusp14 probe | TGACTTTCAGCGATGCCAAGTGTCCA | |
| 2q11.2 | Gnaz fw | GCCTGGTAGAGAGGTCTGTCTTG |
| Gnaz rev | GGGAAATCACTTGGGCAGAA | |
| Gnaz probe | ACAGCTGAGCCCCTGACCGGC | |
| 2q11.2 | BCR fw | GATCCTGCACCCGAACAAA |
| BCR rev | CCAATTCCATTCCAAACACTAACA | |
| BCR probe | CCATCCCCTCCTCCTTCCTGAATGC | |
| 2p24.124.3 | Vsnl1 fw | CTCAGAGAGAAGTCACCCATCAAC |
| Vsnl1 rev | AATGAGAGGGTGTGCAAGTGAA | |
| Vsnl1 probe | CCCTGCCTGGGAAGCTGGCC | |
| 2p24.124.3 | GDF7 fw | GATGGGACTTTTGGCTTGCTAA |
| GDF7 rev | CAGAGCAGCGGACGTCTTC | |
| GDF7 probe | CCAAAGCTCGGTTCGGATATCCCG | |
| 15q21.1 | 15q21 fw | GCTGATTATAAACGGAGCCATATTC |
| h15q21 rev | CCTGGCTGCTTTTGACATCAT | |
| h15q21 probe | TTGAGACCAGGCCTTCACTTTCTCGGAA | |
| 18q23 | hSall3 fw | GGCTTGGGCAAGTGAAGGA |
| hSall3 rev | TGGCCACGCAGAGAATGTT | |
| hSall3-probe | AGACCCGGACCCTTCGAGCTCCA | |
| control | Beta-actin fw | AGGTGCACAGTAGGTCTGAACAGA |
| Beta-Actin rev | AAGTGCAAAGAACACGGCTAAGT | |
| Beta-Actin probe | TCCCCATCCCAAGACCCCAGC |
Probes were dual labelled with FAM (5′) and TAMRA (3′ ).
RESULTS
Our study demonstrated a strikingly high incidence of recurrent copy number variants, with nine (14%) of the 63 samples studied having a copy number variant of this type (table 2 and figures 1 and 2). These copy number variants were confirmed by qPCR (figure 3). For isolated Müllerian aplasia, this applied to 4/38 (10 5%) patients, and for syndromic Müllerian aplasia, to 5/25 (20%) patients. The results are summarised in tables 2–4. In isolated Müllerian aplasia, we identified two microdeletions at 17q12, and two at 16p11.2. Of the five syndromic cases, two had microdeletions at 17q12, two had microdeletions at 16p11.2, and one had a ‘distal’ 22q11.2 microdeletion. A sixth case had a microduplication at 2q11.2, a locus recently reported to represent a novel recurrent copy number variant, though as yet phenotypic data for this new disorder have not been provided.27 Interpretation in this case is made additionally difficult by the presence of a large (4.6 Mb) deletion at 2p24.3. Either, or conceivably both, of these imbalances may have contributed to the phenotype in this case. There were two instances of microdeletion at 16p11.2 in the control cohort; no instances of microdeletion at 17q12 and 22q11.2 (distal) were identified.
Table 2.
Genomic disorder type copy number variants in isolated and syndromic Müllerian aplasia
| Case | Locus | Size | Copy number |
Co-ordinates (Hg18) | Phenotype | Platform/confirmation |
|---|---|---|---|---|---|---|
| 1 | 16p11.2 | 0.55 Mb | Del | 16:29487535–30085308 | MURCS | Agilent, Affymetrix 6.0, qPCR |
| 2 | 16p11.2 | 0.60 Mb | Del | 16:29561000–3010700 | MA | Agilent, Affymetrix 6.0, qPCR |
| 3 | 16p11.2 | 0.55 Mb | Del | 16:29560500–30106808 | MURCS | Agilent, qPCR |
| 4 | 16p11.2 | 0.55 Mb | Del | 16:29487535–30085308 | MA | Agilent, qPCR |
| 5 | 17q12 | 1.4 Mb | Del | 17:31889000–33322000 | MA | Agilent, Affymetrix 6.0, qPCR |
| 6 | 17q12 | 1.4 Mb | Del | 17:31889000–33322000 | MURCS | Agilent, Affymetrix 6.0, qPCR |
| 7 | 17q12 | 1.4 Mb | Del | 17:31889297–33322972 | MURCS | Agilent, qPCR |
| 8 | 17q12 | 1.4 | Mb | Del 17:31889000–33322000 | MURCS | Affymetrix 6.0, qPCR |
| 9 | 22q11.2 | 0.39 Mb | Del | 22:21588000–21973000 | MURCS | Agilent, Affymetrix 6.0, qPCR |
| 10 | 2q11.2 | 1.30 Mb | Dup | 2:96052862–97390919 | MURCS | Agilent, qPCR |
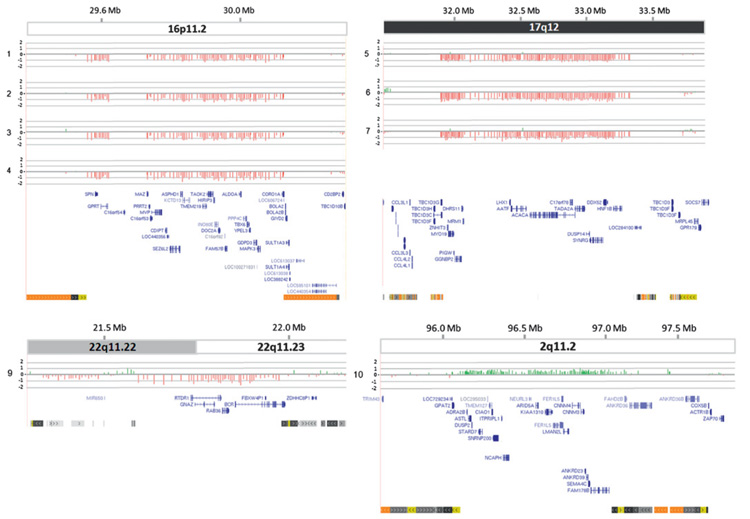
Figure 1.
Copy number variants associated with genomic disorders in patients with isolated and syndromic Müllerian aplasia. Each figure presents microarray data for one of four genomic loci, at 16p11.2, 17q12, 22q11.2 (distal) and 2q11.2. Presented from top down are: scale showing distance from tip of ‘p’ arm in megabases (UCSC genome browser March 2006 Hg18/NCBI Build 36); chromosome ideogram showing chromosome band; array comparative genomic hybridisation (CGH) result showing value for each probe (log2 ratio), where zero corresponds to diploid copy number, −1 to a heterozygous deletion and +0.58 to a heterozygous duplication. Patient ID is shown to the left of each graphic. DNA from case 8 was assayed on the Affymetrix 6.0 platform only; data for this case are given in figure 2. Gene content of the region is shown below. For simplicity of presentation, alternately spliced isoforms are not shown. Finally, segmental duplications of >1 Kb of non-repeat masked sequence are shown. Light to dark grey, 90–98% similar; light to dark yellow, 98–99% similar; light to dark orange, >99% similar; are indicated.
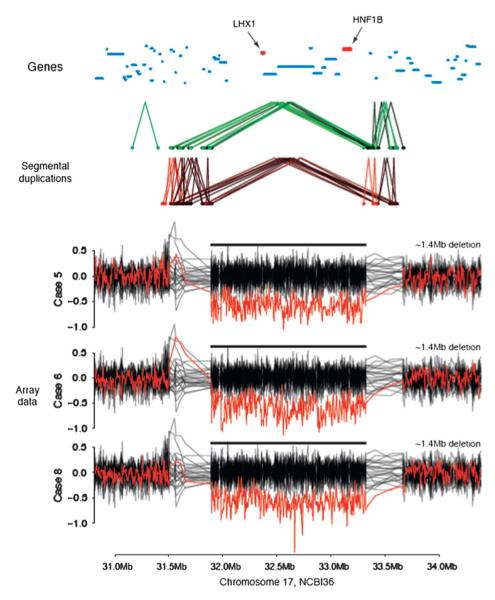
Figure 2.
Affymetrix 6.0 data for three patients with deletions at 17q12 (case 7 was not run on this platform). Log2 ratios for the three samples are highlighted in dark red, with the other samples from the same genotyping plate shown in black. The segmental duplication structure, taken from the UCSC genome browser, is shown. Protein coding genes are indicated by dark blue lines, with LHX1 and HNF1B highlighted in red.
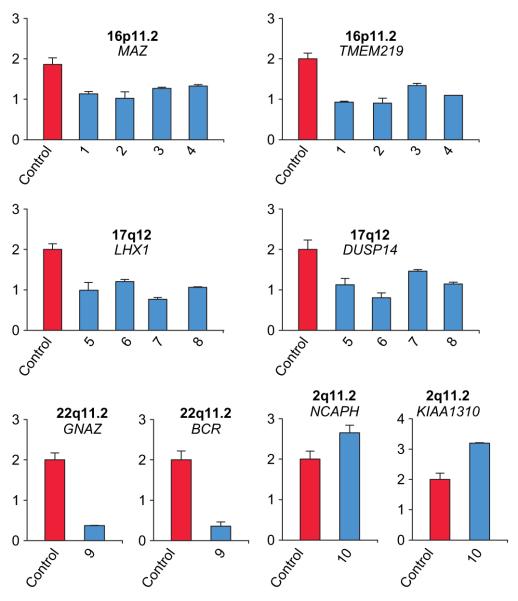
Figure 3.
Quantitative assessment of deletions and amplifications at indicated genomic loci. Quantification was performed on DNA of 4e6 control persons (red bars) and affected individuals (blue bars). Primer and probe sequences were chosen to amplify intronic regions of the indicated genes. Data were normalised to results obtained at the ACTB locus and are presented as relative values.
Table 3.
Summary of the clinical findings in patients with copy number variants of genomic disorder type and isolated or syndromic Müllerian aplasia
| Case | Locus of deletion |
Age (years) |
Classification | Findings | Isolated/ syndromic |
Renal | Cervical/vertebral | Craniofacial | Cognitive | Growth | Other |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 16p11.2 | 30 | V5b C2b U4b A0 MSN Type II/ MURCS |
Blind ending vagina, rudimentary uterus, tubes and ovaries normal | Syndromic | Normal (CT and laparotomy) |
Hypoplasia of the wrist | No clinical indications |
Moderate disturbed psychomotor development |
Normal (no exact measures) |
Epilepsy, moderate bilateral hearing loss |
| 2 | 16p11.2 | 20 | V5b C2b U4b A0 M0 Type I |
Blind ending vagina, rudimentary uterus, tubes and ovaries normal | Isolated | Normal (CT and laparotomy) |
Normal | No clinical indications |
Normal psychomotor development |
Height 146 cm Weight 47 kg |
|
| 3 | 16p11.2 | 32 | V5b C2b U4b A0 MR Type II |
Blind ending vagina, rudimentary uterus, long uterus horns, tubes and ovaries normal | Syndromic | Left atrophic kidney |
Scoliosis | No clinical indications |
Normal psychomotor development |
Normal (no exact measures) |
|
| 4 | 16p11.2 | 18 | V5b C2b U4b A0 M0 Type I |
Blind ending vagina, rudimentary uterus, tubes and ovaries normal | Isolated | Normal (laparoscopy and laparotomy) |
Normal | No clinical indications |
Normal psychomotor development |
Height 161 cm Weight 74 kg |
|
| 5 | 17q12 | 24 | V5b C2b U4b A0 M0 Type I |
Blind ending vagina, rudimentary uterus, tubes and ovaries normal | Isolated | Normal (CT and laparotomy) |
Normal | No clinical indications |
Normal psychomotor development |
Height 171 cm Weight 56 kg |
|
| 6 | 17q12 | 37 | V5b C2b U4b A0 MRSC Type II/ MURCS |
Blind ending vagina, rudimentary uterus, tubes and ovaries normal | Syndromic | Left kidney agenesis with absent ureter | Pelvic misalignment resulting in different leg length | No clinical indications |
Normal psychomotor development |
Height 173 cm Weight 70 kg |
Diabetes type 2, aortic-pulmonary septal defect |
| 7 | 17q12 | 31 | V5b C2b U4b A0 MR Type II |
Rudimentary uterus, tubes normal, ovaries normal | Syndromic | Absent right kidney, left pelvic kidney | Right convex kyphoscoliosis |
No clinical indications |
Normal psychomotor development |
Height 159 cm Weight 40 kg |
Left kidney transplantation |
| 8 | 17q12 | 25 | V5b C2b U4b A0 M0 Type I |
Blind ending vagina, rudimentary uterus, tubes and ovaries normal | Isolated | Normal (CT and laparotomy) |
Mild scoliosis | No clinical indications |
Normal psychomotor development |
Height 154 cm Weight 58 kg |
|
| 9 | 22q11.2 | 16 | V5b C2b U4b A2a MRSC Type II/ MURCS |
1 cm blind ending vagina. Rudimentary uterus. Normal right ovary, streak left ovary. | Syndromic | Fused pelvic kidney |
Short neck; hemivertebrae C1 and C3, C5, C6, ventral and dorsal fusion C2 and C3; cleft vertebral arch C4, C5, C6; scoliosis thoracic vertebral column; hypoplastic first ribs, flattened sacrum |
Dysplastic auricles, right sided cleft lip, cleft palate | Normal psychomotor development |
Height 153 cm (3rd–10th) Weight 48.5 kg (10th–25th) |
Atrial septal defect, persistent left superior vena cava, unroofed coronary sinus, patent ductus arteriosus; multiple nevi; left hypoplastic thumb, hypoplastic middle phalanx, absent distal phalanx second digit, absent middle and distal phalanges, fifth digit |
| 10 | 2q11.2 and 2p24.24.3 |
28 | V5b C2b U4b A1b MR Type II/ MURCS |
Blind ending vagina, no uterus, no uterus horns, very short left tube, tube right agenesis, ovaries normal | Syndromic | Agenesis of right kidney | Normal | No clinical indications |
Normal psychomotor development |
Height 165 cm Weight 51 kg |
Bilateral inguinal hernias |
Please refer to Opelt et al8 for explanation of VCUAM classification of Müllerian abnormalities.
Briefly, V5b, complete atresia of the vagina; C2b, bilateral aplasia of the cervix; U4b, aplasia of the uterus; A, adnex malformations with A1b, bilateral tubal malformation but normal ovaries; A2a, unilateral tubal hypoplasia; M, associated malformation, R, renal system; S, skeleton; C, cardiac; N, neurological; 0, normal.
Table 4.
Non-genomic disorder type copy number variants in isolated and syndromic Müllerian aplasia
| Case | Locus | Size | Copy number | Co-ordinates (Hg18) | Phenotype | Overlap | Gene content | Platform/confirmation |
|---|---|---|---|---|---|---|---|---|
| 9 | 14q32.33 | 0.46 Mb | Del | 14:104840347–105295569 | MURCS | 0.53 | Multiple | Agilent, Affymetrix 6.0 |
| 10 | 2p24.124.3 | 4.6 Mb | Del | 2:16875751–21460570 | MURCS | 0.12 | Agilent, qPCR | |
| 11 | 1q31.1 | 0.40 Mb | Dup | 1:187058991–187457103 | MA | 0.46 | Affymetrix 6.0 | |
| 12 | 2p23.1 | 0.21 Mb | Dup | 2:31493349–31706481 | MA | 0 | SRD5A2 | Affymetrix 6.0 |
| 13 | 5p11 | 0.4 Mb | Del | 5:45989457–46401198 | MURCS | 0.28 | Affymetrix 6.0 | |
| 13 | 11p11.12 | 0.76 Mb | Del | 11:50334299–51095288 | MURCS | 0.29 | Affymetrix 6.0 | |
| 14 | 5q14.3 | 0.4 Mb | Del | 5:91827411–92261197 | MA | 0 | Agilent, Affymetrix 6.0 | |
| 15 | 6q11.1 | 0.41 Mb | Dup | 6:62969555–63392084 | MA | 0.41 | KHDRBS2 | Affymetrix 6.0 |
| 16 | 15q21.1 | 0.28 Mb | Del | 15:45521438–45801152 | MURCS | 0 | SEMA6D | Agilent, qPCR |
| 17 | 16q11.2 | 0.2 Mb | Dup | 16:45210000–45414000 | MA | 0 | Agilent | |
| 18 | 18q23 | 0.2 Mb | Dup | 18:74729993–74935915 | MA | 0 | SALL3 | Agilent, qPCR |
We observed that 4/63 (6%) of cases in our series had microdeletions at 16p11.2, the first time that this locus has been associated with Müllerian aplasia. Recent reports have identified microdeletions at this locus in cohorts of patients with autism spectrum disorder19 and obesity.22 These patients often have syndromic features such as dysmorphic facial features and some congenital malformations including vertebral anomalies,20,28 and in one case, micropenis,20 but not female reproductive tract malformations. The enrichment of this locus in patients with Müllerian aplasia compared with controls (4/63 patients compared with 2/7366 controls) is highly statistically significant (p=6.96e-8, Fisher’s exact test) and suggests a hitherto unappreciated role for genes within the deletion interval in the development of the Müllerian derivatives.
In keeping with previously published studies12,14 we found microdeletions at 17q12 in patients with both syndromic and apparently isolated Müllerian aplasia. This finding is in line with previous reports in the literature linking this locus to syndromic Müllerian aplasia14 and, in a single case, to apparently isolated Müllerian aplasia.12
Several case reports have demonstrated an association between the velocardiofacial syndrome (VCFS) associated microdeletions at 22q11.2 and Müllerian aplasia.13,14,29 Recently, a novel genomic disorder was reported due to microdeletions at an adjacent, telomeric, locus, and this disorder was given the name ‘22q11.2 distal deletion’.30 The complex genomic architecture at this locus gives rise to ‘nested’ microdeletions within the critical interval. In the original report of this syndrome,30 attention was drawn to two cases with distal nested microdeletions, one of which had an isolated congenital heart defect. We now describe a case with syndromic Müllerian aplasia/MURCS and the same, distal nested microdeletion.
We identified a microduplication at 2q11.2 in a patient with features of MURCS association. Copy number variation at this locus has recently been reported and the suggestion has been made that this constitutes a novel genomic disorder.27 However, no phenotypic data concerning this duplication are available, and the interpretation in our case is further confounded by the co-existence of a previously undescribed 4.6 Mb deletion on chromosome 2p. Clarification of the possible contribution of these two individual copy number variants to abnormalities of Müllerian development must await further examples.
The intervals delineated by these copy number variants harbour in some cases interesting candidate genes. The approximately 0.55 Mb interval at 16p11.2 contains TBX6, a gene previously implicated in the development of paraxial mesoderm31 but with no known role in formation of the Müllerian ducts. There are no other compelling developmental candidate genes in the critical interval. The 1.4 Mb interval at 17q12 harbours two genes with known roles in reproductive tract development: HNF1B, in which mutations have been described in patients with renal cysts and diabetes32; these patients also have genital tract malformations such as bicornuate uterus and uterus didelphys, but an absence of uterus and fallopian tubes has been reported.32 Second, this region harbours LHX1. Mutations in this gene have not been described in humans, but mice with targeted knockout of Lhx1 have an absence of uterus and oviducts.33 Sequencing of LHX1 in patients with Müllerian aplasia has to date not revealed any mutations (Bernardini et al,12 and our unpublished data). The ‘distal 22q11.29 locus harbours just four genes, RTDR1, RAB36, GNAZ, and BCR. Germline mutations in none of these four genes have been described in humans, and developmental malformations have not been reported in BCR-null34 or GNAZ-null mice,35 for which mouse data are available.
Eleven non-genomic disorder type copy number variants were identified in the case cohort which were absent in controls, but none of these occurred in more than one patient, and the absence of parental samples is an additional factor limiting our interpretation of their significance. Table 4 lists copy number variants occurring in a single patient in the case cohort and not in the control cohort. Three of these, at 2p24.1–24.3, 15q21.1, and 18q23, were selected for validation by qPCR; all three were confirmed (data not shown). The deletion of 4.6 Mb at 2p24.1–24.3 occurred in a patient with a double segment imbalance, the other copy number variant being a microduplication at 2q11.2, discussed above. One imbalance harbouring a potentially interesting candidate gene was noted, a 200 Kb duplication at 18q23 encompassing SALL3, a member of the SALL gene family, of as yet unknown function. Replication of this study on larger cohorts will be needed in order to determine whether these single occurrence copy number variants are significant.
DISCUSSION
Previous reports have identified copy number variants in Müllerian aplasia,12,14 but these have been case reports and small series, and studies of medium or large scale series have to date not been carried out. Here, we give results of copy number analysis of a series of 63 patients, identifying a strikingly high incidence of 9/63 (14%) of previously characterised microdeletions. Microdeletions at 16p11.2 and 17q12 in particular are highly enriched in the case population in comparison to the control population. Both of these microdeletions, and the 22q11.2 distal microdeletion, have previously been associated with congenital malformations: of the spine in the case of 16p11.2,28 the genitourinary tract in the case of 17q12,36 and cardiovascular system in the case of the distal 22q11.2 deletion.30 Thus, although data on inheritance for these cases are lacking, there can be little doubt that these microdeletions are contributing to the phenotypes of isolated and syndromic Müllerian aplasia.
The apparent contribution of genomic disorder type copy number variants to congenital malformations occurring in isolation is of particular interest. A recent study of non-syndromic tetralogy of Fallot, a complex congenital heart malformation, gives weight to the idea that genomic disorders may be associated with isolated congenital malformations.37 Copy number variants corresponding to known genomic disorders were identified at 22q11.2 (two, both loss), and at 1q21.1 (four gain, one loss) in 7/512 (1.4%) of individuals. In our study, copy number variants associated with genomic disorders were identified in 4/43 (9%) of isolated Müllerian aplasia cases, a significantly higher figure than for isolated tetralogy of Fallot.
Our study extends the phenotypic variability associated with microdeletions at 16p11.2. The previously known phenotypic consequences of microdeletions at this locus are: autism spectrum disorder18; epilepsy21; developmental delay/learning disability and dysmorphism/congenital anomalies20,21; and obesity.22 Both isolated and syndromic forms of Müllerian aplasia can now be added to this list. Increasingly, these findings raise the question of which factor or factors are responsible for determining the phenotypic outcome in patients with 16p11.2 microdeletions in particular and genomic disorders in general. It can be hypothesised that this microdeletion, and others like it, provide an early and general developmental perturbation, the ultimate and precise consequence of which is independent of the perturbation itself, but depends on other factors. Future studies may begin to address this question, through sequencing, identification of ‘second hits’,23 and other modalities.
Our results have implications for genetic counselling in Müllerian aplasia. At present, few women with the isolated form of this malformation are referred to clinical geneticists. Müllerian aplasia results in infertility, and so it might be argued that the need for genetic counselling is less (although it might be important in cases where egg donation is being considered). More critically, though, the diagnosis of a microdeletion at 16p11.2, 17q12 or 22q11.2 (distal) has potential implications for other family members. Copy number variants associated with genomic disorders may be inherited from a phenotypically normal parent and transmitted to other family members. We anticipate that families would wish to be made aware of the associations with autism (16p11.2) or young onset diabetes and renal cysts (17q12) and cardiac malformations (22q11.2 (distal)) as well as with Müllerian aplasia, notwithstanding that our poor understanding of the penetrance and variable expressivity of these disorders makes genetic counselling difficult. In conclusion, our data support the contention that detailed copy number assays should be carried out in the assessment of both isolated and syndromic forms of Müllerian aplasia. We feel that the high incidence of recurrent copy number variants in these patients makes it reasonable to recommend that they should be referred to a clinical geneticist for assessment, and, where appropriate, for genetic testing and family counselling.
Acknowledgements
The authors thank all of the patients who participated in this study and Departments of Obstetrics and Gynaecology in Tübingen (Professor D Wallwiener), Heidelberg (Professor T Strowitzky) and Wuppertal (Professor Hucke), Germany for help in recruitment of patients. CS-S, MS, VM, TF, MH, NH, and NPC are funded by the Wellcome Trust. CS-S is the recipient of an Intermediate Clinical Fellowship from the Wellcome Trust. RS was funded by the Deutsche Forschungsgemeinschaft (DFG-STR923/2-2). The authors wish to thank Cordelia Langford and Sarah Widaa of the Microarray facility, and Sarah Edkins and Emma Gray of the DNA collections group at the Sanger Institute for assistance with sample processing and genotyping.
Funding Other funders: Wellcome Trust; Deutsche Forschungsgemeinschaft.
Footnotes
SN-Z and RS contributed equally to this work.
Competing interests None.
Ethics approval This study was conducted with the approval of the Ethics Review Board of Friedrich-Alexander-University, Erlangen-Nuremberg, Germany.
Provenance and peer review Not commissioned; externally peer reviewed.
REFERENCES
- 1.Raga F, Bauset C, Remohi J, Bonilla-Musoles F, Simon C, Pellicer A. Reproductive impact of congenital Mullerian anomalies. Hum Reprod. 1997;12:2277–81. doi: 10.1093/humrep/12.10.2277. [DOI] [PubMed] [Google Scholar]
- 2.Nahum GG. Uterine anomalies. How common are they, and what is their distribution among subtypes? J Reprod Med. 1998;43:877–87. [PubMed] [Google Scholar]
- 3.Buttram VC, Jr, Gibbons WE. Mullerian anomalies: a proposed classification. (An analysis of 144 cases) Fertil Steril. 1979;32:40–6. doi: 10.1016/s0015-0282(16)44114-2. [DOI] [PubMed] [Google Scholar]
- 4.Folch M, Pigem I, Konje JC. Mullerian agenesis: etiology, diagnosis, and management. Obstet Gynecol Surv. 2000;55:644–9. doi: 10.1097/00006254-200010000-00023. [DOI] [PubMed] [Google Scholar]
- 5.Morcel K, Camborieux L. Programme de Recherches sur les Aplasies Mulleriennes, Guerrier D. Mayer-Rokitansky-Kuster-Hauser (MRKH) syndrome. Orphanet J Rare Dis. 2007;14:2–13. doi: 10.1186/1750-1172-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strubbe EH, Willemsen WN, Lemmens JA, Thijn CJ, Rolland R. Mayer-Rokitansky-Kuster-Hauser syndrome: distinction between two forms based on excretory urographic, sonographic, and laparoscopic findings. AJR Am J Roentgenol. 1993;160:331–4. doi: 10.2214/ajr.160.2.8424345. [DOI] [PubMed] [Google Scholar]
- 7.Oppelt P, Renner SP, Kellermann A, Brucker S, Hauser GA, Ludwig KS, Strissel PL, Strick R, Wallwiener D, Beckmann MW. Clinical aspects of Mayer-Rokitansky-Kuester-Hauser syndrome: recommendations for clinical diagnosis and staging. Hum Reprod. 2006;21:792–7. doi: 10.1093/humrep/dei381. [DOI] [PubMed] [Google Scholar]
- 8.Oppelt P, von Have M, Paulsen M, Strissel PL, Strick R, Brucker S, Wallwiener D, Beckmann MW. Female genital malformations and their associated abnormalities. Fertil Steril. 2007;87:335–42. doi: 10.1016/j.fertnstert.2006.07.1501. [DOI] [PubMed] [Google Scholar]
- 9.Tiker F, Yildirim SV, Barutcu O, Bagis T. Familial mullerian agenesis. Turk J Pediatr. 2000;42:322–4. [PubMed] [Google Scholar]
- 10.Shokeir MH. Aplasia of the Mullerian system: evidence for probable sex-limited autosomal dominant inheritance. Birth Defects Orig Artic Ser. 1978;14:147–65. [PubMed] [Google Scholar]
- 11.Biason-Lauber A, Konrad D, Navratil F, Schoenle EJ. A WNT4 mutation associated with Mullerian-duct regression and virilization in a 46, XX woman. N Engl J Med. 2004;351:792–8. doi: 10.1056/NEJMoa040533. [DOI] [PubMed] [Google Scholar]
- 12.Bernardini L, Gimelli S, Gervasini C, Carella M, Baban A, Frontino G, Barbano G, Divizia MT, Fedele L, Novelli A, Béna F, Lalatta F, Miozzo M, Dallapiccola B. Recurrent microdeletion at 17q12 as a cause of Mayer-Rokitansky-Kuster-Hauser (MRKH) syndrome: two case reports. Orphanet J Rare Dis. 2009;4:25. doi: 10.1186/1750-1172-4-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sundaram UT, McDonald-McGinn DM, Huff D, Emanuel BS, Zackai EH, Driscoll DA, Bodurtha J. Primary amenorrhea and absent uterus in the 22q11.2 deletion syndrome. Am J Med Genet A. 2007;143A:2016–18. doi: 10.1002/ajmg.a.31736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheroki C, Krepischi-Santos AC, Szuhai K, Brenner V, Kim CA, Otto PA, Rosenberg C. Genomic imbalances associated with mullerian aplasia. J Med Genet. 2008;45:228–32. doi: 10.1136/jmg.2007.051839. [DOI] [PubMed] [Google Scholar]
- 15.Klopocki E, Schulze H, Strauss G, Ott CE, Hall J, Trotier F, Fleischhauer S, Greenhalgh L, Newbury-Ecob RA, Neumann LM, Habenicht R, König R, Seemanova E, Megarbane A, Ropers HH, Ullmann R, Horn D, Mundlos S. Complex inheritance pattern resembling autosomal recessive inheritance involving a microdeletion in thrombocytopenia-absent radius syndrome. Am J Hum Genet. 2007;80:232–40. doi: 10.1086/510919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ryan AK, Goodship JA, Wilson DI, Philip N, Levy A, Seidel H, Schuffenhauer S, Oechsler H, Belohradsky B, Prieur M, Aurias A, Raymond FL, Clayton-Smith J, Hatchwell E, McKeown C, Beemer FA, Dallapiccola B, Novelli G, Hurst JA, Ignatius J, Green AJ, Winter RM, Brueton L, Brøndum-Nielsen K, Scambler PJ. Spectrum of clinical features associated with interstitial chromosome 22q11 deletions: a European collaborative study. J Med Genet. 1997;34:798–804. doi: 10.1136/jmg.34.10.798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mefford HC, Eichler EE. Duplication hotspots, rare genomic disorders, and common disease. Curr Opin Genet Dev. 2009;19:196–204. doi: 10.1016/j.gde.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar RA, KaraMohamed S, Sudi J, Conrad DF, Brune C, Badner JA, Gilliam TC, Nowak NJ, Cook EH, Jr, Dobyns WB, Christian SL. Recurrent 16p11.2 microdeletions in autism. Hum Mol Genet. 2008;17:628–38. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- 19.Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, Saemundsen E, Stefansson H, Ferreira MA, Green T, Platt OS, Ruderfer DM, Walsh CA, Altshuler D, Chakravarti A, Tanzi RE, Stefansson K, Santangelo SL, Gusella JF, Sklar P, Wu BL, Daly MJ. Autism Consortium. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–75. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 20.Fernandez BA, Roberts W, Chung B, Weksberg R, Meyn S, Szatmari P, Joseph-George AM, Mackay S, Whitten K, Noble B, Vardy C, Crosbie V, Luscombe S, Tucker E, Turner L, Marshall CR, Scherer SW. Phenotypic spectrum associated with de novo and inherited deletions and duplications at 16p11.2 in individuals ascertained for diagnosis of autism spectrum disorder. J Med Genet. 2010;47:195–203. doi: 10.1136/jmg.2009.069369. [DOI] [PubMed] [Google Scholar]
- 21.Shinawi M, Liu P, Kang SH, Shen J, Belmont JW, Scott DA, Probst FJ, Craigen WJ, Graham BH, Pursley A, Clark G, Lee J, Proud M, Stocco A, Rodriguez DL, Kozel BA, Sparagana S, Roeder ER, McGrew SG, Kurczynski TW, Allison LJ, Amato S, Savage S, Patel A, Stankiewicz P, Beaudet AL, Cheung SW, Lupski JR. Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. J Med Genet. 2010;47:332–41. doi: 10.1136/jmg.2009.073015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walters RG, Jacquemont S, Valsesia A, de Smith AJ, Martinet D, Andersson J, Falchi M, Chen F, Andrieux J, Lobbens S, Delobel B, Stutzmann F, El-Sayed Moustafa JS, Chèvre JC, Lecoeur C, Vatin V, Bouquillon S, Buxton JL, Boute O, Holder-Espinasse M, Cuisset JM, Lemaitre MP, Ambresin AE, Brioschi A, Gaillard M, Giusti V, Fellmann F, Ferrarini A, Hadjikhani N, Campion D, Guilmatre A, Goldenberg A, Calmels N, Mandel JL, Le Caignec C, David A, Isidor B, Cordier MP, Dupuis-Girod S, Labalme A, Sanlaville D, Béri-Dexheimer M, Jonveaux P, Leheup B, Ounap K, Bochukova EG, Henning E, Keogh J, Ellis RJ, Macdermot KD, van Haelst MM, Vincent-Delorme C, Plessis G, Touraine R, Philippe A, Malan V, Mathieu-Dramard M, Chiesa J, Blaumeiser B, Kooy RF, Caiazzo R, Pigeyre M, Balkau B, Sladek R, Bergmann S, Mooser V, Waterworth D, Reymond A, Vollenweider P, Waeber G, Kurg A, Palta P, Esko T, Metspalu A, Nelis M, Elliott P, Hartikainen AL, McCarthy MI, Peltonen L, Carlsson L, Jacobson P, Sjöström L, Huang N, Hurles ME, O’Rahilly S, Farooqi IS, Mánnik K, Jarvelin MR, Pattou F, Meyre D, Walley AJ, Coin LJ, Blakemore AI, Froguel P, Beckmann JS. A new highly penetrant form of obesity due to deletions on chromosome 16p11.2. Nature. 2010;463:671–5. doi: 10.1038/nature08727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Girirajan S, Rosenfeld JA, Cooper GM, Antonacci F, Siswara P, Itsara A, Vives L, Walsh T, McCarthy SE, Baker C, Mefford HC, Kidd JM, Browning SR, Browning BL, Dickel DE, Levy DL, Ballif BC, Platky K, Farber DM, Gowans GC, Wetherbee JJ, Asamoah A, Weaver DD, Mark PR, Dickerson J, Garg BP, Ellingwood SA, Smith R, Banks VC, Smith W, McDonald MT, Hoo JJ, French BN, Hudson C, Johnson JP, Ozmore JR, Moeschler JB, Surti U, Escobar LF, El-Khechen D, Gorski JL, Kussmann J, Salbert B, Lacassie Y, Biser A, McDonald-McGinn DM, Zackai EH, Deardorff MA, Shaikh TH, Haan E, Friend KL, Fichera M, Romano C, Gécz J, DeLisi LE, Sebat J, King MC, Shaffer LG, Eichler EE. A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat Genet. 2010;42:203–9. doi: 10.1038/ng.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stankiewicz P, Sen P, Bhatt SS, Storer M, Xia Z, Bejjani BA, Ou Z, Wiszniewska J, Driscoll DJ, Maisenbacher MK, Bolivar J, Bauer M, Zackai EH, McDonald-McGinn D, Nowaczyk MM, Murray M, Hustead V, Mascotti K, Schultz R, Hallam L, McRae D, Nicholson AG, Newbury R, Durham-O’Donnell J, Knight G, Kini U, Shaikh TH, Martin V, Tyreman M, Simonic I, Willatt L, Paterson J, Mehta S, Rajan D, Fitzgerald T, Gribble S, Prigmore E, Patel A, Shaffer LG, Carter NP, Cheung SW, Langston C, Shaw-Smith C. Genomic and genic deletions of the FOX gene cluster on 16q24.1 and inactivating mutations of FOXF1 cause alveolar capillary dysplasia and other malformations. Am J Hum Genet. 2009 doi: 10.1016/j.ajhg.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bochukova EG, Huang N, Keogh J, Henning E, Purmann C, Blaszczyk K, Saeed S, Hamilton-Shield J, Clayton-Smith J, O’Rahilly S, Hurles ME, Farooqi IS. Large, rare chromosomal deletions associated with severe early-onset obesity. Nature. 2010;463:666–70. doi: 10.1038/nature08689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rad R, Brenner L, Bauer S, Schwendy S, Layland L, da Costa CP, Reindl W, Dossumbekova A, Friedrich M, Saur D, Wagner H, Schmid RM, Prinz C. CD25+/Foxp3+ T cells regulate gastric inflammation and Helicobacter pylori colonization in vivo. Gastroenterology. 2006;131:525–37. doi: 10.1053/j.gastro.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 27.Rudd MK, Keene J, Bunke B, Kaminsky EB, Adam MP, Mulle JG, Ledbetter DH, Martin CL. Segmental duplications mediate novel, clinically relevant chromosome rearrangements. Hum Mol Genet. 2009;18:2957–62. doi: 10.1093/hmg/ddp233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimojima K, Inoue T, Fujii Y, Ohno K, Yamamoto T. A familial 593-kb microdeletion of 16p11.2 associated with mental retardation and hemivertebrae. Eur J Med Genet. 2009;52:433–5. doi: 10.1016/j.ejmg.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 29.Devriendt K, Moerman P, Van Schoubroeck D, Vandenberghe K, Fryns JP. Chromosome 22q11 deletion presenting as the Potter sequence. J Med Genet. 1997;34:423–5. doi: 10.1136/jmg.34.5.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ben-Shachar S, Ou Z, Shaw CA, Belmont JW, Patel MS, Hummel M, Amato S, Tartaglia N, Berg J, Sutton VR, Lalani SR, Chinault AC, Cheung SW, Lupski JR, Patel A. 22q11.2 distal deletion: a recurrent genomic disorder distinct from DiGeorge syndrome and velocardiofacial syndrome. Am J Hum Genet. 2008;82:214–21. doi: 10.1016/j.ajhg.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.White PH, Farkas DR, McFadden EE, Chapman DL. Defective somite patterning in mouse embryos with reduced levels of Tbx6. Development. 2003;130:1681–90. doi: 10.1242/dev.00367. [DOI] [PubMed] [Google Scholar]
- 32.Lindner TH, Njolstad PR, Horikawa Y, Bostad L, Bell GI, Sovik O. A novel syndrome of diabetes mellitus, renal dysfunction and genital malformation associated with a partial deletion of the pseudo-POU domain of hepatocyte nuclear factor-1beta. Hum Mol Genet. 1999;8:2001–8. doi: 10.1093/hmg/8.11.2001. [DOI] [PubMed] [Google Scholar]
- 33.Kobayashi A, Shawlot W, Kania A, Behringer RR. Requirement of Lim1 for female reproductive tract development. Development. 2004;131:539–49. doi: 10.1242/dev.00951. [DOI] [PubMed] [Google Scholar]
- 34.Voncken JW, van Schaick H, Kaartinen V, Deemer K, Coates T, Landing B, Pattengale P, Dorseuil O, Bokoch GM, Groffen J. Increased neutrophil respiratory burst in bcr-null mutants. Cell. 1995;80:719–28. doi: 10.1016/0092-8674(95)90350-x. [DOI] [PubMed] [Google Scholar]
- 35.Hendry IA, Kelleher KL, Bartlett SE, Leck KJ, Reynolds AJ, Heydon K, Mellick A, Megirian D, Matthaei KI. Hypertolerance to morphine in G(z alpha)-deficient mice. Brain Res. 2000;870:10–19. doi: 10.1016/s0006-8993(00)02387-8. [DOI] [PubMed] [Google Scholar]
- 36.Mefford HC, Clauin S, Sharp AJ, Moller RS, Ullmann R, Kapur R, Pinkel D, Cooper GM, Ventura M, Ropers HH, Tommerup N, Eichler EE, Bellanne-Chantelot C. Recurrent reciprocal genomic rearrangements of 17q12 are associated with renal disease, diabetes, and epilepsy. Am J Hum Genet. 2007;81:1057–69. doi: 10.1086/522591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greenway SC, Pereira AC, Lin JC, DePalma SR, Israel SJ, Mesquita SM, Ergul E, Conta JH, Korn JM, McCarroll SA, Gorham JM, Gabriel S, Altshuler DM, Quintanilla-Dieck Mde L, Artunduaga MA, Eavey RD, Plenge RM, Shadick NA, Weinblatt ME, De Jager PL, Hafler DA, Breitbart RE, Seidman JG, Seidman CE. De novo copy number variants identify new genes and loci in isolated sporadic tetralogy of Fallot. Nat Genet. 2009;41:931–5. doi: 10.1038/ng.415. [DOI] [PMC free article] [PubMed] [Google Scholar]