Abstract
TRIM5 is a restriction factor that blocks retrovirus infection soon after the virion core enters the cell cytoplasm. Restriction activity is targeted to the virion core via recognition of the capsid protein lattice that encases the viral genomic RNA. In common with all of the many TRIM family members, TRIM5 has RING, B-box, and coiled-coil domains. As an E3 ubiquitin ligase TRIM5 cooperates with the heterodimeric E2, UBC13/UEV1A, to activate the TAK1 (MAP3K7) kinase, NF-κB and AP-1 signaling, and the transcription of inflammatory cytokines and chemokines. TAK1, UBC13, and UEV1A all contribute to TRIM5-mediated retrovirus restriction activity. Interaction of the carboxy-terminal PRYSPRY or cyclophilin domains of TRIM5 with the retroviral capsid lattice stimulates the formation of a complementary lattice by TRIM5, with greatly increased TRIM5 E3 activity, and host cell signal transduction. Structural and biochemical studies on TRIM5 have opened a much needed window on how the innate immune system detects the distinct molecular features of HIV-1 and other retroviruses.
The tripartite motif (TRIM) family of proteins
TRIMs are multi-domain proteins defined by an N-terminal RING finger domain, one or two B-box domains, and a coiled-coil domain (Figure 1A). A large proportion of the TRIM proteins possess a C-terminal PRYSPRY domain that interacts with target proteins. Nearly 100 human genes encode TRIM proteins, and many of these are synthesized as multiple isoforms [1–3]. This enormous family of cellular proteins are involved in diverse cellular processes, including cell proliferation, differentiation, development, apoptosis, oncogenesis, and innate immunity. Of the many TRIM genes, several exhibit anti-retroviral activity, including TRIM11, 15, and 31 [4], TRIM1 [5,6], TRIM28 [7] and TRIM22 [8,9,9]. Among TRIM family members that inhibit HIV-1, TRIM5 is the best-studied.
Figure 1.
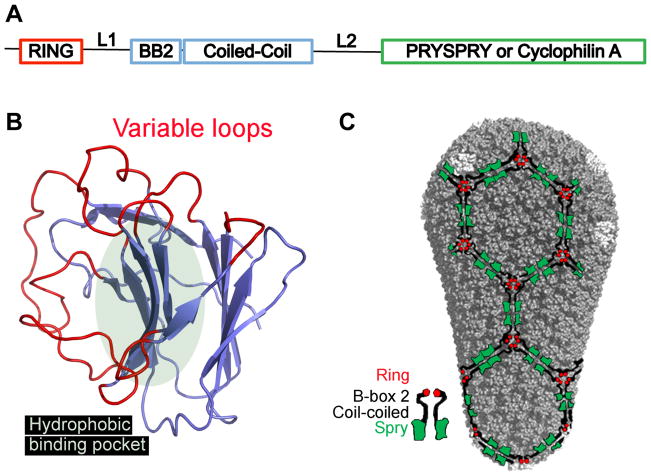
Figure 1A: Schematic representation of the domains within TRIM5, with relative positions of the domains along the linear sequence indicated. RING, really interesting new gene; L1, linker 1; BB2, B box 2; L2, linker 2.
Figure 1B. Ribbon diagram of a model of the human TRIM5α PRYSPRY (B30.2) domain. Variable loops extending from the β-sandwich are marked in red, the putative hydrophobic binding pocket is indicated by the light green oval.
Fig. 1C. Model of the HIV-1 capsid fullerene cone with hexagonal TRIM5α lattice superimposed, illustrating a putative recognition mode of capsid by TRIM5α, as in reference [40]. The color code for the TRIM5α domains is indicated in the schematic model of the TRIM5α dimer at the bottom left of the panel.
TRIM5 and retrovirus restriction
TRIM5 is a cytoplasmic protein that blocks HIV-1 infection soon after the virus enters the target cell cytoplasm. It was discovered to be an HIV-1 restriction factor in functional, expression screens of cDNA libraries from macaque and owl monkey cells [10,11]. Cells from these species were targeted for study because they had particularly strong, well-characterized blocks to HIV-1 infection [12,13].
Once TRIM5 was cloned, it was found that, as compared with other species, the macaque and owl monkey TRIM5 orthologues associated relatively strongly with the HIV-1 virion core [14]. Interspecies variation in strength of TRIM5 binding to the capsid protein lattice of the virion core correlated with the ability of the TRIM5 orthologue from any given host species to block a given retrovirus. Lab strains of HIV-1 are weakly recognized by the human TRIM5 orthologue, which inhibits these viruses only 2-fold in single-cycle assays [10,15]. Compared to lab strains, though, some primary isolates are 10-fold more sensitive to restriction by human TRIM5 [16].
The clinical significance of TRIM5 for HIV-1 infection and disease progression in people is supported by several observations. TRIM5 polymorphisms and differences in expression influence rates of HIV-1 acquisition or disease progression [17–20]. HIV-1 variants that are highly sensitive to restriction by human TRIM5 appear to have been selected in vivo by pressure to escape from potent CTL targeting overlapping capsid determinants [21]. Additionally, a growing body of evidence indicates that TRIM5 in non-human primates plays an important role in limiting transmission of SIVs, or in controlling the outcome of infection with these viruses [22–26].
Capsid recognition by the PRYSPRY domain
Major determinants for capsid recognition are found in the C-terminus of TRIM5 (Figure 1A). In most species, TRIM5-mediated antiviral activity is associated with the alpha isoform. The C-terminus of this protein is a PRYSPRY (or B30.2) domain. PRYSPRY domains are found in over 500 different proteins and structures of PRYSPRY domains from the proteins sRFLPL1, TRIM21, GUSTAVUS, and PYRIN have been determined [27–30]. The common structure is a seven-stranded and a six-stranded antiparallel β-sheet, arranged in a β sandwich (Figure 1B). The loops that connect the β-strands form a surface that has been proposed to be the target specificity determinant.
The highly polymorphic PRYSPRY domain of the TRIM5α isoform is a capsid-specificity determinant. This was demonstrated experimentally by testing the specificity of retrovirus restriction after swapping PRYSPRY domains among orthologues, as well as with phylogenetic comparisons [31,32]. In those cells that have been examined, 50% of TRIM5 transcripts encode alternative isoforms that lack a PRYSPRY domain, lack restriction activity, and act in trans to block the restriction activity of the alpha isoform [33]; in order of decreasing abundance, the mRNAs encoding these isoforms are called iota, gamma, kappa, or delta.
Direct, capsid-binding studies with TRIM5α have been elusive because TRIM5α recognizes the retroviral capsid protein lattice, not free capsid protein [14,34]. Efforts to detect CA binding to TRIM5α have resorted to co-sedimentation with detergent-stripped virion cores mixed together in vitro [34], virion cores pelleted from the cytoplasm of infected cells [14], or tube assemblies generated in vitro from recombinant capsid protein [35]. Among other efforts to pinpoint sites of protein-protein interaction between TRIM5 and CA some investigators have attempted to stabilize minimal CA assemblies via oxidation of cysteine residues substituted at critical capsid interfaces [36,37]. These methods were critical to solving the atomic structure of the capsid hexamer and pentamer subunits that form the virion core [38]. Biochemical studies with these cross-linked components suggest that TRIM5 interacts with the trimer interface in the capsid lattice, not with the hexameric interface [35,39].
When visualized by electron microscopy after negative staining, recombinant rhesus TRIM5α forms two dimensional crystals arranged in a hexagonal lattice [40]. TRIM5 mutants that block homo-dimerization or higher-order assembly, either of which disrupt restriction activity, prevented lattice formation. The efficiency of lattice formation was greatly increased when soluble TRIM5 protein was added to a planar lattice of HIV-1 capsid protein on an EM grid (Figure 1C). These fascinating observations suggest that complementary lattice formation plays an important role in TRIM5 restriction activity. Buttressing these biochemical observations, live-microscopy experiments detected accumulation of TRIM5 on virion cores as they arrive in the cytoplasm of a newly-infected cell [41].
Capsid recognition by cyclophilin A
All species of New World owl monkeys have a cyclophilin A domain at the C-terminus that was put there by LINE1-mediated retrotransposition of a cyclophilin A cDNA into the TRIM5 locus [11]. A similar TRIM5-cyclophilin A fusion gene in some species of Asian macaques was created by an independent retrotransposition event [42–45]. Prior to the discovery of TRIM5, free cyclophilin A protein had been demonstrated to be an HIV-1 capsid binding protein [46,47]. Consistent with this, disruption of the cyclophilin domain destroyed HIV-1 restriction by owl monkey TRIM5Cyp [11] and a potent, HIV-1-specific inhibitor was engineered by fusing human TRIM5 to human cyclophilin A [48].
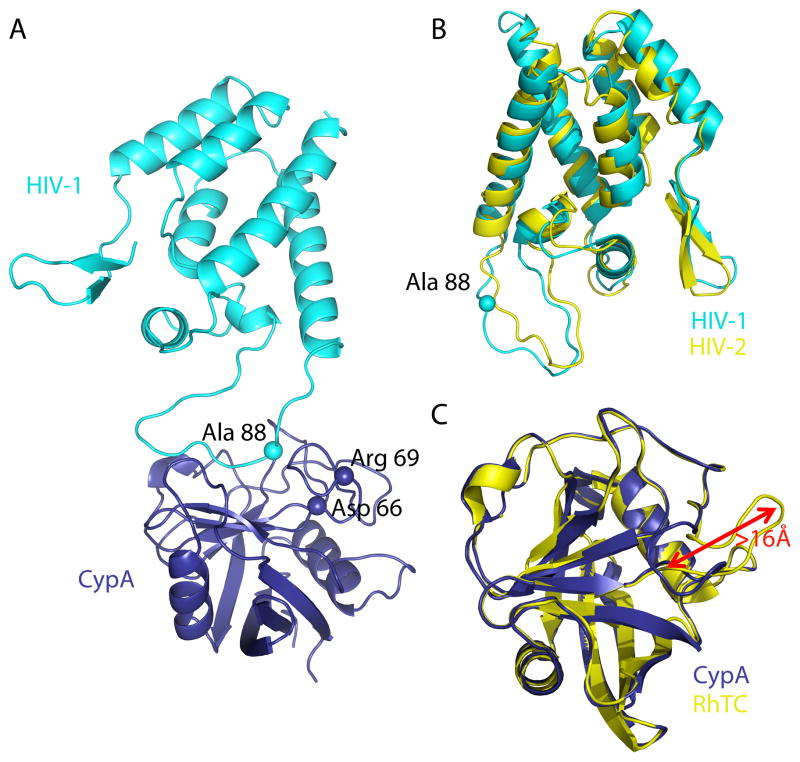
Cyclophilin A orthologues from Saccharomyces cerevisiae to Homo sapiens are functionally interchangeable [49] and orthologues from all species that have been examined retain the ability to bind HIV-1 capsid, but not HIV-2 capsid [11,50]. The owl monkey TRIM5-cyclophilin A fusion protein restricts HIV-1, but not HIV-2 [11,51]. It was therefore quite surprising when the TRIM5-cyclophilin A fusion protein from macaques was found to neither bind HIV-1 capsid nor restrict HIV-1 replication [43,44]; instead, it restricts HIV-2. The overall topology of the HIV-1 and HIV-2 capsids is the same, but the orientation of the critical residues linking helix 4 and 5 is very different [52]. X-ray crystallography, in combination with functional analysis of mutants, showed that a single amino acid change in the cyclophilin domain, D66N, confers these specificity differences [52]. Loss of constraining interactions from the aspartate residue frees a loop of 10 residues, such that its orientation is shifted by 16Å (Figure 2). This demonstrates that cyclophilin has an extraordinary capacity to alter its mode of substrate binding and perhaps explains the utility and widespread presence of this modular unit in many different cellular proteins.
Figure 2.
Figure 2A: Cartoon representation of HIV-1-CypA complex (PDB 1AK4, Chains A and D) in which HIV-1 is in cyan and CypA is in dark blue. The proximity of HIV-1 CA residue Ala 88 to the CypA residues Asp 66 and Arg 69 (spheres) is shown.
Figure 2B: Cartoon representation of the N-terminal capsid domains of HIV-1 (PWB 1GWP, first NMR model) in cyan and of HIV-2 (PDB 2WLV, chain A) in yellow after SSM superposition. The cyan sphere indicates the position of Ca Ala 88 in HIV-1, which is deleted in HIV-2.
Figure 2C: Cartoon representation of CypA (PDB 2CPL) in dark blue SSM superimposed on RhTC (PDB 2WLW) in yellow. Loop 64–74, which contains amino acid replacements D66N and R69H has undergone a rearrangement of >16Å.
The RING domain
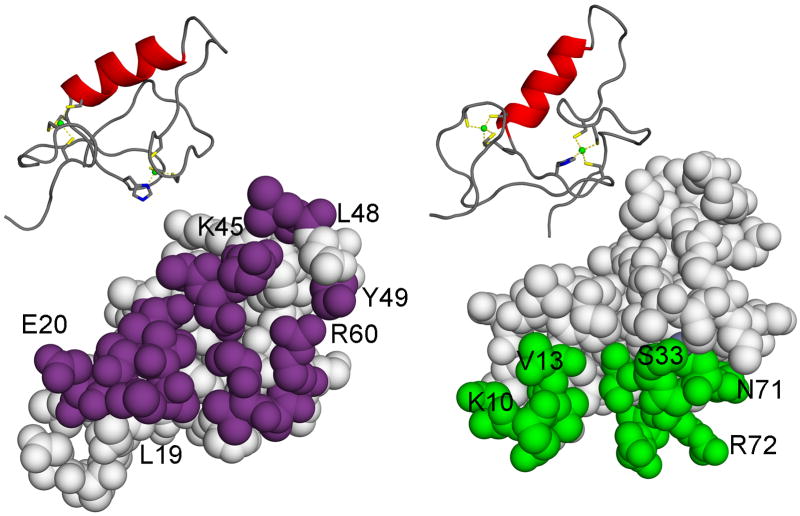
The RING domain of TRIM5 functions as an E3 ubiquitin ligase [39,53–55]. The NMR solution structure was determined for a fragment of TRIM5 that comprises amino acid residues 1 to 78 (Figure 3), encompassing the RING domain [56]. It reveals a compact ββα RING-fold core, with residues 10 to 61 interacting with residues 62 to 70. Two Zn2+ ions are coordinated by zinc-fingers, one of which is coordinated by 4 cysteines and the other by a histidine and 3 cysteines. Hydrophobic residues are mostly packed in the core of the structure. However, some of the hydrophobic residues are solvent exposed. Two hydrophobic surface patches have been defined, the RING-RING interaction region and the E2 binding region of TRIM5. Mutations have been identified in the latter that disrupt retrovirus restriction activity and TRIM5 auto-ubiquitination in the presence of E2 UBCH5B, without consequence to other biochemical functions essential for restriction, such as TRIM5 association with capsid or with itself [56]. These structure-function studies indicate that RING E3 ubiquitin ligase activity is necessary for retrovirus restriction activity.
Figure 3.
NMR solution structure of human TRIM5α, amino acid residues 1 to 78, encompassing the RING domain [56].
Top: ribbon diagrams with Zn2+ ions indicated as green balls. Coordinating residues are highlighted.
Bottom: surface representations.
Left: the RING-RING interaction region is indicated in magenta.
Right: the E2 binding region is indicated in green.
The B-box domain
All TRIM proteins have either one or two B box domains [1,2]. TRIM5 has only one. Generally, B-box domains are involved in protein-protein interactions. In TRIM proteins, these domains cooperate with the coiled-coil domain, to form higher-order oligomers and cytoplasmic bodies. Mutations in the TRIM5α B-box-2 domain prevent the higher-order multimerization required for efficient capsid binding and restriction activity [57,58]. Amino acid residues in the linker domain between the coiled-coil and PRYSPRY domains also contribute to these activities [59].
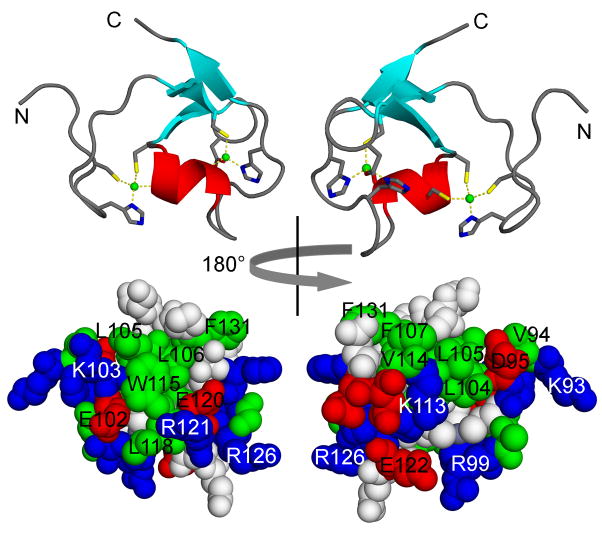
The NMR solution structure of a fragment of the TRIM5 protein (residues 86–131) that comprises the B-box 2 domain showed a ββα RING-like fold with two Zn2+ ions in a CHCDC2H2 coordination mode [58] (Figure 4). An additional short β-strand precedes the helix. TRIM B-box domains commonly have two hydrophobic surface patches. Cluster 1 is located on the outer surface of the helical part and the first β-strand (TRIM5α residues L103, L104, W115, and L116) and is surrounded by charged residues. Cluster 2 is adjacent to cluster 1, and includes residues L102, V112, F105, F129. Mutations within cluster 1, in particular of Arg121, indicate that this surface region is responsible for most of the biological effects attributed to the B-box 2 domain in TRIM5.
Figure 4.
NMR solution structure of human TRIM5α amino acid residues 86 to 131, encompassing the B-box 2 domain [58].
Top: ribbon diagrams with Zn2+ ions indicated as green balls, in a CHCDC2H2 coordination mode. Coordinating residues are highlighted.
Bottom: surface representations.
Left: hydrophobic surface cluster 1.
Right: hydrophobic surface cluster 2.
The coiled-coil domain
Sequence comparisons revealed a likely coiled-coil domain in the region between the B-box 2 domain and the PRYSPRY domain of TRIM5 (Figure 1A). From the standpoint of structure, this domain is perhaps the least well explored and the exact boundaries are not well defined due to lack of sequence homology with other coiled-coil proteins. The coiled-coil domain in TRIM proteins is responsible for dimerization [35,55]. A structure of this domain and its interaction with the other domains of TRIM proteins would be extremely useful for understanding the function of TRIM5, especially considering that the coiled-coil is absolutely essential for restriction activity.
Structure of full-length TRIM5
As discussed above, the domain composition and a series of biochemical experiments suggest that TRIM5 exists as a dimer (via the coiled-coil domain) with a tendency to form higher oligomeric forms (via the B box 2 domain and the linker 2 region). Despite significant progress on the structure of the isolated TRIM5 domains, full-length TRIM5 protein has proved difficult to express and purify in a soluble form. Therefore, the spatial arrangement of the domains within the context of the full-length protein is unknown. Recombinant rhesus TRIM5α in which the N-terminal RING domain is replaced by the TRIM21 RING domain (TRIM5-21R) is better behaved in vitro than the wild-type protein [55]. Since TRIM5-21R retains retroviral restriction activity like that of rhesus TRIM5α [53] it has been used as a clever surrogate for full-length TRIM5. Recently, full-length, dimeric TRIM5 was expressed and purified in sufficient quantities for biochemical analysis, in vitro assessment of its enzymatic activity, and protein-protein interaction studies with capsid and other cellular proteins [39]. To date, though, there is no three-dimensional structural information for the full-length protein.
Mechanism of retrovirus restriction
The exact mechanism of retrovirus restriction by TRIM5 is not known. Pairing of select TRIM5 orthologues with retroviruses for which capsid avidity is highest - for example, rhesus macaque TRIM5α with HIV-1, or human TRIM5α with N-tropic MLV – results in disassembly of the incoming capsid before completion of reverse transcription [32]. This block before reverse transcription requires TRIM5 E3 ubiquitin ligase activity [39,56], and possibly recruitment of proteasomes via direct contact of TRIM5 with the proteasome component PSMC2 [60], or via the adaptor protein p62/sequestosome 1 (SQTM-1) [61]. TRIM5 is autoubiquinated and degraded in response to restriction sensitive virus [53,56,62]. Despite these observations, TRIM5-mediated ubiquitination of HIV-1 capsid has not been detected. Understanding the restriction mechanism is confounded further by the fact that, if the block to reverse transcription is bypassed, replication steps after reverse transcription are also blocked by TRIM5 [63,64]. Particularly intriguing is the report that leptomycin traps the human and rhesus orthologues of TRIM5α in the nucleus [65], indicating that these proteins shuttle from the nucleus to the cytoplasm in a CRM1-dependent manner.
Innate immune signaling and TRIM5-mediated restriction
TRIM5 restriction activity is cell-type specific [66,67], and the isolation of restriction-defective, TRIM5-positive cell lines suggested that TRIM5 restriction requires specific host co-factors [68]. Additional clues about the TRIM5 restriction mechanism came from the finding that retroviruses are rescued from restriction by drugs that block signal transduction [67,69,70]. Indeed, TRIM5 promotes AP-1 and NF-κB signaling [39,71] (Figure 4), and knockdown of TRIM5 in dendritic cells prevents innate immune signaling downstream of LPS and other pathogen-associated molecular patterns [39].
Dissection of the mechanism by which TRIM5 promotes signaling showed that it acts with the E2 enzymes UBC13/UEV1A (also known as UBE2N–UBE2V1) to catalyze the synthesis of unattached, K63-linked ubiquitin chains that activate the TAK1 kinase complex and, consequently, the transcription of inflammatory cytokines [39] (Figure 5). It was then demonstrated that TAK1, UBC13 and UEV1A contribute to the restriction mechanism. Reciprocally, interaction with HIV-1 capsid lattice tubes, but not with free capsid protein or crosslinked capsid hexamers, greatly enhances the UBC13–UEV1A-dependent E3 activity of TRIM5 and the induction of inflammatory cytokines [39]. These findings demonstrate that TRIM5 is a pattern recognition receptor (PRR) specific for the protein lattice of the retrovirus capsid.
Figure 5.
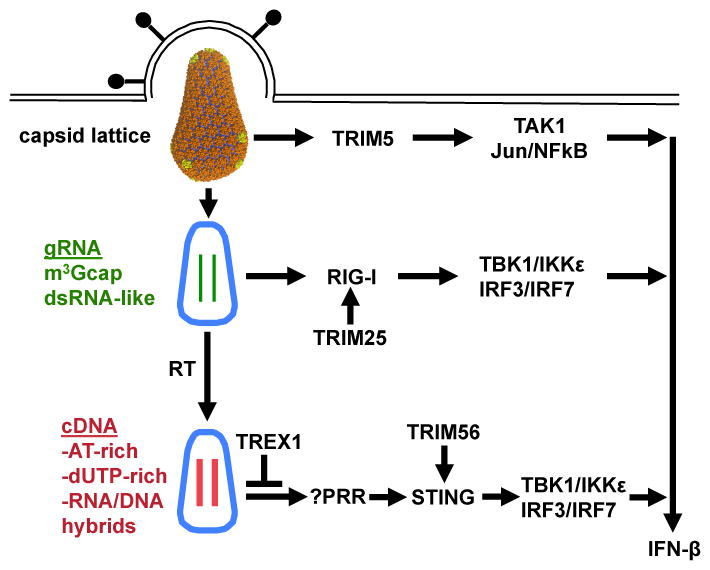
Schematic diagram showing involvement in the innate immune response to HIV-1 of some of the many TRIM proteins. The role of TRIM5 has been confirmed, and, based on the evidence, the roles of TRIM25 and TRIM56 are likely.
TRIM5 recognition of the capsid lattice stimulates its E3 ubiquitin ligase activity [39]. This activates TAK1 and downstream NF-κB and AP-1 factors. Such signalling is required for, but not sufficient to activate, interferon-beta transcription.
TRIM25 is required for signaling by the RIG-I pattern recognition receptor [81,82]. RIG-I responds to the highly, structured HIV-1 genomic RNA [79,80].
TRIM56 is required for STING-dependent signalling from the cytoplasmic pattern recognition receptor for DNA [75], that is activated when HIV-1 infects TREX1-deficient cells [74].
It is becoming clear that many TRIM family members regulate innate immune signaling, and that many of these TRIMs will be important HIV-1 regulators [72]. Recently it was shown that, when the TREX1 nuclease is disrupted, the viral cDNA of HIV-1 and other retroviruses is detected by an innate immune receptor in the host cell cytoplasm, resulting in strong induction of type 1 interferon [73,74]. The identity of the DNA receptor has not been determined, but the signaling pathway downstream of this DNA receptor requires K63-linked ubiquitination of STING by TRIM56 [74,75] (Figure 5).
HIV-1 genomic RNA is highly structured [76,77], and bears an unusual trimethylguanosine cap [78]. One can speculate that, under conditions analogous to those where TREX1 is disrupted, HIV-1 RNA would be detected by an innate immune sensor. Some studies suggest that RIG-I senses HIV-1 RNA and that the HIV-1 protease blocks innate immune signaling in response to HIV-1 RNA by binding and sequestering RIG-I [79,80]. Interestingly, RIG-I signaling is also activated by K63-linked ubiquitin, and this requires TRIM25 [81,82] (Figure 5).
Concluding remarks
The complementarity of the TRIM5 lattice for the capsid suggests that TRIM5 recognition involves low-affinity binding sites distributed across the capsid hexameric lattice [40]. This would explain how given TRIM5 orthologues restrict retroviruses bearing capsids of heterogeneous amino acid sequence and geometry. A full understanding of the restriction mechanism will necessitate solution of the full TRIM5 structure in complex with the capsid lattice. Restriction activity may involve premature disassembly of the capsid lattice with blockade of reverse transcription [10] and targeting of the TRIM5-capsid complex to the proteasome [60,62]. Alternatively, the TRIM5 lattice might stabilize the capsid and prevent uncoating or reverse transcription, or block capsid interaction with host machinery essential for subsequent steps in infection such as nuclear entry [63,64]. With either scenario, free K63 Ub chains generated by UBC13, UEV1A and capsid-hexamerized TRIM5 contribute to restriction activity by activating TAK1 [39]. Identification of TAK1 substrates, whether viral proteins or host effector proteins, will inform future attempts to clarify the mechanism of restriction. It will be interesting to determine if other HIV-1 restriction factors such as APOBEC3G and TETHERIN also signal in response to HIV-1 infection, or if restriction activity by these factors also depends upon any of the hundreds of other TRIM family members.
Highlights.
TRIM5, with UBC13/UEV1A, catalyzes the synthesis of free K63-linked ubiquitin chains.
TRIM5 activates the TAK1 (MAP3K7) complex
TAK1, UBC13, and UEV1A are required for TRIM5-mediated restriction
TRIM5 spontaneously forms a two-dimensional paracrystalline hexagonal lattice
HIV-1 capsid lattice promotes TRIM5 assembly into a lattice
Capsid lattice stimulates TRIM5 E3 activity and signal transduction
Acknowledgments
The authors thank Christophe Briand for generation of the figures. This work was supported by NIH grant RO1AI59159 and by Swiss National Science Foundation grants 3100A0-128655 and 310030-122342.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, Riganelli D, Zanaria E, Messali S, Cainarca S, et al. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nisole S, Stoye JP, Saïb A. TRIM family proteins: retroviral restriction and antiviral defence. Nat Rev Microbiol. 2005;3:799–808. doi: 10.1038/nrmicro1248. [DOI] [PubMed] [Google Scholar]
- 3•.Han K, Lou DI, Sawyer SL. Identification of a Genomic Reservoir for New TRIM Genes in Primate Genomes. PLoS Genet. 2011;7:e1002388. doi: 10.1371/journal.pgen.1002388. Identification of new human TRIM genes, bringing total number to nearly 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4•.Uchil PD, Quinlan BD, Chan W-T, Luna JM, Mothes W. TRIM E3 ligases interfere with early and late stages of the retroviral life cycle. PLoS Pathog. 2008;4:e16. doi: 10.1371/journal.ppat.0040016. Demonstration that many TRIM genes restrict retroviruses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matloubian M, Concepcion RJ, Ahmed R. CD4+ T cells are required to sustain CD8+ cytotoxic T-cell responses during chronic viral infection. J Virol. 1994;68:8056–8063. doi: 10.1128/jvi.68.12.8056-8063.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yap MW, Nisole S, Lynch C, Stoye JP. Trim5alpha protein restricts both HIV-1 and murine leukemia virus. Proc Natl Acad Sci USA. 2004;101:10786–10791. doi: 10.1073/pnas.0402876101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allouch A, Di Primio C, Alpi E, Lusic M, Arosio D, Giacca M, Cereseto A. The TRIM family protein KAP1 inhibits HIV-1 integration. Cell Host Microbe. 2011;9:484–495. doi: 10.1016/j.chom.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 8.Kajaste-Rudnitski A, Marelli SS, Pultrone C, Pertel T, Uchil PD, Mechti N, Mothes W, Poli G, Luban J, Vicenzi E. TRIM22 inhibits HIV-1 transcription independently of its E3 ubiquitin ligase activity, Tat, and NF-kappaB-responsive long terminal repeat elements. J Virol. 2011;85:5183–5196. doi: 10.1128/JVI.02302-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barr SD, Smiley JR, Bushman FD. The interferon response inhibits HIV particle production by induction of TRIM22. PLoS Pathog. 2008;4:e1000007. doi: 10.1371/journal.ppat.1000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10••.Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. Screen that identified TRIM5 as an HIV-1 restriction factor. [DOI] [PubMed] [Google Scholar]
- 11••.Sayah DM, Sokolskaja E, Berthoux L, Luban J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature. 2004;430:569–573. Screen that identified TRIM5 as an HIV-1 restriction factor. [Google Scholar]
- 12.Hofmann W, Schubert D, LaBonte J, Munson L, Gibson S, Scammell J, Ferrigno P, Sodroski J. Species-specific, postentry barriers to primate immunodeficiency virus infection. J Virol. 1999;73:10020–10028. doi: 10.1128/jvi.73.12.10020-10028.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Towers GJ, Hatziioannou T, Cowan S, Goff SP, Luban J, Bieniasz PD. Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat Med. 2003;9:1138–1143. doi: 10.1038/nm910. [DOI] [PubMed] [Google Scholar]
- 14••.Stremlau M, Perron M, Lee M, Li Y, Song B, Javanbakht H, Diaz-Griffero F, Anderson DJ, Sundquist WI, Sodroski J. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci USA. 2006;103:5514–5519. doi: 10.1073/pnas.0509996103. Biochemical demonstration that rhesusTRIM5 associates with HIV-1 capsid and decreases the amount of capsid protein that pellets from the cytoplasm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sokolskaja E, Berthoux L, Luban J. Cyclophilin A and TRIM5alpha independently regulate human immunodeficiency virus type 1 infectivity in human cells. J Virol. 2006;80:2855–2862. doi: 10.1128/JVI.80.6.2855-2862.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16•.Battivelli E, Lecossier D, Matsuoka S, Migraine J, Clavel F, Hance AJ. Strain-specific differences in the impact of human TRIM5alpha, different TRIM5alpha alleles, and the inhibition of capsid-cyclophilin A interactions on the infectivity of HIV-1. J Virol. 2010;84:11010–11019. doi: 10.1128/JVI.00758-10. Demonstration that some primary HIV-1 isolates are inhibited quite strongly by human TRIM5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sewram S, Singh R, Kormuth E, Werner L, Mlisana K, Karim SSA, Ndung’u T CAPRISA Acute Infection Study Team. Human TRIM5alpha expression levels and reduced susceptibility to HIV-1 infection. J INFECT DIS. 2009;199:1657–1663. doi: 10.1086/598861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Manen D, Rits MAN, Beugeling C, van Dort K, Schuitemaker H, Kootstra NA. The effect of Trim5 polymorphisms on the clinical course of HIV-1 infection. PLoS Pathog. 2008;4:e18. doi: 10.1371/journal.ppat.0040018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu F-L, Qiu Y-Q, Li H, Kuang Y-Q, Tang X, Cao G, Tang NLS, Zheng Y-T. An HIV-1 resistance polymorphism in TRIM5α gene among Chinese intravenous drug users. J Acquir Immune Defic Syndr. 2011;56:306–311. doi: 10.1097/QAI.0b013e318205a59b. [DOI] [PubMed] [Google Scholar]
- 20.Price H, Lacap P, Tuff J, Wachihi C, Kimani J, Ball TB, Luo M, Plummer FA. A TRIM5alpha exon 2 polymorphism is associated with protection from HIV-1 infection in the Pumwani sex worker cohort. AIDS. 2010;24:1813–1821. doi: 10.1097/QAD.0b013e32833b5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Battivelli E, Migraine J, Lecossier D, Yeni P, Clavel F, Hance AJ. Gag cytotoxic T lymphocyte escape mutations can increase sensitivity of HIV-1 to human TRIM5alpha, linking intrinsic and acquired immunity. J Virol. 2011;85:11846–11854. doi: 10.1128/JVI.05201-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reynolds MR, Sacha JB, Weiler AM, Borchardt GJ, Glidden CE, Sheppard NC, Norante FA, Castrovinci PA, Harris JJ, Robertson HT, et al. The TRIM5{alpha} genotype of rhesus macaques affects acquisition of simian immunodeficiency virus SIVsmE660 infection after repeated limiting-dose intrarectal challenge. J Virol. 2011;85:9637–9640. doi: 10.1128/JVI.05074-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yeh WW, Rao SS, Lim S-Y, Zhang J, Hraber PT, Brassard LM, Luedemann C, Todd JP, Dodson A, Shen L, et al. The TRIM5 gene modulates penile mucosal acquisition of simian immunodeficiency virus in rhesus monkeys. J Virol. 2011;85:10389–10398. doi: 10.1128/JVI.00854-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lim S-Y, Chan T, Gelman RS, Whitney JB, O’Brien KL, Barouch DH, Goldstein DB, Haynes BF, Letvin NL. Contributions of Mamu-A*01 status and TRIM5 allele expression, but not CCL3L copy number variation, to the control of SIVmac251 replication in Indian-origin rhesus monkeys. PLoS Genet. 2010;6:e1000997. doi: 10.1371/journal.pgen.1000997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25•.Kirmaier A, Wu F, Newman RM, Hall LR, Morgan JS, O’Connor S, Marx PA, Meythaler M, Goldstein S, Buckler-White A, et al. TRIM5 suppresses cross-species transmission of a primate immunodeficiency virus and selects for emergence of resistant variants in the new species. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000462. Demonstration of the capsid-specific effects of TRIM5 on SIV infectivity in rhesus macaques. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim S-Y, Rogers T, Chan T, Whitney JB, Kim J, Sodroski J, Letvin NL. TRIM5alpha Modulates Immunodeficiency Virus Control in Rhesus Monkeys. PLoS Pathog. 2010;6:e1000738. doi: 10.1371/journal.ppat.1000738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grütter C, Briand C, Capitani G, Mittl PRE, Papin S, Tschopp J, Grütter MG. Structure of the PRYSPRY-domain: implications for autoinflammatory diseases. FEBS Lett. 2006;580:99–106. doi: 10.1016/j.febslet.2005.11.076. [DOI] [PubMed] [Google Scholar]
- 28.James LC, Keeble AH, Khan Z, Rhodes DA, Trowsdale J. Structural basis for PRYSPRY-mediated tripartite motif (TRIM) protein function. Proc Natl Acad Sci USA. 2007;104:6200–6205. doi: 10.1073/pnas.0609174104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woo J-S, Imm J-H, Min C-K, Kim K-J, Cha S-S, Oh B-H. Structural and functional insights into the B30. 2/SPRY domain. EMBO J. 2006;25:1353–1363. doi: 10.1038/sj.emboj.7600994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weinert C, Grütter C, Roschitzki-Voser H, Mittl PRE, Grütter MG. The crystal structure of human pyrin b30. 2 domain: implications for mutations associated with familial Mediterranean fever. J Mol Biol. 2009;394:226–236. doi: 10.1016/j.jmb.2009.08.059. [DOI] [PubMed] [Google Scholar]
- 31.Sawyer SL, Wu LI, Emerman M, Malik HS. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc Natl Acad Sci USA. 2005;102:2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luban J. Cyclophilin A, TRIM5, and resistance to human immunodeficiency virus type 1 infection. J Virol. 2007;81:1054–1061. doi: 10.1128/JVI.01519-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Battivelli E, Migraine J, Lecossier D, Matsuoka S, Perez-Bercoff D, Saragosti S, Clavel F, Hance AJ. Modulation of TRIM5alpha activity in human cells by alternatively spliced TRIM5 isoforms. J Virol. 2011;85:7828–7835. doi: 10.1128/JVI.00648-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34••.Sebastian S, Luban J. TRIM5alpha selectively binds a restriction-sensitive retroviral capsid. Retrovirology. 2005;2:40. doi: 10.1186/1742-4690-2-40. First demonstration of capsid-specific binding by the PRYSPRY domain of TRIM5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35•.Zhao G, Ke D, Vu T, Ahn J, Shah VB, Yang R, Aiken C, Charlton LM, Gronenborn AM, Zhang P. Rhesus TRIM5α disrupts the HIV-1 capsid at the inter-hexamer interfaces. PLoS Pathog. 2011;7:e1002009. doi: 10.1371/journal.ppat.1002009. Demonstration that TRIM5 disrupts the HIV-1 capsid lattice at the trimer interface. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36••.Pornillos O, Ganser-Pornillos BK, Kelly BN, Hua Y, Whitby FG, Stout CD, Sundquist WI, Hill CP, Yeager M. X-ray structures of the hexameric building block of the HIV capsid. Cell. 2009;137:1282–1292. doi: 10.1016/j.cell.2009.04.063. Atomic resolution structure of the HIV-1 capsid hexamer. Demonstration of the utility of cysteine crosslinking technique for studies of HIV-1 capsid structure. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37•.Byeon I-JL, Meng X, Jung J, Zhao G, Yang R, Ahn J, Shi J, Concel J, Aiken C, Zhang P, et al. Structural convergence between Cryo-EM and NMR reveals intersubunit interactions critical for HIV-1 capsid function. Cell. 2009;139:780–790. doi: 10.1016/j.cell.2009.10.010. Identification of the CTD-CTD interface at the three-fold axis of symmetry in the HIV-1 capsid lattice. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38••.Pornillos O, Ganser-Pornillos BK, Yeager M. Atomic-level modelling of the HIV capsid. Nature. 2011;469:424–427. doi: 10.1038/nature09640. Complete, atomic-resolution model of the HIV-1 capsid protein lattice in the virion core. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39••.Pertel T, Hausmann S, Morger D, Züger S, Guerra J, Lascano J, Reinhard C, Santoni FA, Uchil PD, Chatel L, et al. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature. 2011;472:361–365. doi: 10.1038/nature09976. Showed that TRIM5 regulates innate immune signaling, that it is a pattern recognition receptor specific for the retroviral capsid lattice, that E3 ubiquitin ligase activity, TAK1, UBC13, and UEV1A are required for restriction activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40••.Ganser-Pornillos BK, Chandrasekaran V, Pornillos O, Sodroski JG, Sundquist WI, Yeager M. Hexagonal assembly of a restricting TRIM5alpha protein. Proceedings of the National Academy of Sciences. 2011;108:534–539. doi: 10.1073/pnas.1013426108. Showed that TRIM5 forms a lattice that is complementary to the retroviral capsid lattice. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Campbell EM, Perez O, Anderson JL, Hope TJ. Visualization of a proteasome-independent intermediate during restriction of HIV-1 by rhesus TRIM5alpha. J Cell Biol. 2008;180:549–561. doi: 10.1083/jcb.200706154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Newman RM, Hall L, Kirmaier A, Pozzi L-A, Pery E, Farzan M, O’Neil SP, Johnson W. Evolution of a TRIM5-CypA splice isoform in old world monkeys. PLoS Pathog. 2008;4:e1000003. doi: 10.1371/journal.ppat.1000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Virgen CA, Kratovac Z, Bieniasz PD, Hatziioannou T. Independent genesis of chimeric TRIM5-cyclophilin proteins in two primate species. Proceedings of the National Academy of Sciences. 2008;105:3563–3568. doi: 10.1073/pnas.0709258105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson SJ, Webb BLJ, Ylinen LMJ, Verschoor E, Heeney JL, Towers GJ. Independent evolution of an antiviral TRIMCyp in rhesus macaques. Proceedings of the National Academy of Sciences. 2008;105:3557–3562. doi: 10.1073/pnas.0709003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brennan G, Kozyrev Y, Hu S-L. TRIMCyp expression in Old World primates Macaca nemestrina and Macaca fascicularis. Proceedings of the National Academy of Sciences. 2008;105:3569–3574. doi: 10.1073/pnas.0709511105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luban J, Bossolt KL, Franke EK, Kalpana GV, Goff SP. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell. 1993;73:1067–1078. doi: 10.1016/0092-8674(93)90637-6. [DOI] [PubMed] [Google Scholar]
- 47.Gamble TR, Vajdos FF, Yoo S, Worthylake DK, Houseweart M, Sundquist WI, Hill CP. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell. 1996;87:1285–1294. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- 48.Neagu MR, Ziegler P, Pertel T, Strambio-De-Castillia C, Grütter C, Martinetti G, Mazzucchelli L, Grütter M, Manz MG, Luban J. Potent inhibition of HIV-1 by TRIM5-cyclophilin fusion proteins engineered from human components. J Clin Invest. 2009;119:3035–3047. doi: 10.1172/JCI39354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ansari H, Greco G, Luban J. Cyclophilin A peptidyl-prolyl isomerase activity promotes ZPR1 nuclear export. Mol Cell Biol. 2002;22:6993–7003. doi: 10.1128/MCB.22.20.6993-7003.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yin L, Boussard S, Allan J, Luban J. The HIV type 1 replication block in nonhuman primates is not explained by differences in cyclophilin A primary structure. AIDS Res Hum Retroviruses. 1998;14:95–97. doi: 10.1089/aid.1998.14.95. [DOI] [PubMed] [Google Scholar]
- 51.Nisole S, Lynch C, Stoye JP, Yap MW. A Trim5-cyclophilin A fusion protein found in owl monkey kidney cells can restrict HIV-1. Proc Natl Acad Sci USA. 2004;101:13324–13328. doi: 10.1073/pnas.0404640101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52••.Price AJ, Marzetta F, Lammers M, Ylinen LMJ, Schaller T, Wilson SJ, Towers GJ, James LC. Active site remodeling switches HIV specificity of antiretroviral TRIMCyp. Nat Struct Mol Biol. 2009;16:1036–1042. doi: 10.1038/nsmb.1667. Demonstration that a single amino acid change in cyclophilin A completely changes the mode of capsid binding. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Diaz-Griffero F, Li X, Javanbakht H, Song B, Welikala S, Stremlau M, Sodroski J. Rapid turnover and polyubiquitylation of the retroviral restriction factor TRIM5. Virology. 2006;349:300–315. doi: 10.1016/j.virol.2005.12.040. [DOI] [PubMed] [Google Scholar]
- 54.Kar AK, Diaz-Griffero F, Li Y, Li X, Sodroski J. Biochemical and biophysical characterization of a chimeric TRIM21-TRIM5alpha protein. J Virol. 2008;82:11669–11681. doi: 10.1128/JVI.01559-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55•.Langelier CR, Sandrin V, Eckert DM, Christensen DE, Chandrasekaran V, Alam SL, Aiken C, Olsen JC, Kar AK, Sodroski JG, et al. Biochemical characterization of a recombinant TRIM5alpha protein that restricts human immunodeficiency virus type 1 replication. J Virol. 2008;82:11682–11694. doi: 10.1128/JVI.01562-08. First biochemical characterization of a full-length, soluble, TRIM5-surrogate protein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56•.Lienlaf M, Hayashi F, Di Nunzio F, Tochio N, Kigawa T, Yokoyama S, Diaz-Griffero F. Contribution of E3-ubiquitin ligase activity to HIV-1 restriction by TRIM5alpha(rh): structure of the RING domain of TRIM5alpha. J Virol. 2011;85:8725–8737. doi: 10.1128/JVI.00497-11. NMR structure of the E3 RING domain and demonstration that E3 ubiquitin ligase activity is required for restriction activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li X, Sodroski J. The TRIM5alpha B-box 2 domain promotes cooperative binding to the retroviral capsid by mediating higher-order self-association. J Virol. 2008;82:11495–11502. doi: 10.1128/JVI.01548-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58•.Diaz-Griffero F, Qin X-R, Hayashi F, Kigawa T, Finzi A, Sarnak Z, Lienlaf M, Yokoyama S, Sodroski J. A B-box 2 surface patch important for TRIM5alpha self-association, capsid binding avidity, and retrovirus restriction. J Virol. 2009;83:10737–10751. doi: 10.1128/JVI.01307-09. NMR structure and function of the TRIM5 B-box 2 domain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sastri J, O’Connor C, Danielson CM, McRaven M, Perez P, Diaz-Griffero F, Campbell EM. Identification of residues within the L2 region of rhesus TRIM5alpha that are required for retroviral restriction and cytoplasmic body localization. Virology. 2010;405:259–266. doi: 10.1016/j.virol.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60•.Lukic Z, Hausmann S, Sebastian S, Rucci J, Sastri J, Robia SL, Luban J, Campbell EM. TRIM5alpha associates with proteasomal subunits in cells while in complex with HIV-1 virions. Retrovirology. 2011;8:93. doi: 10.1186/1742-4690-8-93. Demonstration that TRIM5 associates with proteasomes via the proteasome protein PSMC2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O’Connor C, Pertel T, Gray S, Robia SL, Bakowska JC, Luban J, Campbell EM. p62/sequestosome-1 associates with and sustains the expression of retroviral restriction factor TRIM5alpha. J Virol. 2010;84:5997–6006. doi: 10.1128/JVI.02412-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rold CJ, Aiken C. Proteasomal degradation of TRIM5alpha during retrovirus restriction. PLoS Pathog. 2008;4:e1000074. doi: 10.1371/journal.ppat.1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Berthoux L, Sebastian S, Sokolskaja E, Luban J. Lv1 inhibition of human immunodeficiency virus type 1 is counteracted by factors that stimulate synthesis or nuclear translocation of viral cDNA. J Virol. 2004;78:11739–11750. doi: 10.1128/JVI.78.21.11739-11750.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu X, Anderson JL, Campbell EM, Joseph AM, Hope TJ. Proteasome inhibitors uncouple rhesus TRIM5alpha restriction of HIV-1 reverse transcription and infection. Proc Natl Acad Sci USA. 2006;103:7465–7470. doi: 10.1073/pnas.0510483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Diaz-Griffero F, Gallo DE, Hope TJ, Sodroski J. Trafficking of some old world primate TRIM5α proteins through the nucleus. Retrovirology. 2011;8:38. doi: 10.1186/1742-4690-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bérubé J, Bouchard A, Berthoux L. Both TRIM5alpha and TRIMCyp have only weak antiviral activity in canine D17 cells. Retrovirology. 2007;4:68. doi: 10.1186/1742-4690-4-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sebastian S, Sokolskaja E, Luban J. Arsenic counteracts human immunodeficiency virus type 1 restriction by various TRIM5 orthologues in a cell type-dependent manner. J Virol. 2006;80:2051–2054. doi: 10.1128/JVI.80.4.2051-2054.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sayah DM, Luban J. Selection for loss of Ref1 activity in human cells releases human immunodeficiency virus type 1 from cyclophilin A dependence during infection. J Virol. 2004;78:12066–12070. doi: 10.1128/JVI.78.21.12066-12070.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Berthoux L, Towers GJ, Gurer C, Salomoni P, Pandolfi PP, Luban J. As(2)O(3) enhances retroviral reverse transcription and counteracts Ref1 antiviral activity. J Virol. 2003;77:3167–3180. doi: 10.1128/JVI.77.5.3167-3180.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kapahi P, Takahashi T, Natoli G, Adams SR, Chen Y, Tsien RY, Karin M. Inhibition of NF-kappa B activation by arsenite through reaction with a critical cysteine in the activation loop of Ikappa B kinase. J Biol Chem. 2000;275:36062–36066. doi: 10.1074/jbc.M007204200. [DOI] [PubMed] [Google Scholar]
- 71.Tareen SU, Emerman M. Human Trim5α has additional activities that are uncoupled from retroviral capsid recognition. Virology. 2011;409:113–120. doi: 10.1016/j.virol.2010.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kawai T, Akira S. Regulation of innate immune signalling pathways by the tripartite motif (TRIM) family proteins. EMBO Mol Med. 2011;3:513–527. doi: 10.1002/emmm.201100160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74••.Yan N, Regalado-Magdos AD, Stiggelbout B, Lee-Kirsch MA, Lieberman J. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat Immunol. 2010;11:1005–1013. doi: 10.1038/ni.1941. First clear demonstration that HIV-1 replication activates cytoplasmic pattern recognition receptors and type 1 interferon. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tsuchida T, Zou J, Saitoh T, Kumar H, Abe T, Matsuura Y, Kawai T, Akira S. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity. 2010;33:765–776. doi: 10.1016/j.immuni.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 76.Lu K, Heng X, Garyu L, Monti S, Garcia EL, Kharytonchyk S, Dorjsuren B, Kulandaivel G, Jones S, Hiremath A, et al. NMR detection of structures in the HIV-1 5′-leader RNA that regulate genome packaging. Science. 2011;334:242–245. doi: 10.1126/science.1210460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Swanstrom R, Burch CL, Weeks KM. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yedavalli VSRK, Jeang K-T. Trimethylguanosine capping selectively promotes expression of Rev-dependent HIV-1 RNAs. Proc Natl Acad Sci USA. 2010;107:14787–14792. doi: 10.1073/pnas.1009490107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Solis M, Nakhaei P, Jalalirad M, Lacoste J, Douville R, Arguello M, Zhao T, Laughrea M, Wainberg MA, Hiscott J. RIG-I-mediated antiviral signaling is inhibited in HIV-1 infection by a protease-mediated sequestration of RIG-I. J Virol. 2011;85:1224–1236. doi: 10.1128/JVI.01635-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Berg RK, Melchjorsen J, Rintahaka J, Diget E, Søby S, Horan KA, Gorelick RJ, Matikainen S, Larsen CS, Ostergaard L, et al. Genomic HIV RNA Induces Innate Immune Responses through RIG-I-Dependent Sensing of Secondary-Structured RNA. PLoS ONE. 2012;7:e29291. doi: 10.1371/journal.pone.0029291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81••.Gack MU, Shin YC, Joo C-H, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. Critical demonstration that TRIM25 regulates RIG-I. [DOI] [PubMed] [Google Scholar]
- 82••.Zeng W, Sun L, Jiang X, Chen X, Hou F, Adhikari A, Xu M, Chen ZJ. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–330. doi: 10.1016/j.cell.2010.03.029. Development of in vitro assay for RIG-I activation by K63-linked ubiquitin chains synthesized by TRIM25. [DOI] [PMC free article] [PubMed] [Google Scholar]