Abstract
Recent investigations of archaeal viruses have revealed novel features of their structures and life cycles when compared to eukaryotic and bacterial viruses, yet there are structure-based unifying themes suggesting common ancestral relationships among dsDNA viruses in the three kingdoms of life. Sulfolobus solfataricus and the infecting virus Sulfolobus turreted icosahedral virus (STIV) is one of the well-established model systems to study archaeal virus replication and viral-host interactions. Reliable laboratory conditions to propagate STIV and available genetic tools allowed structural characterization of the virus and viral components that lead to the proposal of common capsid ancestry with PRD1 (bacteriophage), Adenovirus (eukaryotic virus) and PBCV (chlorellavirus). Micro-array and proteomics approaches systematically analyzed viral replication and the corresponding host responses. Cellular cryo-electron tomography and thin-section EM studies uncovered the assembly and maturation pathway of STIV and revealed dramatic cellular ultra-structure changes upon infection. The viral induced pyramid-like protrusions on cell surfaces represent a novel viral release mechanism and previously uncharacterized functions in viral replication.
Keywords: Sulfolobus turreted icosahedral virus (STIV), Archaea, virus
Introduction
Archaea are the third domain of life. They exist in a broad diversity of environments and dominate in extreme environments, such as low pH, high temperature, and high salinity. The archaeal viruses isolated so far show a wide diversity in morphology and encode genes sharing little similarity to other known genes in eukaryotes and prokaryotes [1–4]. Archaeal virus studies have revealed important insights regarding the fundamentals of their life cycles, their diversity and the relationship among viruses in different domains of life. Sulfolobus turreted icosahedral virus (STIV) is a well-characterized archaeal virus. Defined culture systems and robust genetic tools allowed genomic, proteomic and structural studies, relating STIV to its host and to other viruses [5,6].
Host and Viral Replication Cycle
STIV infects Sulfolobus solfataricus, which belongs to Crenarchaeota. Cells were originally isolated from hot springs and grow optimally at 80°C and pH ~3. STIV infection induces dramatic host morphological changes with characteristic pyramid-like structures protruding from cell surfaces (Fig 2) [4,7]. S. solfataricus cells have an oval shape about 1 µm in diameter. Exterior to the cytoplasmic membrane is a surface protein layer (S-layer) that is formed by a single glycosylated protein arranged with hexagonal surface packing[8]. The STIV replication cycle can be followed in a near synchronized, single cycle infection of S. solfataricus. Microarray analysis demonstrated that the transcription of the STIV genes was first detected at 8 hr post infection and peaked at ~24 hrs post infection [9]. Cells containing assembled viral particles became dominant at ~32 hrs post infection. During STIV infection, 124 host genes were up-regulated, which were mainly associated with DNA replication and repair and a number of unassigned function. Fifty-three genes were down regulated in expression. Most of these were detected at 32 hr post infection and were related with energy production and metabolism, likely associated with forthcoming cell lysis and virus release.
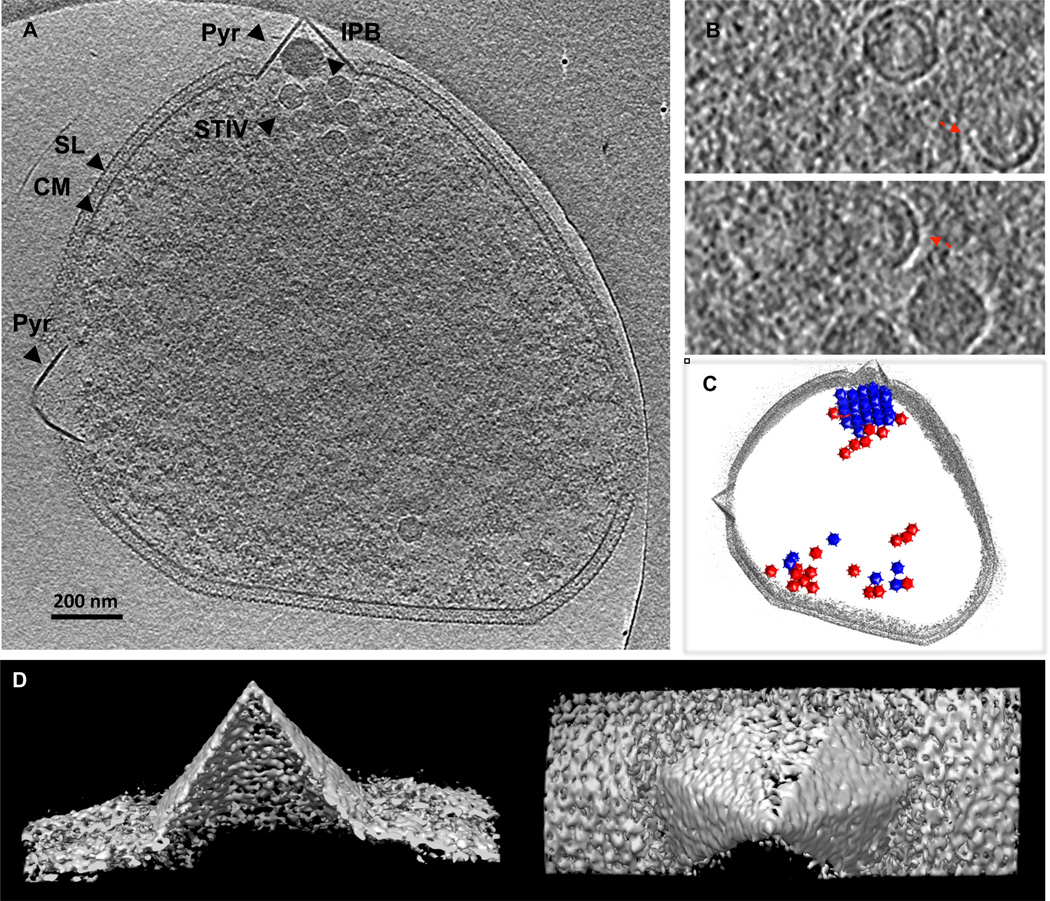
Fig 2. Whole cell cryo electron tomography of Sulfolubs infected with STIV.
(A) A computationally isolated tomographic slice of Sulfolubs infected with STIV. SL, s-layer; PS, periplasmic space; CM, cytoplasmic membrane; Pyr, pyramid like protrusion; STIV, STIV particles. (B) Subtomographic slices displaying STIV virions, procapsids and partially assembled particles that contain parts of capsid and membrane viewed perpendicular to the direction of the beam (x-y plane). (C) The model representation of the viral distribution in a cell with cell periphery outlined in gray, virions in blue and procapsids in red. (D) Surface representations of a pyramid in 3D viewed from the side and the top of the structure.
STIV Structure, Assembly and Maturation
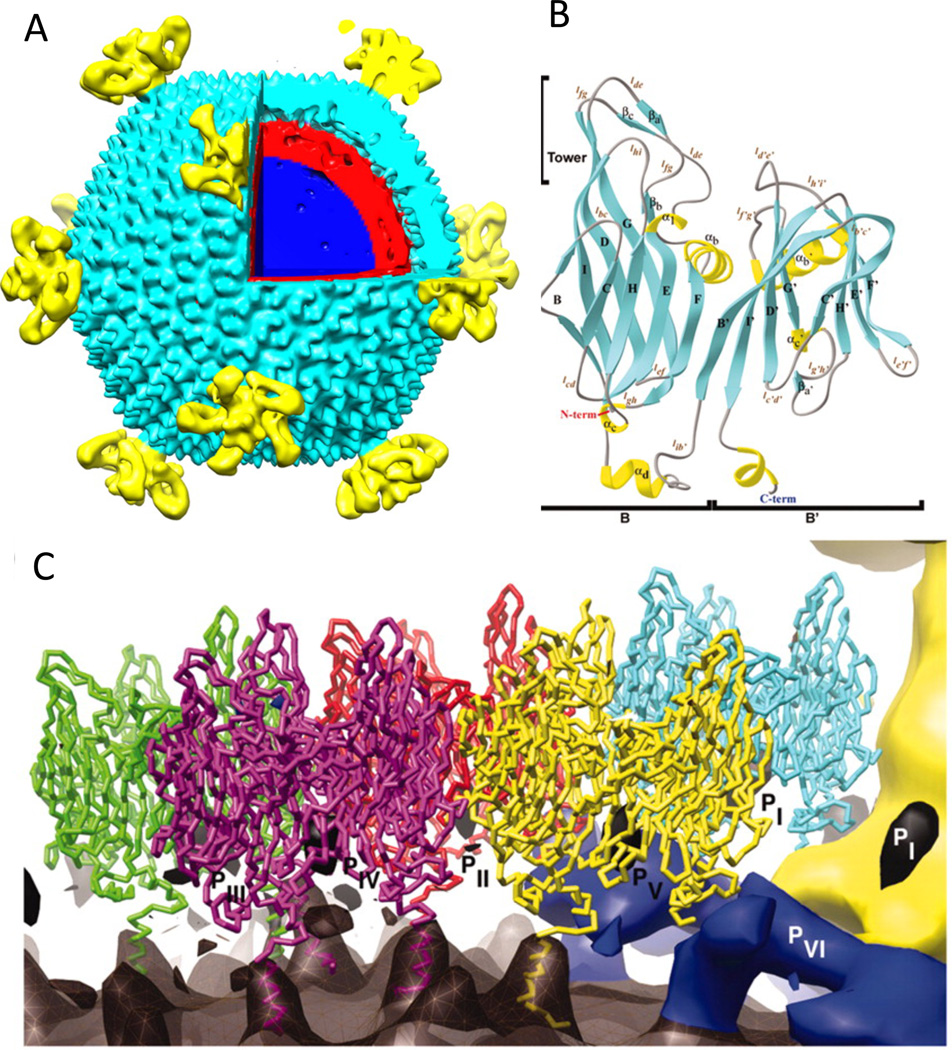
STIV has an icosahedral capsid, 74 nm in diameter, that contains an inner membrane enclosing a 17.3 kbp ds circular DNA genome (Fig 1a) [10–12]. The twelve five-fold vertices have turret-like appendages extending 13 nm above the capsid shell. The structure of the major capsid protein was determined by X-ray crystallography and consists of two eight-stranded β-sandwiches, referred as viral jelly rolls, formed by a single polypeptide chain (Fig 1b) [13]. Trimeric capsomers display pseudo-hexagonal symmetry with each subunit contributing two jelly rolls. The pseudo hexagons form an icosahedral shell with triangular number T= 31d. The coordinates of the crystal structure of the major capsid protein fit the cryo-electron microscopy (CryoEM) reconstructed density of the STIV virion extremely well. The pseudo-atomic model shows that the major capsid proteins contact the viral membrane via a basic C-terminal helix (Fig 1c). The jelly roll fold is widely adopted in viral capsid subunits with both a single copy per subunit (e.g. picornaviruses, polyomaviruses and many icosahedral plant viruses) and with two copies per subunit as in the capsid proteins of the PRD1-Adeno viral lineage. The latter is conserved through viruses infecting the three domains of life, including bacteriophages PRD1, PM2, the alga virus PBCV-1, the mammalian adenovirus, and vaccinia virus [14–19].
Fig 1.
The structures of STIV. (A) The surface representation of STIV single particle cryo-EM reconstruction. The turret-like vertices are colored in yellow, the capsid shell in cyan, the lipid layer in red, and the genome in blue. (B) The crystal structure of the STIV major capsid protein. α-helices, β-strands and coils are colored in yellow, in cyan, and in gray respectively. (C) Cα- trace of the major capsid protein overlaid onto the difference map of the STIV cryoEM reconstruction. The outer viral membrane leaflet closely follows the capsid shell is shown in dark gray, the minor capsid proteins (PVI) are in blue, and the vertex appendage is in yellow. The proposed C-terminal helices for the major capsid protein subunits are modeled into the difference map to show the interaction with the outer leaflet.
The mechanism of inner-membrane incorporation into viruses is biologically and biochemically an intriguing problem. The crystal structures of PRD1 and PM2 revealed the protein and membrane organization in the virions and suggested that the membrane-associated proteins may function as tape measures or scaffolding proteins to control assembly size and fidelity [15,20,21]. Proteomics studies of purified STIV virions and membranes show the presence of viral gene products in the viral inner membrane [22]. These proteins may be the constituents of the “feet-like” densities, seen in the cryo EM reconstruction, at the base of the turret vertices and making contact with the viral membrane, the vertices, and the capsid shell [10]. The observation of partially built particles in the cellular cryo electron tomograms of STIV infected Sulfolobus supports the hypothesis that assembly of the STIV capsid shell and the membrane are tightly coupled (Fig 2) [23]. The curvature and layer spacing of partial shells resembled those of fully assembled capsids, implying defined local interactions between capsid and membrane and between capsid subunits occur as assembly proceeds. Open membrane crescents were also observed as assembly intermediates of immature vaccina virus particles. Vaccinia virus membrane crescents are recruited by hexagonal arrays of the vacinia protein D13 and it has the same double jelly roll fold observed in STIV [18,24] relating, again, eukaryotic viruses and archaeal viruses to a common ancestor. STIV cellular tomograms suggested that the viral lipid membrane is likely derived de novo in the host, which does not have membrane-containing organelles, since partially assembled particles do not appear to bud off the cytoplasmic membrane. The viral lipids likely are synthesized de novo and associated through their hydrophobic nature and organized by trans-membrane proteins and capsid proteins. Mass spectrometry studies concluded that the lipid compositions of viral and cellular membranes are different [22].
The STIV life cycle involves assembly of DNA-free procapsids with subsequent genome packaging to form DNA-filled virions (Fig 2). Comparing the sub-tomographic reconstructions of virions and procapsids demonstrated closely similar particle size and morphology indicating that large-scale reorganization of the capsid or membrane did not occur with DNA packaging in STIV [23,25]. Similar post assembly dsDNA filling is observed in PRD1, herpesviruses, and dsDNA bacteriophages where, in all cases, a specialized vertex serves as a conduit for DNA packaging and allows the association of packaging enzyme complexes with the particle. On the other hand, some RNA viruses package their genome through one of the twelve identical vertices, which have ATPase activities to power the genome encapsidation. The proteins involved in STIV genome packaging and the packaging mechanism are largely unknown. In PSI-BLAST searches, the viral gene B164 displays significant homology to a large class of P-loop ATPases and to poxvirus A32, which is thought to be involved in viral DNA packaging [22]. Whether any of the twelve turrets seen in the symmetric EM reconstruction of STIV functions in DNA packaging or if there is a specialized vertex for DNA packaging remains to be determined.
Cell Biology of STIV Infection
Viral Quasicrystalline Arrays
Virus factories or viroplasms have been reported in eukaryotic and prokaryotic viral systems where proteins and newly synthesized genomes are confined within specific compartments for efficient viral replication and assembly [26–28]. The cellular rearrangements during virus infections generate sophisticated platforms and microenvironments for viral replication. Little is known if archaeal viruses use similar strategy. Cellular tomography studies showed that STIV particles are arranged as strikingly well organized quasi-crystalline arrays [23,25]. Viral arrays, with extremely tight packing, contained the majority of STIV particles in the cell. Such an organization may offer an important advantage in that it accommodates the greatest number of viruses in the limited cellular space. Procapsids and virions have different cellular distribution patterns. DNA-free procapsids were scattered throughout the cell but DNA-filled virions were localized in the arrays, with an occasional empty particle also seen in the arrays. It appeared that DNA-packaging caused the particles to reorganize into an array. The apparent non-random localization might reflect a cellular microenvironment for DNA replication and packaging. Alternatively, particles may assemble outside the arrays and traffic to specific sites where viral genome packaging takes place with the formation of tightly packed clusters. More study is required to discern and verify different models.
Viral Infection Induced Pyramid-Like Protrusions
Cells infected by STIV develop unique pyramid-like structural protrusions from the cell surfaces (Fig 2) [7,23]. The novel structures were also observed in viral infections of Sulfolobus, S. islandicus, infected by Sulfolobus islandicus rod-shaped virus 2 (SIRV2) [29–31]. 3-D isosurfaces of STIV infected cells showed that the seven-sided pyramids have remarkably sharply defined facets and apex [23]. Pyramids lack the S-layer and have thicker cross-section than the cytoplasmic membrane, indicating different protein and/or lipid composition. Several proteins were shown to be up-regulated in the membrane fractions of Sulfolobus infected by viruses [29,32]. A 9.8 kDa membrane bound viral protein, C92, was highly expressed. The secondary structure of C92 was predicted to have high helical content with a trans-membrane helix in the N-terminal region. Sulfolobus over-expressing C92 proteins develop pyramid protrusions in the absence of viral infection, implying that C92 is sufficient to build pyramids and/or recruit other necessary cellular or viral factors. The molecular organization and interactions of C92 and possible involvements of other factors are yet to be characterized. Cellular tomograms of STIV infected cells also captured some pyramids in the process of protruding out of the rigid S-layer. The pyramids could form by either mechanically protruding and/or enzymatically digesting through the S-layer, perturbing its integrity and detaching it from the pyramid membrane.
The pyramids were shown to be the sites that cells break to release viruses at the late infection stage (40 hr. post-infection.) It represents a novel mechanism of viral egress. In addition, they are also essential for viral replication [7,30]. In fact, pyramids started to appear before viral particles became visible in the cytoplasm in time course studies of viral infection. It is likely that the cellular domains formed by pyramid proteins contain binding sites to recruit or localize precise cellular/viral factors to carry out specific viral replication steps. Several proteins have been shown to be up-regulated in the membrane fractions of Sulfolobus infected by viruses, including among these are the ESCRT (endosomal sorting complex required for transport)-like proteins, which are important in Sulfolobus cell division and are utilized by eukaryotic viruses to facilitate intracellular virus transport and viral release [33]. Microarray studies observed the up-regulation of a hypothetical Vps4 (vacuolar protein sorting 4) AAA+ (ATPase associated with various cellular activities) and ESCRT-III. Vps4 also co-purified with STIV particles. These observations strongly suggest a role for ESCRT-like proteins in the STIV replication cycle.
CRISPR/Cas Viral Defense Immune System
CRISPR/Cas immune systems were recently discovered in archaea and bacteria as RNA-based immune systems that defend against invasions of viruses and plasmids [11,34,35]. CRISPRs (clusters of regularly interspaced palindromic repeats) are identical repeats separated by unique spacer sequences of constant length. They originate from extra-chromosomal gene elements and are integrated to chromosomes and present in almost all archaea and about 40% of bacteria. Small RNAs transcribed from the CRISPR loci lead Cas proteins to recognize and degrade the invading nucleic acids. Almost 100% immunity was reported in S. solfataricus against the recombinant SSV1 virus when the chromosomal CRISPR spacer matched perfectly to the proto-spacer [36]. STIV genes were shown to be biased for matching non-coding regions or the anti-sense strands to the CRISPR spacers in the S. solfataricus genome [37]. More insights are sure to emerge and reveal the fundamentals of the inhibitory or regulatory roles of CRISPR/Cas systems in propagating archaeal viruses.
Conclusions
STIV infection causes significant remodeling and alteration of sulfolobus host. The genomics, proteomics, cellular and structural studies have taught us surprising and fascinating lessons of the life from hot springs. As our knowledge keeps evolving, more insights regarding viral replication and viral-host interactions are sure to be uncovered. The fundamental mechanisms underlying formation of the unique viral-induced pyramid-like cellular protrusions and their roles in viral replication and viral release, as well as the viral and cellular factors involved in viral replication and cellular overtaking will undoubtedly advance our understanding of the interplay between viruses and hosts in archaea.
Highlights.
STIV major capsid protein structure and capsid architecture were conserved in PRD1-Adeno lineage.
Replication, assembly and maturation of STIV were characterized by transcriptom, EM and cryo-ET.
Infection induced cellular pyramid protrusions mediate a novel viral release mechanism.
Acknowledgements
This work was supported by grant from the National Institutes of Health GM54076.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Prangishvili D, Forterre P, Garrett RA. Viruses of the Archaea: a unifying view. Nature reviews. Microbiology. 2006;4:837–848. doi: 10.1038/nrmicro1527. [DOI] [PubMed] [Google Scholar]
- 2.Snyder JC, Stedman K, Rice G, Wiedenheft B, Spuhler J, Young MJ. Viruses of hyperthermophilic Archaea. Res Microbiol. 2003;154:474–482. doi: 10.1016/S0923-2508(03)00127-X. [DOI] [PubMed] [Google Scholar]
- 3.Rice G, Stedman K, Snyder J, Wiedenheft B, Willits D, Brumfield S, McDermott T, Young MJ. Viruses from extreme thermal environments. Proc Natl Acad Sci U S A. 2001;98:13341–13345. doi: 10.1073/pnas.231170198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ortmann AC, Wiedenheft B, Douglas T, Young M. Hot crenarchaeal viruses reveal deep evolutionary connections. Nat Rev Microbiol. 2006;4:520–528. doi: 10.1038/nrmicro1444. [DOI] [PubMed] [Google Scholar]
- 5.Lawrence CM, Menon S, Eilers BJ, Bothner B, Khayat R, Douglas T, Young MJ. Structural and functional studies of archaeal viruses. The Journal of biological chemistry. 2009;284:12599–12603. doi: 10.1074/jbc.R800078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Snyder JC, Young MJ. Advances in understanding archaea-virus interactions in controlled and natural environments. Current opinion in microbiology. 2011;14:497–503. doi: 10.1016/j.mib.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 7.Brumfield SK, Ortmann AC, Ruigrok V, Suci P, Douglas T, Young MJ. Particle assembly and ultrastructural features associated with replication of the lytic archaeal virus sulfolobus turreted icosahedral virus. J Virol. 2009;83:5964–5970. doi: 10.1128/JVI.02668-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engelhardt H, Peters J. Structural research on surface layers: a focus on stability, surface layer homology domains, and surface layer-cell wall interactions. J Struct Biol. 1998;124:276–302. doi: 10.1006/jsbi.1998.4070. [DOI] [PubMed] [Google Scholar]
- 9. Ortmann AC, Brumfield SK, Walther J, McInnerney K, Brouns SJ, van de Werken HJ, Bothner B, Douglas T, van de Oost J, Young MJ. Transcriptome analysis of infection of the archaeon Sulfolobus solfataricus with Sulfolobus turreted icosahedral virus. J Virol. 2008;82:4874–4883. doi: 10.1128/JVI.02583-07.. Microarray analysis of STIV infection revealed insights of the timing and extent of virus transcription, as well as differential regulation of host genes.
- 10.Khayat R, Fu CY, Ortmann AC, Young MJ, Johnson JE. The architecture and chemical stability of the archaeal sulfolobus turreted icosahedral virus. J Virol. 2010;84:9575–9583. doi: 10.1128/JVI.00708-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Happonen LJ, Redder P, Peng X, Reigstad LJ, Prangishvili D, Butcher SJ. Familial relationships in hyperthermo- and acidophilic archaeal viruses. J Virol. 2010;84:4747–4754. doi: 10.1128/JVI.02156-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rice G, Tang L, Stedman K, Roberto F, Spuhler J, Gillitzer E, Johnson JE, Douglas T, Young M. The structure of a thermophilic archaeal virus shows a double-stranded DNA viral capsid type that spans all domains of life. Proc Natl Acad Sci U S A. 2004;101:7716–7720. doi: 10.1073/pnas.0401773101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Khayat R, Tang L, Larson ET, Lawrence CM, Young M, Johnson JE. Structure of an archaeal virus capsid protein reveals a common ancestry to eukaryotic and bacterial viruses. Proc Natl Acad Sci U S A. 2005;102:18944–18949. doi: 10.1073/pnas.0506383102.. The study reported the x-ray crystal structure of the major capsid protein of STIV and revealed the evolutionary link between three domains of life in PRD1-Adeno viral lineage.
- 14.Benson SD, Bamford JK, Bamford DH, Burnett RM. Viral evolution revealed by bacteriophage PRD1 and human adenovirus coat protein structures. Cell. 1999;98:825–833. doi: 10.1016/s0092-8674(00)81516-0. [DOI] [PubMed] [Google Scholar]
- 15.Abrescia NG, Grimes JM, Kivela HM, Assenberg R, Sutton GC, Butcher SJ, Bamford JK, Bamford DH, Stuart DI. Insights into virus evolution and membrane biogenesis from the structure of the marine lipid-containing bacteriophage PM2. Molecular cell. 2008;31:749–761. doi: 10.1016/j.molcel.2008.06.026. [DOI] [PubMed] [Google Scholar]
- 16.Nandhagopal N, Simpson AA, Gurnon JR, Yan X, Baker TS, Graves MV, Van Etten JL, Rossmann MG. The structure and evolution of the major capsid protein of a large, lipid-containing DNA virus. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:14758–14763. doi: 10.1073/pnas.232580699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bazinet C, King J. The DNA translocating vertex of dsDNA bacteriophage. Annu Rev Microbiol. 1985;39:109–129. doi: 10.1146/annurev.mi.39.100185.000545. [DOI] [PubMed] [Google Scholar]
- 18.Bahar MW, Graham SC, Stuart DI, Grimes JM. Insights into the evolution of a complex virus from the crystal structure of vaccinia virus D13. Structure. 2011;19:1011–1020. doi: 10.1016/j.str.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krupovic M, Bamford DH. Virus evolution: how far does the double beta-barrel viral lineage extend? Nature reviews. Microbiology. 2008;6:941–948. doi: 10.1038/nrmicro2033. [DOI] [PubMed] [Google Scholar]
- 20.Abrescia NG, Cockburn JJ, Grimes JM, Sutton GC, Diprose JM, Butcher SJ, Fuller SD, San Martin C, Burnett RM, Stuart DI, et al. Insights into assembly from structural analysis of bacteriophage PRD1. Nature. 2004;432:68–74. doi: 10.1038/nature03056. [DOI] [PubMed] [Google Scholar]
- 21.Cockburn JJ, Abrescia NG, Grimes JM, Sutton GC, Diprose JM, Benevides JM, Thomas GJ, Jr, Bamford JK, Bamford DH, Stuart DI. Membrane structure and interactions with protein and DNA in bacteriophage PRD1. Nature. 2004;432:122–125. doi: 10.1038/nature03053. [DOI] [PubMed] [Google Scholar]
- 22.Maaty WS, Ortmann AC, Dlakic M, Schulstad K, Hilmer JK, Liepold L, Weidenheft B, Khayat R, Douglas T, Young MJ, et al. Characterization of the archaeal thermophile Sulfolobus turreted icosahedral virus validates an evolutionary link among double-stranded DNA viruses from all domains of life. J Virol. 2006;80:7625–7635. doi: 10.1128/JVI.00522-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fu CY, Wang K, Gan L, Lanman J, Khayat R, Young MJ, Jensen GJ, Doerschuk PC, Johnson JE. In vivo assembly of an archaeal virus studied with whole-cell electron cryotomography. Structure. 2010;18:1579–1586. doi: 10.1016/j.str.2010.10.005.. The whole cell cryo-electron tomography revealed vivid images of STIV assembly, maturation, particle distribution and viral induced host pyramid protrusions and ultra-structural changes in close to native sample preservation.
- 24.Chlanda P, Carbajal MA, Cyrklaff M, Griffiths G, Krijnse-Locker J. Membrane rupture generates single open membrane sheets during vaccinia virus assembly. Cell Host Microbe. 2009;6:81–90. doi: 10.1016/j.chom.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 25.Wang K, Fu CY, Khayat R, Doerschuk PC, Johnson JE. In vivo virus structures: Simultaneous classification, resolution enhancement, and noise reduction in whole-cell electron tomography. J Struct Biol. 2011 doi: 10.1016/j.jsb.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Netherton C, Moffat K, Brooks E, Wileman T. A guide to viral inclusions, membrane rearrangements, factories, and viroplasm produced during virus replication. Adv Virus Res. 2007;70:101–182. doi: 10.1016/S0065-3527(07)70004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bravo A, Serrano-Heras G, Salas M. Compartmentalization of prokaryotic DNA replication. FEMS Microbiol Rev. 2005;29:25–47. doi: 10.1016/j.femsre.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 28.Cook PR. The organization of replication and transcription. Science. 1999;284:1790–1795. doi: 10.1126/science.284.5421.1790. [DOI] [PubMed] [Google Scholar]
- 29.Quax TE, Krupovic M, Lucas S, Forterre P, Prangishvili D. The Sulfolobus rod-shaped virus 2 encodes a prominent structural component of the unique virion release system in Archaea. Virology. 2010;404:1–4. doi: 10.1016/j.virol.2010.04.020. [DOI] [PubMed] [Google Scholar]
- 30. Quax TE, Lucas S, Reimann J, Pehau-Arnaudet G, Prevost MC, Forterre P, Albers SV, Prangishvili D. Simple and elegant design of a virion egress structure in Archaea. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:3354–3359. doi: 10.1073/pnas.1018052108.. The study characterized the structure and the protein component of pyramid-like protrusions formed in Sulfolobus infected by Sulfolobus islandicus rod- shaped virus 2.
- 31.Bize A, Karlsson EA, Ekefjard K, Quax TE, Pina M, Prevost MC, Forterre P, Tenaillon O, Bernander R, Prangishvili D. A unique virus release mechanism in the Archaea. Proc Natl Acad Sci U S A. 2009;106:11306–11311. doi: 10.1073/pnas.0901238106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Snyder JC, Brumfield SK, Peng N, She Q, Young MJ. Sulfolobus turreted icosahedral virus c92 protein responsible for the formation of pyramid-like cellular lysis structures. J Virol. 2011;85:6287–6292. doi: 10.1128/JVI.00379-11.. This study investigated the STIV gene products required for pyramid formation in the host and demonstrates its essential roles in virus replication besides in pyramid-mediated viral release.
- 33.Snyder JC, Young MJ. Potential role of cellular ESCRT proteins in the STIV life cycle. Biochemical Society transactions. 2011;39:107–110. doi: 10.1042/BST0390107. [DOI] [PubMed] [Google Scholar]
- 34.Snyder JC, Bateson MM, Lavin M, Young MJ. Use of cellular CRISPR (clusters of regularly interspaced short palindromic repeats) spacer-based microarrays for detection of viruses in environmental samples. Applied and environmental microbiology. 2010;76:7251–7258. doi: 10.1128/AEM.01109-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garrett RA, Shah SA, Vestergaard G, Deng L, Gudbergsdottir S, Kenchappa CS, Erdmann S, She Q. CRISPR-based immune systems of the Sulfolobales: complexity and diversity. Biochemical Society transactions. 2011;39:51–57. doi: 10.1042/BST0390051. [DOI] [PubMed] [Google Scholar]
- 36.Manica A, Zebec Z, Teichmann D, Schleper C. In vivo activity of CRISPR-mediated virus defence in a hyperthermophilic archaeon. Molecular microbiology. 2011;80:481–491. doi: 10.1111/j.1365-2958.2011.07586.x. [DOI] [PubMed] [Google Scholar]
- 37.Shah SA, Hansen NR, Garrett RA. Distribution of CRISPR spacer matches in viruses and plasmids of crenarchaeal acidothermophiles and implications for their inhibitory mechanism. Biochemical Society transactions. 2009;37:23–28. doi: 10.1042/BST0370023. [DOI] [PubMed] [Google Scholar]