Abstract
A membrane alignment technique has been used to measure the distance between two TOAC nitroxide spin labels on the membrane-spanning M2δ, peptide of the nicotinic acetylcholine receptor (AChR), via CW-EPR spectroscopy. The TOAC-labeled M2δ peptides were mechanically aligned using DMPC lipids on a planar quartz support, and CW-EPR spectra were recorded at specific orientations. Global analysis in combination with rigorous spectral simulation was used to simultaneously analyze data from two different sample orientations for both single- and double-labeled peptides. We measured an internitroxide distance of 14.6 Å from a dual TOAC-labeled AChR M2δ peptide at positions 7 and 13 that closely matches with the 14.5 Å distance obtained from a model of the labeled AChR M2δ peptide. In addition, the angles determining the relative orientation of the two nitroxides have been determined, and the results compare favorably with molecular modeling. The global analysis of the data from the aligned samples gives much more precise estimates of the parameters defining the geometry of the two labels than can be obtained from a randomly dispersed sample.
Keywords: Dipolar Broadening CW-EPR, Aligned DMPC Bicelles, Distance measurement
Introduction
Site-directed spin labeling (SDSL) in conjunction with electron paramagnetic resonance (EPR) spectroscopy can be used to study the structural and dynamic properties of peptides, proteins, and nucleic acids 1-4. Distance measurements between two strategically placed nitroxide spin labels can provide pertinent structural information on complicated biological systems such as membrane proteins. Distances of approximately 8 Å - 25 Å can be determined by measuring line broadening due to dipolar coupling using continuous wave (CW)-EPR 5-7. Longer distances (20 Å-80 Å) can be determined using the pulsed EPR technique Double Electron Electron Resonance (DEER) 8-13. For the dipolar line broadening experiments, the distances can be estimated by spectral analysis based on Fourier convolution/deconvolution methods 7,14-16 or by rigorous spectral simulations taking into account both interspin distance and the relative orientation of the nitroxide probes 5,6,17,18.
Typically, the distance measurements are conducted using a flexible methanethiosulfonate (MTSL) in a frozen unoriented powder sample. Due to distributions in the rotameric states of the MTSL labels, both CW-EPR spectra 14 and DEER data 8 are usually fit assuming an isotropic distribution of the relative orientation between the two labels and the distance distribution obtained has significant width. On the other hand, the TOAC (2,2,6,6-tetramethylpiperidine-1-oxyl-4-amino-4-carboxylic acid) label which is directly coupled to the protein backbone has been shown to give a unique distance and orientation between the two nitroxides without disturbing the structure and function of helices in the case of a soluble peptide 5. The use of TOAC labels has the potential to provide more powerful constraints for the building of structural models.
Previous studies have also demonstrated the advantages of aligning a TOAC spin labeled peptide within a lipid bilayer at different orientations with respect to the static magnetic field 19,20. Using this approach, the orientation of a spin-labeled peptide with respect to the membrane normal can be determined. This EPR technique is comparable to 15N solid-state NMR techniques that are able to probe membrane protein/peptide topology. Understanding the location or orientation of a membrane protein with respect to the lipid bilayer is important to characterize structure and function. The novel approach of this study is the combination of doubly TOAC-labeled peptides in aligned lipid bilayers to investigate the dipolar coupling between labels. The aligned anisotropic EPR spectra reveal linewidths that are sharper than unoriented powder spectra, allowing distance and orientation information to be obtained more precisely when data from two different sample orientations are globally analyzed.
The AChR M2δ peptide was used as a model membrane peptide system to demonstrate this TOAC alignment distance approach. AChR M2δ is a transmembrane peptide that forms a pore-lining region of the ligand-gated ion channel nicotinic acetylcholine receptor (AChR). It is a 23-residue peptide, which forms an α-helical structure in phospholipid bilayers with a helical tilt of 12° with respect to the membrane normal 21,22. AChR is a 290 kDa protein that forms a cation-specific ion channel in neurons or muscles by arranging its five subunits (α2βγδ) symmetrically around a central pore 23,24. Each of these five subunits consists of four transmembrane segments (M1-M4) and a large extracellular N-terminal domain of nearly 200 amino acids 24,25. The M2 segment of the protein from each five subunits contributes to ion channel pore formation.
We measured a mid-range (< 25 Å) distance between two TOAC spin labels on a membrane peptide using mechanically aligned glass plates. The rigorous spectral simulation method pioneered by Hustedt and Beth was used to analyze data to give the distance and relative orientation between two TOAC nitroxides 5,6,26. The experimental distance and the relative orientations of the TOAC nitroxides were compared with unoriented spectra and molecular modeling studies of dual TOAC-labeled AChR M2δ peptide. The results reveal that TOAC SL-SL distances and relative orientations can be measured more accurately and precisely when the samples are aligned with respect to the magnetic field.
Experimental Methods
Materials
1,2-Dimyristoyl-sn-glycero-3-phosphocholine (DMPC) pre-dissolved in chloroform was obtained from Avanti Polar Lipids (Alabaster, AL). Deuterium-depleted water was purchased from Isotec (Miamisburg, OH). Fmoc-Arg (Pbf)-Nova Syn TGA resin (0.23 mmol/g), 9-Fluorenylmethyl-oxycarbonyl-O-succinimide (Fmoc-OSu) and all amino acids were purchased from Novabiochem, San Diego, CA. Triisopropylsilane (TIS), anisole, hexafluro-2-propanol (HFIP), 30% ammonium hydroxide [NH3 (aq.)], piperidine were purchased from Sigma-Aldrich, St. Louis, MO. 2,2,6,6-Tetramethylpiperidine-1-oxyl-4-carboxylic acid (TOAC) was purchased from Acros Organics, Belgium. Trifluoroacetic acid (TFA), 1-hydroxybenzo-triazole (HOBt), (2-(7-Aza-1H-benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HATU), diisopropylethylamine (DIEA), M-methyl-pyrrolidone (NMP), and dichloromethane (DCM) were purchased from Applied Biosystems, Foster City, CA. Acetic anhydride (Ac2O) and methyl-tert-butyl ether were purchased from Fisher Scientific, Pittsburgh, PA.
Peptide synthesis, purification and characterization
Solid-phase peptide synthesis was carried out on a ABI 433A peptide synthesizer (Applied Biosystems Inc., Foster City, CA) using Fmoc-protection chemistry. The amino acid sequences of the pore-lining transmembrane region of acetylcholine receptor (AChR M2 domain, δ subunit peptide) from Torpedo californica which were used in this study are
| 1EKMSTAISVLLAQAVCLLLTSQ23R | wild-type (WT) |
| 1EKMSTA7XSVLLAQAVCLLLTSQ23R | singly-labeled TOAC7 |
| 1EKMSTAISVLLA13XAVCLLLTSQ23R | singly-labeled TOAC13 |
| 1EKMSTA7XSVLLA13XAVCLLLTSQ23R | doubly-labeled |
where, X represents the location of the TOAC spin label.
The 433A peptide synthesizer connected with a UV-detector (wavelength 301 nm) was used to monitor the removal of the Fmoc-group from the N-terminus of the growing peptide. A modified version of 0.1 mmol Fmoc chemistry protocol in the SynthAssist 2.0 software from Applied Biosystems was used for peptide synthesis. The software modifications were made to include special functions such as double coupling, increased reaction times, and incorporation of unnatural amino acids in order to facilitate the optimal synthesis of peptides. All amino acids were purchased as Fmoc-protected and side-chain-protected form to minimize the unnecessary reactions during peptide synthesis. For site-specific TOAC labeling by solid-phase peptide synthesis, the amino terminal of the TOAC was protected with a Fmoc group. The synthesis of Fmoc-TOAC was carried out following standard procedures from the literature 27,28. The TOAC-labeled AChR M2δ peptides were synthesized following the standard protocol published in the literature 29-31. To optimize the peptide synthesis, the superior coupling agent HATU/HOAt 32 was used for attaching the TOAC spin label and surrounding amino acids (2 preceding the TOAC and 3-4 subsequent) with extended coupling times. This step was necessary to ensure the attachment of the bulky TOAC spin label and to prevent the peptide truncation just after the TOAC attachment due to low nucleophilicity reaction of the TOAC amino group in a peptide chain 29,33. The rest of the amino acids were attached using HBTU/HOBt coupling reagent.
About 150 mg of peptide resin was treated with a cleavage mixture [8.5 mL TFA (trifluoroacetic acid), 0.5 mL TIS (triisopropyl silane), 0.5 mL anisole, and 0.5 mL distilled water] for 3 hours to remove the side chains and detach peptides from the resin beads. The peptide mixture was filtered, and the resins were rinsed with TFA and removed. The filtrate was concentrated and precipitated in ice-cold methyl tert-butyl ether. The peptides were collected by centrifugation and vacuum dried overnight. The crude peptides were purified on Amersham Pharmacia Biotech AKTA Explorer 10S HPLC system using a reversed phase C-18 semiprep column (Vydac cat. # 218TP1010, 10 μm, 10 mm × 250 mm). The HPLC solvent system for peptide purification contained H2O with 0.1% TFA as solvent A and the mixture of 90% acetonitrile (MeCN), and 10% water with 0.1% TFA as solvent B. The TOAC7 peptide was double purified with a slightly modified solvent system on a reversed phase C-4 semiprep column (Vydac cat. # 214TP1010, 10 μm, 10mm × 250 mm) to obtain high purity. Since trifluoroacetic acid converts nitroxide spin label into hydroxylamine form during peptide cleavage and purification, the purified peptides were dissolved in 70:30 n-propanol:water mixture and treated with a few drops of 10% aq. ammonia (pH=9.5) for 3 hours to regenerate the radical signal of the TOAC nitroxide. The purity of the peptides were confirmed by matrix assisted laser desorption ionization time of flight (MALDI-TOF) mass spectrometry to be ~ 95%.
Sample Preparation
The mechanically aligned glass plate samples were made following the standard procedures described in the literature 19,21. Briefly, about 75 μg of labeled peptide (diluted 9-times with the wild-type peptide) was dissolved in 50 μL, 1:1 mixture of TFE and chloroform and mixed with 5 mg of DMPC phospholipid pre-dissolved in chloroform (250 μL, conc. 20 mg/mL). The molar peptide to lipid ratio was 1;250. The spin-labeled peptide was diluted with wild-type to minimize the effect of inter-monomer dipolar couplings. The lipid peptide mixture was subjected to a slow stream of N2-gas to reduce the final volume to 50-60 μL and then 10 μL of this peptide lipid mixture was uniformly plated on 5 glass plates (dimension: 5.7 × 12 mm, Marienfeld GmbH & Co.KG, Germany). The lipids on the glass plates were air-dried for approximately 10 minutes and then vacuum dried overnight in a desiccator. The glass plates were then hydrated with 4 μL of deuterium-depleted water and mounted in a stack. The stacked glass plates were kept for 12 hr in a hydration chamber of saturated ammonium phosphate monobasic solution (temperature 42°C, relative humidity 93%). The next day, the stacked glass plates were covered by double-sided tape and mounted on a glass rod with a flat quartz end and inserted into an EPR cavity to record CW-EPR spectra. Multilamellar vesicles were prepared from the same composition of peptide-lipid mixture in a glass tube (12 × 75 mm) and drying the sample completely in a stream of nitrogen gas followed by overnight drying in vacuum pump desiccators. Subsequently, 100 μL of deuterium-depleted water was added to the tube, and the sample was vortexed constantly. Periodic sample heating was applied by immersing the tube into a hot water bath at 60 °C to achieve complete dissolution of the lipids. The multilamellar vesicles suspension was drawn into a capillary tube, sealed and CW-EPR spectra were recorded in standard quartz EPR tube (Wilmad, 707-SQ-250M).
EPR Spectroscopy
EPR experiments were performed on a Bruker EMX X-band CW-EPR spectrometer consisting of ER041xG microwave bridge and ER4119-HS cavity coupled with a BVT 3000 nitrogen gas temperature controller (temperature stability ± 0.2K) at 9.434 GHz microwave frequency, 10 mW microwave power, 100 kHz modulation frequency, and 1.0 G modulation amplitude. EPR spectra were recorded aligning the glass plate samples with the membrane normal, π, either parallel or perpendicular to the direction of the static magnetic field, B0. A 42-s field sweep scan with a central field of 3400 G and sweep-width of 100 G was applied. All EPR spectra were collected at 318 K. At 318 K, DMPC exists in the liquid crystalline phase and the alignment of the DMPC bilayers is optimal 30.
Analysis of EPR Spectra
The EPR spectra of randomly-dispersed singly-labeled peptides were analyzed using a previously described approach 34 to determine the values of the diagonalized A- and g-tensors for the TOAC label at the two sites. These tensor values were then used in all subsequent analyses.
A simulation algorithm was developed for calculating the spectrum of a spin label on a helix aligned in a bilayer. In this algorithm, the orientation dependent elements of the nitroxide A- and g-tensors are calculated using Equation 1,
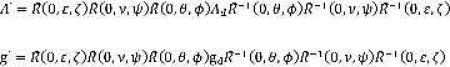 |
(1) |
where the angles θ and φ determine the orientation of the nitroxide with respect to the helix axis, the angle ν determines the helix tilt with respect to the membrane normal, the angle ψ determines the rotation about the helix axis in the membrane, the angle ε is the angle between the magnetic field and the membrane normal (the sample orientation), and ζ determines the rotation of the tilted helix about the membrane normal. The Euler rotation matrix R̂ used here was previously defined 17. In principle, any of these angles can either assume a single value or adopt a distribution of values. In practice, for the transmembrane helix in this study, the best fits to the data were obtained using a single value for all of the angles except for ζ and ψ. For ν = 0°, the angles ζ and ψ are degenerate. In this work, it was assumed that ν = 12° 21 breaking this degeneracy. Best-fits were obtained for the angle ζ uniformly distributed between 0° and 360° indicating that there is no net grain in the alignment of the slightly tilted peptides. The disorder in angle ψ is modeled by a Gaussian distribution with mean given by ψ0 and standard deviation given by σψ. The elements of the A- and g-tensors were used to calculate EPR spectra using a standard approach 35.
A second simulation algorithm was developed for calculating the spectrum of dipolar-coupled spin labels on a helix aligned in a bilayer. Using an approach similar to that used previously 6, the A- and g-tensors for nitroxide 1 were calculated using Equation 1 above. The A- and g-tensors for nitroxide 2 were calculated using Equation 2,
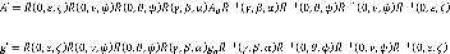 |
(2) |
where the angles α, β, and γ determine the orientation of nitroxide 2 with respect to nitroxide 1. The unique element of the dipolar coupling tensor is given by
| (3) |
where μ is the angle between the interelectron vector and the DC magnetic field,
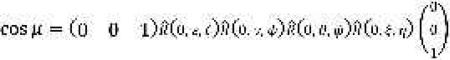 |
(4) |
and the angles ξ and η determine the orientation of with respect to nitroxide 1.
These algorithms were incorporated into a global analysis program for the non-linear least-squares analysis of multiple data sets utilizing the Marquardt-Levenberg algorithm to find the optimal set of parameters to minimize χ2 5,6,17,35. This approach allows the simultaneous fitting of data sets obtained at two different sample orientations (ε = 0° and ε= 90°).
For the case of singly-labeled aligned peptides the fit parameters are the angles φ, θ, ψ0, and σψ. For the case of the doubly-labeled peptides, the values of these parameters were fixed to those obtained for one of the singly-labeled peptides, while the values of the angles α, β, γ, ξ, and η together with the interelectron distance R were varied to optimize the fit to the data. In both cases, the Gaussian and Lorentzian components of the linewidths are allowed to vary independently for the two different sample orientations. A variable fraction, fiso, of a non-aligned isotropic component is included in each fit. Finally, there is an additional splitting evident in the experimental spectra that is presumed to be due to a fraction of the nitroxides which are hydrogen bonded 36,37. This additional splitting is treated by, in effect, convolving the simulation with a stick spectrum with three peaks at 0 and ±δ Gauss with amplitudes of 1-2κ and κ respectively.
Confidence intervals for the various fit parameters were obtained by fixing each particular parameter to a linear sequence of values and allowing all of the other fit parameters to vary to find the optimal fit. This determines how χ2 varies as a function of a particular parameter and thus gives a measure of the uncertainty in the parameter value.
Modeling studies of TOAC labeled peptides
The NMR structure of the AChR M2δ peptide (PDB ID: 1EQ8) was used as the starting coordinates for molecular modeling studies 21. The TOAC spin label was incorporated into the specific sites of the peptide using AMBER94 force field parameters within the MolMol program 38. The TOAC-labeled AChR M2δ peptides were modeled using visual molecular dynamics modeling (VMD) software 39. From these models, estimates of the angles and interelectron distance were obtained for comparison with the experimental results.
First, the models of the singly-labeled helices were rotated so that the helix axis was aligned with the z-axis of the laboratory frame, and then the nitroxide nitrogen was translated to the origin of the coordinate system. The vector was defined to be the coordinates of the nitroxide oxygen, and the vectors and were defined to be the coordinates of the two adjacent carbon atoms bonded to the nitroxide nitrogen 40. The X-, Y-, and Z-axes of the nitroxide were then calculated according to Equations 5 40,41.
| (5) |
Using the coordinates of these three axes, the angles θ and φ were calculated from Equations 6.
| (6) |
For the doubly-labeled peptide, the interelectron distance was calculated from the midpoints of the nitrogen-oxygen bonds for the two nitroxides. For simplicity the nitroxide nitrogen was used as the origin of all coordinate systems used for determining the various angles. The nitrogen of the label at residue 13 (nitroxide 1) was translated to the origin of the coordinate system, and the axes of nitroxide 1 were determined from Equation 5. The entire system was then transformed using a rotation R̂' so that the axes of nitroxide 1 were coincident with the X-, Y-, and Z- axes of the laboratory frame. The angles ξ and η were determined from the transformed coordinates, , of the nitrogen of nitroxide 2 at residue 7.
| (7) |
The coordinates of the nitrogen, oxygen, and two adjacent carbon atoms of nitroxide 2 were then transformed so that the nitrogen was at the origin. The coordinates of the , , and axes of nitroxide 2 were then calculated from these atomic coordinates using Equations 5 and the angles α, β, γ determined as follows.
| (8) |
All of these calculations were performed using a Mathematica 42 script. In order to more directly compare in Cartesian space the results from fitting the EPR data to the model of the doubly-labeled peptide, additional scripts were developed to reverse these procedures so that for a given set of R, ξ, η α, β, and γ values the corresponding x-,y-, and z-axes of nitroxide 2 can be calculated relative to nitroxide 1.
Results and Discussion
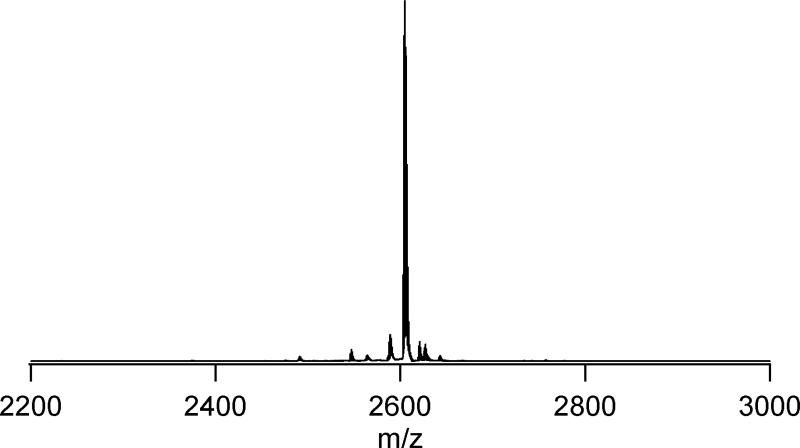
Figure 1 shows the MALDI-TOF spectra of the SL-TOAC7 AChR M2δ peptide which is representative of those obtained for other peptides. The peak at 2605 Da corresponds to the molar mass (M+1).
Figure 1.
A MALDI-TOF mass spectrum of single labeled TOAC-7 AChR M2δ peptide
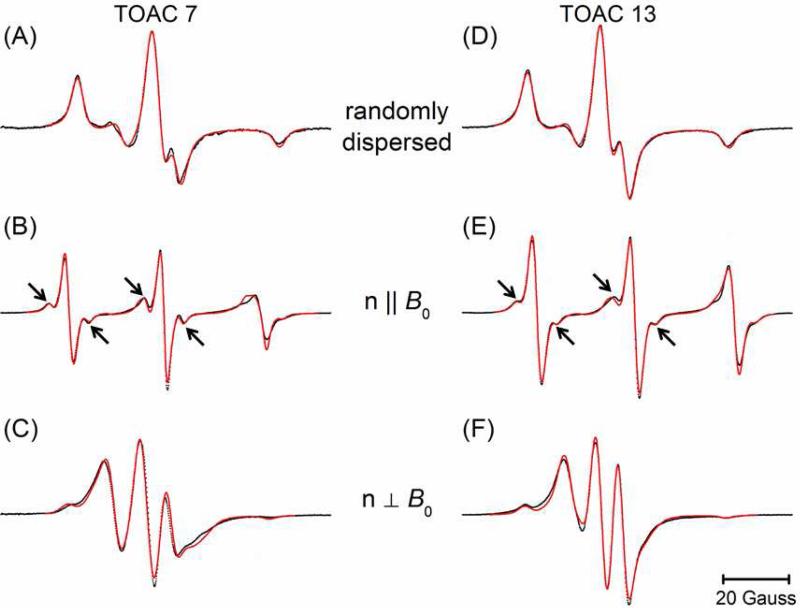
Figure 2 shows the CW-EPR spectra of singly-labeled AChR M2δ peptides labeled with TOAC at residue 7 (TOAC7) and residue 13 (TOAC13). The upper panels contain the spectra (black) of the randomly dispersed samples which are well-fit as rigid-limit powder patterns (red lines) to give the g- and A-tensor values (Table 1) used in all subsequent analyses. Even at the elevated temperature (318 K) used in this study, these data are successfully modeled without including dynamic effects.
Figure 2.
CW-EPR spectra (black dots) of TOAC7 (left) and TOAC13 (right) AChR M2δ peptides in DMPC bilayers at 318 K together with the best-fit simulations (red lines). The molar peptide to lipid ratio was 1:250. The upper panels (A) and (D) contain spectra of randomly dispersed samples from which the best-fit g- and A-tensor values (Table 1) were determined. The middle panels (B) and (E) contain spectra from parallel-aligned samples. The lower panels (C) and (F) contain spectra from the perpendicular-aligned samples. For each site, the n||B0 and  were simultaneously analyzed to determine the best fit values of ψ0, σψ, θ, and φ (Table 2).
were simultaneously analyzed to determine the best fit values of ψ0, σψ, θ, and φ (Table 2).
Table 1.
Parameters for the best-fit simulation of randomly-dispersed singly-labeled AChR M2δ peptides.
| site | gxx | gyy | gzz | Axx (G) | Ayy (G) | Azz (G) | |
|---|---|---|---|---|---|---|---|
| a | 7 | 2.00775 | 2.00608 | 2.00268 | 8.7 | 3.4 | 30.6 |
| b | 13 | 2.00780 | 2.00627 | 2.00244 | 7.7 | 4.2 | 30.5 |
The middle panels of Figure 2 show the spectra (black) of the mechanically aligned samples in the parallel orientation, while the lower panels are the spectra of the perpendicularly-orientated samples. The aligned EPR spectra clearly show the orientation dependent hyperfine splitting, where the hyperfine splitting is at a minimum (10.50 G for TOAC7 and 10.35 G for TOAC13) at the perpendicular orientation (n⊥Bo), and becomes maximum (29.97 G for TOAC7 and 30.5 G for TOAC13) at the parallel orientation (n||Bo) 19. This type of orientation dependent hyperfine splitting arises because the molecular axis i.e. the z-axis of the TOAC nitroxide lies nearly parallel to the direction of the static magnetic field (Bo) when the glass plates are aligned parallel, which results in a maximal hyperfine splitting value. However, the z-axis of the TOAC nitroxide orients nearly perpendicular to the static magnetic field when the glass plates are aligned perpendicularly, producing a minimal hyperfine splitting value.
The fits obtained from the non-linear least-squares analyses of the data from the singly-labeled aligned samples are shown in Figure 2 (red lines) and the best-fit parameters are given in Table 2. These fits were obtained by simultaneously fitting the two spectra for a given sample obtained at different sample alignments. A helix tilt angle, ν, of 12° was assumed as reported in the previous study 21. The rotation of the peptide about the helix axis is determined by a distribution centered at ψ0 with standard deviation σψ. The best-fit angle between the nitroxide Z-axis and the helix axis, θ, was determined to be 21° for TOAC7 and 17° for TOAC13 which agree well and with the values of 20° and 8°, respectively, determined from molecular modeling of the TOAC-labeled AChR M2δ peptides. Additional efforts were made to model these data assuming that there were two populations with different values for the angles θ and φ. These efforts did not result in significantly better fits. It is possible that at the relatively high temperatures employed (318 K), there is rapid intraconversion between two conformers of the TOAC ring. The values of θ obtained in the current study agree well with the value of 21° previously reported for the twist boat conformer of the TOAC label 5.
Table 2.
Parameters for the best-fit simulation of aligned singly-labeled AChR M2δ peptides compared to the parameters from molecular modeling.
| site | ψ 0 | σ ψ | θ | ϕ | δ (G) | κ | isotropic fraction | ||
|---|---|---|---|---|---|---|---|---|---|
| a | Fit | 7 | 112° | 39° | 21° | 219° | 5.1 | 0.08 | 0.22 |
| b | Model | 7 | - | - | 20° | 325° | - | - | - |
| c | Fit | 13 | 159° | 55° | 17° | 261° | 5.3 | 0.06 | 0.19 |
| d | Model | 13 | - | - | 8° | 287° | - | - | - |
The narrow linewidths observed at the parallel orientation reveal the presence of an additional splitting as indicated by the arrows in Figures 2B and 2E which are not evident in the spectra of the randomly dispersed samples. Extensive efforts were made to fit these features using a variety of different approaches. In particular, these features could not be fit using a second population at a different orientation (i.e. different θ and θ). Instead, these features are due to a fraction of the labels experiencing an additional coupling. This additional coupling is similar to the 13C satellite lines seen in very fast motion spectra of nitroxides, but the splitting and amplitude are both too large to be due to 13C. Instead, it appears that the splitting is due to a fraction of the TOAC labels that are hydrogen-bonded as has been previously seen by Marsh and coworkers using electron spin-echo envelope modulation 36.
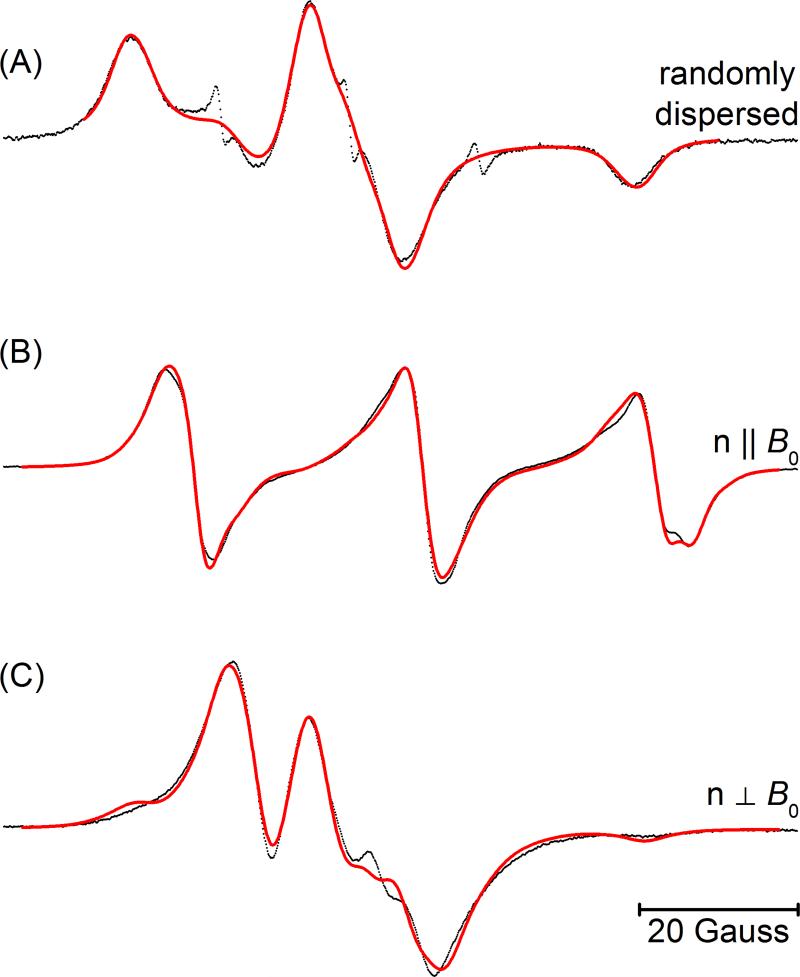
Figure 3 shows the CW-EPR spectra of the doubly TOAC-labeled AChR M2δ peptide. The upper panel (A) shows the spectrum of the randomly-dispersed sample. Figures 3(B) and 3(C) reveal parallel-aligned, and the perpendicular-aligned spectra of doubly-labeled AChR M2δ peptide in planar quartz-supported DMPC bilayers. Line broadening due to electron-electron dipolar interactions is evident in all three spectra in comparison to the corresponding spectra of the singly-labeled peptides in Figure 2.
Figure 3.
CW-EPR spectra for the doubly labeled AChR M2δ peptide in DMPC bilayers at 318 K for the (A) randomly-dipersed sample, n||Bo sample (B), and n⊥Bo sample (C). The molar peptide to lipid ratio was 1:250. The n||B0 and 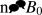 data were simultaneously analyzed to determine the best fit values of α, β, γ, ξ, η, and R (Table 3). A similar analysis was performed on the randomly-dispersed spectrum (Table 3).
data were simultaneously analyzed to determine the best fit values of α, β, γ, ξ, η, and R (Table 3). A similar analysis was performed on the randomly-dispersed spectrum (Table 3).
The fit overlaid on the randomly dispersed sample (Figure 3A) assumes a single distance and orientation between the two TOAC labels. The best-fit distance is R= 15.3 Å. There are a number of different sets of angles that are symmetry-related and give equivalent fits to the data. One such set of angles is given in Table 3a. Alternatively, a fit to this spectrum using a convolution approach which assumes an isotropic distribution in the relative orientation of the two angles (results not shown) was not as good.
The parameters for the best global fit to the spectra of the parallel-aligned (Figure 3B) and the perpendicular-aligned samples (Figure 3C) are also given in Table 3. The global fit was performed with the label at site 13 as a reference and the values of ψ0, σψ, θ, and φ were taken from the fit of the data in Figures 2E and 2F (Table 2c). The best-fit inter-electron distance was found to be 14.3 Å. Again, there are a number of sets of angles which are symmetry related and given equivalent fits to the data. For one such set of angles, the two angles defining the inter-electron vector are ξ = 49° and η = 211°. The Euler angles that orient TOAC7 nitroxide with respect to the TOAC13 (first nitroxide) are α = 132°, β = 14°, and γ = 229°.
Table 3.
Parameters for the best-fit simulation of doubly-labeled AChR M2δ peptides for both the randomly-dispersed and aligned samples compared to the parameters from molecular modeling.
| γ | β | α | ξ | η | R (Å) | δ (G) | κ | isotropic fraction | ||
|---|---|---|---|---|---|---|---|---|---|---|
| a | isotropic | 286° | 17° | 353° | 67° | 251° | 15.3 | - | - | 1.00 |
| b | aligned | 230° | 14° | 132° | 49° | 211° | 14.6 | 4.3 | 0.09 | 0.24 |
| c | model | 224° | 22° | 23° | 39° | 218° | 14.5 | - | - | - |
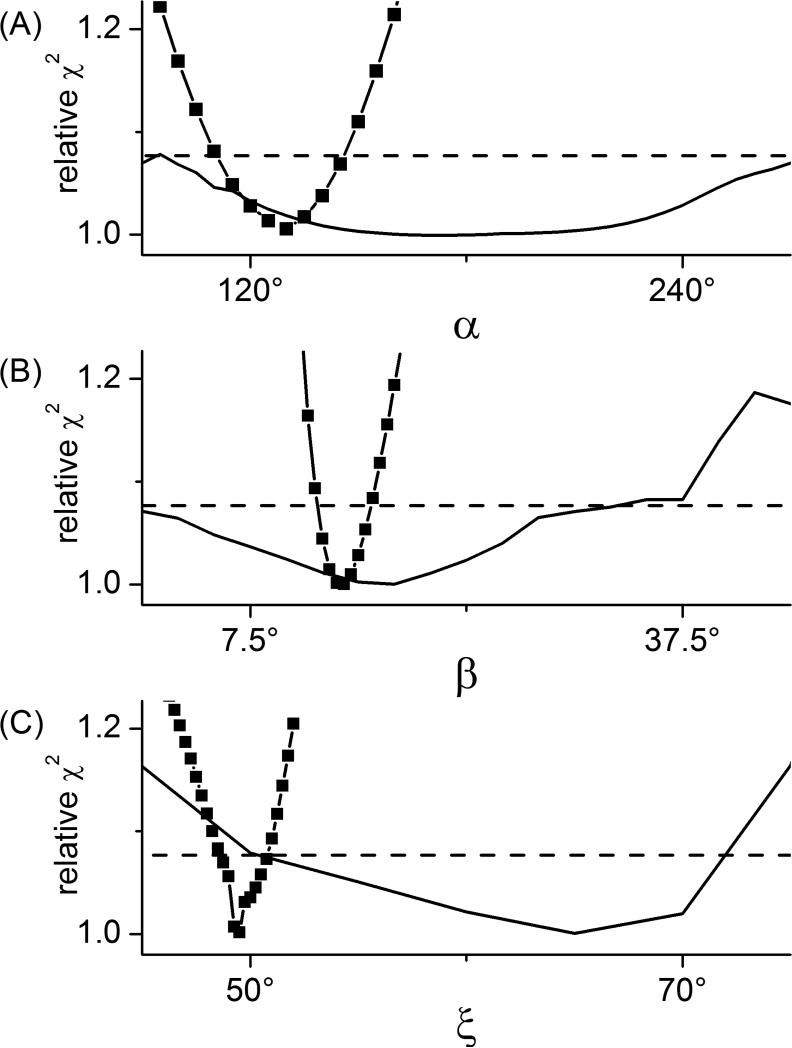
A number of factors influence the precision with which the various parameters defining the geometry between two dipolar-coupled labels can be determined 43. In order to assess whether the global analysis of spectra from aligned samples offers advantages over the analysis of a single spectrum from a randomly dispersed sample, confidence intervals were calculated for the various parameters. Representative confidence intervals for the angles α, β, and ξ are shown in Figure 4. From these plots, the uncertainty in a given parameter can be estimated from the range of values which fall below the horizontal dashed lines. These results demonstrate that the global analysis of the aligned data gives much more precise estimates of the angles α, β, and ξ than can be obtained from the analysis of the randomly dispersed data.
Figure 4. Confidence intervals for the angles γ (A), β (B), and ξ (C).
The best-fit χ2 values obtained from fitting the two oriented spectra (black solid squares) and the isotropic spectrum (black solid line) are plotted for a linear sequence of values for the particular angle. The horizontal dashed lines are drawn at the χ2 value corresponding to an approximate 95% confidence level calculated using the F-statistic.
Molecular modeling studies on the AChR M2δ peptide were conducted using MolMol and Visual Molecular Dynamics programs (see Material and Method section for details). The various parameters that define the geometry and distance of the two TOAC spin probes in AChR M2δ peptides were calculated. The distance between two TOAC nitroxides and their orientation angles obtained from the spectral simulations were found comparable with the molecular modeling studies. Using the coordinates of the nitroxide nitrogen and oxygen atoms in a doubly-labeled AChR M2δ peptide, we calculated an internitroxide distance of 14.5 Å, which matches very well with the 14.6 Å distance obtained from the aligned CW-EPR spectra simulations. Similarly, the two angles, ξ and η, that define the orientation of the internitroxide vector were calculated to be 39° and 218° from modeling studies, which agree well with the 49° and 211° values obtained from the simulations. The rotational Euler angles that define the relative orientation of TOAC13 nitroxide with respect to TOAC7 nitroxide were also agreed well (except the first angle) α = 132°, β = 14°, and γ = 230° from EPR spectra simulations and α = 23°, β = 22°, and γ = 224° from modeling studies.
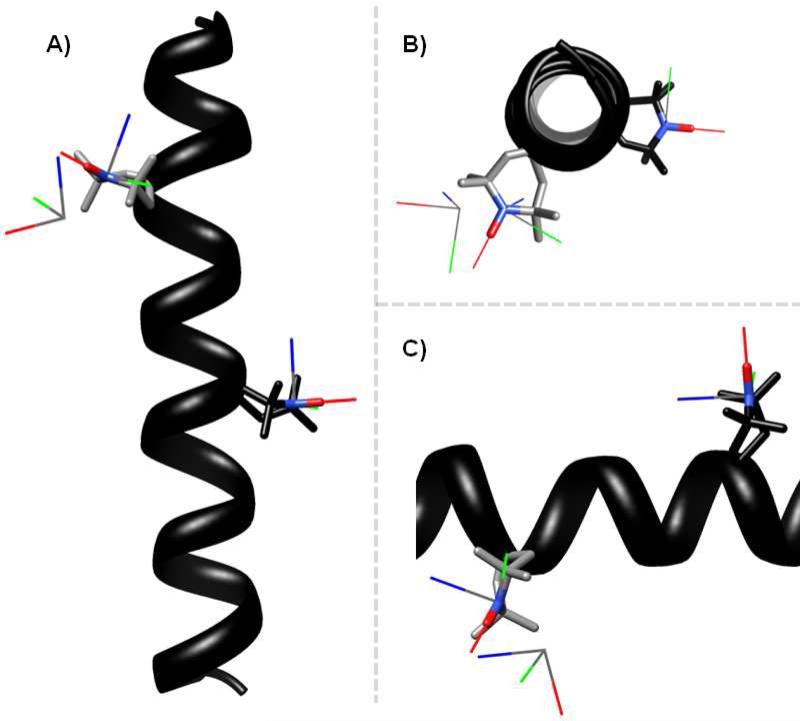
To better appreciate how the results from fitting the aligned EPR spectra compare to those from modeling, a set of x-,y-,and z-axes were back calculated from the distance and angles given in Table 3b and overlaid on the model peptide. The results are shown in three different views in Figure 5. The nitrogen atom of the best-fit axes is shifted by approximately 3 Å from that of the model. The angle between the best-fit z-axis and the model z-axis is 30°.
Figure 5. Three different views comparing the best-fit results to the predicted model of the doubly-labeled peptide.
For the predicted model, the helix is shown in black. The nitrogen, oxygen, and adjacent carbon atoms of the TOAC nitroxides are shown as stick models. The TOACs at residues 7 and 13 are colored in grey and black respectively. Overlaid on the TOAC for the model are the calculated x-, y-, and z-axes of the TOAC nitroxides in red, green, and blue respectively. The set of additional axes represent the position and orientation of TOAC 7 relative to TOAC 13 as back calculated for the best-fit values of the distance and angles determined in Figure 3. Three different views are shown each rotated by 90°.
Conclusions
By orienting dual TOAC-labeled AChR M2δ peptide in mechanically aligned glass plates, applying rigorous spectral simulation techniques, and simultaneously analyzing data at different sample orientations, the internitroxide distance and relative orientation of two TOAC nitroxides have been determined. Distance measurement using mechanically aligned glass plates is a potentially important new technique as samples are easy to prepare and the system provides adequate lipid hydration but still restrains the mobility of the nitroxide side chain leading to more accurate distance measurements. Distance measurements can be performed at room temperature or even higher temperatures (e.g. 318 K), and sample orientations can be adjusted from 0° to 90° or any angles in between to get detailed structural parameters needed for distance measurements. Therefore, utilization of mechanically aligned planar bilayer system and rigorous spectral simulation methods as employed in this study opens the door for distance measurements in other proteins, DNA, or RNA that might prove useful in understanding their structures and conformational dynamics. Also, this approach will be useful for antimicrobial peptides that lay on the surface such as magainin. The significance of this mechanically aligned technique is clearly evident in our study as the internitroxide distance obtained from aligned dual-labeled EPR spectra matches closely the distance obtained from modeling studies (14.6 Å vs 14.5 Å), while fitting of the randomly dispersed gave a distance of 15.3 Å. Additionally, the relative orientation between the labels could be much more precisely determined from the oriented data.
Acknowledgements
This project was supported by NIGMS/NIH (GM60259-01), NSF (CHE-0645709), and MRI-0722403 to GAL and NIHGM080513 to EJH.
References
- 1.Hubbell WL, Altenbach C. Curr. Opinion Struct. Biol. 1994;4:566. [Google Scholar]
- 2.Qin PZ, Hideg K, Feigon J, Hubbell WL. Biochemistry. 2003;42:6772. doi: 10.1021/bi027222p. [DOI] [PubMed] [Google Scholar]
- 3.Fanucci GE, Cafiso DS. Curr. Opinion Struct. Biol. 2006;16:644. doi: 10.1016/j.sbi.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 4.Klug CS, Feix JB. Methods Cell Biol. 2008;84:617. doi: 10.1016/S0091-679X(07)84020-9. [DOI] [PubMed] [Google Scholar]
- 5.Hanson P, Anderson DJ, Martinez G, Millhauser G, Formaggio F, Crisma M, Toniolo C, Vita C. Mol. Phys. 1998;95:957. [Google Scholar]
- 6.Hustedt EJ, Smirnov AI, Laub CF, Cobb CE, Beth AH. Biophys. J. 1997;72:1861. doi: 10.1016/S0006-3495(97)78832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steinhoff HJ, Radzwill N, Thevis W, Lenz V, Brandenburg D, Antson A, Dodson G, Wollmer A. Biophys. J. 1997;73:3287. doi: 10.1016/S0006-3495(97)78353-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeschke G, Koch A, Jonas U, Godt A. J. Magn. Reson. 2002;155:72. doi: 10.1006/jmre.2001.2498. [DOI] [PubMed] [Google Scholar]
- 9.Borbat PP, McHaourab HS, Freed JH. J. Am. Chem. Soc. 2002;124:5304. doi: 10.1021/ja020040y. [DOI] [PubMed] [Google Scholar]
- 10.Cai Q, Kusnetzow AK, Hubbell WL, Haworth IS, Gacho GPC, Van Eps N, Hideg K, Chambers EJ, Qin PZ. Nucleic Acids Research. 2006;34:4722. doi: 10.1093/nar/gkl546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sen KI, Logan TM, Fajer PG. Biochemistry. 2007;46:11639. doi: 10.1021/bi700859p. [DOI] [PubMed] [Google Scholar]
- 12.Schiemann O, Prisner TF. Quarterly Rev. Biophys. 2007;40:1. doi: 10.1017/S003358350700460X. [DOI] [PubMed] [Google Scholar]
- 13.Ghimire H, McCarrick RM, Budil DE, Lorigan GA. Biochemistry. 2009;48:5782. doi: 10.1021/bi900781u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rabenstein MD, Shin YK. Proc. Natl. Acad. Sci U.S.A. 1995;92:8239. doi: 10.1073/pnas.92.18.8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altenbach C, Oh KJ, Trabanino RJ, Hideg K, Hubbell WL. Biochemistry. 2001;40:15471. doi: 10.1021/bi011544w. [DOI] [PubMed] [Google Scholar]
- 16.McHaourab HS, Oh KJ, Fang CJ, Hubbell WL. Biochemistry. 1997;36:307. doi: 10.1021/bi962114m. [DOI] [PubMed] [Google Scholar]
- 17.Hustedt EJ, Stein RA, Sethaphong L, Brandon S, Zhou Z, DeSensi SC. Biophys. J. 2006;90:340. doi: 10.1529/biophysj.105.068544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hustedt EJ, Beth AH. Annu. Rev. Biophys. Biomol. Struct. 1999;28:129. doi: 10.1146/annurev.biophys.28.1.129. [DOI] [PubMed] [Google Scholar]
- 19.Inbaraj JJ, Laryukhin M, Lorigan GA. J. Am. Chem. Soc. 2007;129:7710. doi: 10.1021/ja071587l. [DOI] [PubMed] [Google Scholar]
- 20.Mayo DJ, Inbaraj JJ, Subbaraman N, Grosser SM, Chan CA, Lorigan GA. J. Am. Chem. Soc. 2008;130:9656. doi: 10.1021/ja803590w. [DOI] [PubMed] [Google Scholar]
- 21.Opella SJ, Marassi FM, Gesell JJ, Valente AP, Kim Y, Oblatt-Montal M, Montal M. Nat. Struct. Biol. 1999;6:374. doi: 10.1038/7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montal M, Opella SJ. Biochim. Biophys. Acta. 2002;1565:287. doi: 10.1016/s0005-2736(02)00575-8. [DOI] [PubMed] [Google Scholar]
- 23.Karlin A. Curr. Opin. Neurobiol. 1993;3:299. doi: 10.1016/0959-4388(93)90121-e. [DOI] [PubMed] [Google Scholar]
- 24.Unwin N. Nature. 1995;373:37. doi: 10.1038/373037a0. [DOI] [PubMed] [Google Scholar]
- 25.Corringer PJ, Le Novere N, Changeux JP. Ann. Rev. Pharmacol. Toxicol. 2000;40:431. doi: 10.1146/annurev.pharmtox.40.1.431. [DOI] [PubMed] [Google Scholar]
- 26.Hustedt EJ, Beth AH. Biophysical Journal. 2004;86:3940. doi: 10.1529/biophysj.103.035048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchetto R, Schreier S, Nakaie CR. J. Am. Chem. Soc. 1993;115:11042. [Google Scholar]
- 28.Victor KG. Dissertation. University of Virginia; 2000. An Investigation of the Electrochemical Interactions Between Acidic Phospholipid Membranes and Charged Peptides Using Electron Paramagnetic Resonance. [Google Scholar]
- 29.Martin L, Ivancich A, Vita C, Formaggio F, Toniolo C. J. Pept. Res. 2001;58:424. doi: 10.1034/j.1399-3011.2001.00940.x. [DOI] [PubMed] [Google Scholar]
- 30.Inbaraj JJ, Cardon TB, Laryukhin M, Grosser SM, Lorigan GA. J. Am. Chem. Soc. 2006;128:9549. doi: 10.1021/ja0622204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karim CB, Zhang Z, Thomas DD. Nature Protocols. 2007;2:42. doi: 10.1038/nprot.2007.2. [DOI] [PubMed] [Google Scholar]
- 32.Carpino LA. J. Am. Chem. Soc. 1993;115:4397. [Google Scholar]
- 33.Carpino LA, Elfaham A, Minor CA, Albericio F. J. Chem. Soc., Chem. Commun. 1994:201. [Google Scholar]
- 34.Hustedt EJ, Spaltenstein A, Kirchner JJ, Hopkins PB, Robinson BH. Biochemistry. 1993;32:1774. doi: 10.1021/bi00058a011. [DOI] [PubMed] [Google Scholar]
- 35.Hustedt EJ, Cobb CE, Beth AH, Beechem JM. Biophys. J. 1993;64:614. doi: 10.1016/S0006-3495(93)81420-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bartucci R, Guzzi R, Sportelli L, Marsh D. Biophys. J. 2009;96:997. doi: 10.1016/j.bpj.2008.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Erilov DA, Bartucci R, Guzzi R, Shubin AA, Maryasov AG, Marsh D, Dzuba SA, Sportelli L. J. Phys. Chem. B. 2005;109:12003. doi: 10.1021/jp050886z. [DOI] [PubMed] [Google Scholar]
- 38.Koradi R, BIlleter M, Wuthrich K. J. Mol. Graphics. 1996;14:51. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 39.Humphrey W, Dalke A, Schulten K. J. Mol. Graphics. 1996;14:33. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 40.DeSensi SC, Rangel DP, Beth AH, Lybrand TP, Hustedt EJ. Biophys. J. 2008;94:3798. doi: 10.1529/biophysj.107.125419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Budil DE, Sale KL, Khairy KA, Fajer PG. J. Phys. Chem. A. 2006;110:3703. doi: 10.1021/jp054738k. [DOI] [PubMed] [Google Scholar]
- 42.Mathematica . Wolfram Research Inc.; Champaign, IL: [Google Scholar]
- 43.Hustedt EJ, Beth AH. Structural Information from CW-EPR Spectra of Dipolar Coupled Nitroxide Spin Labels. In: L. J., Eaton SS, Eaton GR, editors. Biological Magnetic Resonance; Berlinger. Vol. 19. Kluwer Academic/Plennum Publishers; New York: 2000. p. 155. [Google Scholar]