Summary
In spite of its recent achievements, the technique of single particle electron cryomicroscopy (cryoEM) has not been widely used to study proteins smaller than 100kDa, although it is a highly desirable application of this technique. One fundamental limitation is that images of small proteins embedded in vitreous ice do not contain adequate features for accurate image alignment. We describe a general strategy to overcome this limitation by selecting a fragment antigen binding (Fab) to form a stable and rigid complex with a target protein, thus providing a defined feature for accurate image alignment. Using this approach, we determined a three-dimensional structure of a ~65 kDa protein by single particle cryoEM. Because Fabs can be readily generated against a wide range of proteins by phage display, this approach is generally applicable to study many small proteins by single particle cryoEM.
INTRODUCTION
Single particle electron cryomicroscopy (cryoEM) has achieved great success in the last decade, and has become a versatile technique for structural analysis of biological macromolecular complexes at high resolution. In recent years, this method has achieved near-atomic resolution for large protein assemblies with high symmetry, such as non-enveloped viruses with icosahedral symmetry (Chen et al., 2009; Wolf et al., 2010; Yu et al., 2008; Zhang et al., 2010; Zhang et al., 2008). It has also attained resolutions of 4 – 5 Å for large macromolecular complexes without symmetry, such as mammalian chaperonin (Cong et al., 2010) and the ribosome (Armache et al., 2010). In single particle cryoEM, purified samples in their native conformations are embedded in vitreous ice and imaged in an electron microscope at liquid nitrogen temperature using a limited electron dose, typically ~ 20 e−/Å2. A large number of images of individual particles representing different views of the same molecule are selected from many electron micrographs and used to calculate a three-dimensional (3D) reconstruction. The resolution of a 3D reconstruction is iteratively improved by refining the orientation parameters of each individual particle, i.e. three Euler angles and two in-plane shifts, and the microscope parameters, including defocus and astigmatism. In addition to homogeneity, the highest achievable resolution of a given sample is greatly dependent on the ability to refine these parameters to high accuracy. In general, large molecules are relatively easy to be recognized in noisy low-dose images of frozen hydrated samples and these particles often have sufficient structural features to facilitate accurate determination of their orientation parameters (Henderson, 1995). So far, all near-atomic resolution structures determined by single particle cryoEM are from molecular complexes with a total molecular weight on the order of a mega-Dalton.
Single particle cryoEM has also been applied to study much smaller proteins, such as the human transferrin receptor-transferrin complex (~300 kDa) (Cheng et al., 2004) and gamma-secretase (~200 kDa) (Osenkowski et al., 2009). However, the process of collecting and processing images of such small proteins is much more difficult than larger targets. Although the precise lower limit to the size of a molecule whose structure can be reconstructed is not known, it is generally acknowledged that using current technologies it is difficult to obtain 3D reconstructions at a resolution better than ~20 Å from proteins smaller than 200 kDa, and nearly impossible to study proteins smaller than 100 kDa. This is in line with predictions made more than fifteen years ago (Henderson, 1995). Due to such a size limitation, we often refrain ourselves from applying this technique to small proteins, despite tremendous needs for such applications. In particular, many integral membrane proteins are smaller than 100 kDa. Yet, examples of structure determination of integral membrane proteins to subnanometer resolution by single particle cryoEM are few. In practice, negative stain single particle EM has often been used to study 3D structures of relatively small proteins, albeit only to a resolution lower than 20 Å (Lederkremer et al., 2001).
There are several physical, as well as technological, challenges in using single particle cryoEM to determine 3D reconstructions of small molecules, especially those smaller than 100kDa. First of all, it is difficult to visualize such small proteins embedded in vitreous ice with a limited electron dose. Nevertheless, in view of major technological developments in the field of electron microscopy during the last decade, visualizing small frozen hydrated protein particles is no longer an impossible task. Taking advantage of a field emission electron source, one can image small proteins with a relatively high defocus. One can also use a lower accelerating voltage and a small objective aperture to increase the image contrast, although these practices have the disadvantage of reducing the achievable resolution (Glaeser et al., 2011). There is great hope that Zernicke-type phase plate technology, currently undergoing intensive development, will make visualization of small proteins easier (Murata et al., 2010). Secondly, even when they appear with sufficient contrast by the above-mentioned methods, small particles often lack well-defined structural features required to facilitate accurate image alignment. Moreover, 3D reconstructions calculated from noisy images of small molecules are strongly influenced by reference models, a phenomenon known as model-induced bias. Therefore, it is difficult to validate the correctness of 3D reconstructions from molecules with unknown structures when the resolution of these reconstructions is not sufficient to resolve secondary structural features. Only when these challenges are overcome, single particle cryoEM studies of small proteins will be possible.
Monoclonal fragments antigen binding (Fabs) could provide a tool to address these challenges by forming stable and rigid complexes with their target proteins. A Fab is a ~50 kDa antibody fragment composed of one constant domain and one variable domain from both heavy and light chains. It retains the full antigen binding capability through its variable domains. Besides having potential in pharmacological applications and drug development for the treatment of human diseases (Chan et al., 2009; Marasco and Sui, 2007), Fabs are also powerful research tools widely used in structural biology. In X-ray crystallography, Fabs have been used to assist crystallization of soluble and integral membrane proteins by stabilizing specific conformations of the target proteins and facilitating crystal packing (Newton et al., 2008; Rasmussen et al., 2011; Ye et al., 2008). In cellular EM, antibodies conjugated with gold cluster are used for “immuno-gold” labeling to localize target proteins (Roth, 1996). Fabs have also been used in negative stain single particle EM to label domains within a complex (Aebi et al., 1977; Nakagawa et al., 2005) and to increase the size of a target protein for better visualization in cryoEM (Jiang et al., 2004). A method of using antibody conjugated with heavy-metal cluster for image alignment was also proposed (Jensen and Kornberg, 1998).
In this study, we introduce a new approach of using a monoclonal Fab to enable single particle cryoEM studies of small proteins. We show that the bound Fab not only increases the effective mass of the target protein but also provides a fiducial marker with specific features to facilitate image alignment and to validate the final 3D reconstruction. We present examples to demonstrate each step of this approach, from the selection of suitable Fabs against a number of small proteins to the determination of a cryoEM 3D reconstruction of a 65 kDa protein in complex with two Fabs. Our experiments have resulted in a methodological strategy that promises to be generally applicable to a wide variety of small proteins, including integral membrane proteins.
RESULTS
Overall strategy to enable single particle cryoEM of small proteins
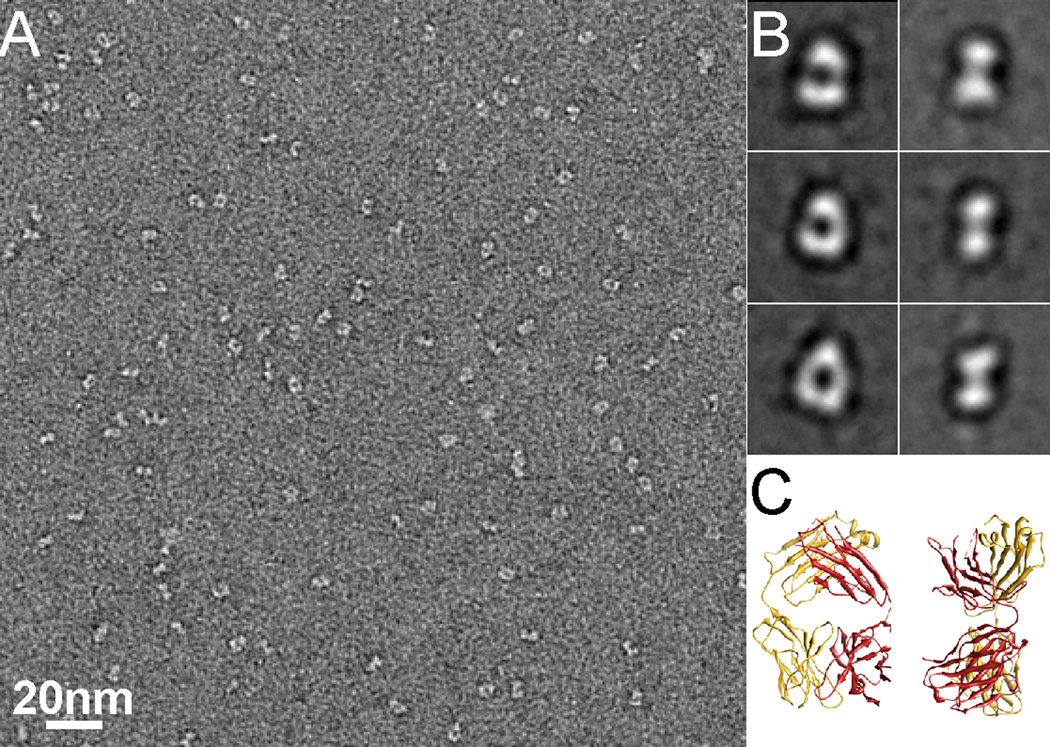
Our overall strategy is to use one or more monoclonal Fabs to form a stable and rigid complex with the target protein. Forming such a complex increases the effective size of the target protein. By virtue of this enlargement, it becomes easier to visualize by cryoEM (Jiang et al., 2004). More importantly, all available atomic structures of Fabs have similar and characteristic shapes, having variable and constant domains connected by a short linker called “elbow” as shown by a representative Fab (pdb code: 1M71) in Figure 1C. These characteristic shapes can also be recognized in raw images as well as two-dimensional class averages, with two typical views: one is characterized by a hole in the middle, and the other by two blobs (Figure 1). Therefore, a Fab bound to a target protein would add a recognizable feature and facilitate image alignment of otherwise featureless small proteins. Furthermore, the presence of a distinct feature with the size and shape of a Fab in the 3D reconstruction provides a strong evidence for the correctness of the structure and serves as a validation criterion. In this study, we demonstrate this strategy by applying it to the following protein examples:
Figure 1. Negative stain EM of Fab.
(A) An EM image of negatively stained Fab J16. The shape of Fab can be easily recognized in the raw images. (B) Representative 2D class averages showing two typical views of negatively stained Fab. (C) Two different views of a typical Fab atomic structure (pdb code: 1M71) in ribbon diagram corresponding to the class averages shown in B. See also Figure S1.
Human immunodeficiency virus (HIV-1) integrase (IN)
Retroviral integrase (IN) is an enzyme that catalyses the incorporation of a viral genome into the chromosome of infected host cells. HIV-1 IN plays a key role in the process of HIV viral replication (Kukolj and Skalka, 1995) and is a potential drug target against HIV infection (Savarino, 2006). During the integration process, IN functions as a tetramer and forms a complex with host and viral DNA. There are a number of crystal structures of truncated HIV-1 IN dimer (Chen et al., 2000; Maignan et al., 1998; Wang et al., 2001) and models of the tetramer IN-DNA complex based on the structure of a prototype foamy virus IN (Krishnan et al., 2010). The full length HIV-1 IN monomer has a molecular weight of only ~32 kDa. Structural studies of IN by single particle EM have been difficult because of its small size (Ren et al., 2007). Examination of purified IN by negative stain EM showed some degree of heterogeneity in particle size and shape. The source of such heterogeneity is not clear, but it may be due to the occurrence of different views of the same molecule or a mixture of IN dimers and tetramers. Our previous attempts to calculate a reliable 3D reconstruction of IN were not successful. In this study, we used IN to demonstrate the entire process of our approach and determined a 3D reconstruction of IN dimer in complex with two Fabs.
Human proprotein convertase subtilisin/kexin type 9 (PCSK9)
Human PCSK9 is a key ~70 kDa regulator of serum hepatocyte low-density lipoprotein receptor (LDL-R) expression (Costet et al., 2008; Horton et al., 2009). Since it interacts directly with the LDL-R and regulates the serum LDL cholesterol level, it is an attractive therapeutic target for treatment of hypercholesterolemia (Steinberg and Witztum, 2009). It has been shown that it is possible to lower plasma LDL levels by blocking the interaction between PCSK9 and LDL-R using a neutralizing antibody (Chan et al., 2009). Analysis of PCSK9 images in negative stain by the standard procedure of multi-reference alignment (MRA) and classification (Shaikh et al., 2008) yielded 2D class averages showing a triangular shaped particle with the expected dimensions for this molecule (Figure S1A). However, it was not possible to recognize different domains of PCSK9 in both raw images and class averages. We determined a negative stain 3D reconstruction of PCSK9 in complex with a neutralizing Fab (J16) and revealed how J16 interacts with PSCK9.
Integral membrane proteins
Furthermore, we used two integral membrane proteins, an E. coli homologue of the mammalian vesicular glutamate transporter (EcoliVGLUT3) and an ATP binding cassette (ABC) transporter, to assess the potential applicability of using Fab to facilitate their structural analysis by single particle cryoEM. Detergent solubilized and purified EcoliVGLUT3 is a ~50kDa monomer, and appears in negatively stain EM as a donut shape with a diameter of about 5 nm (Figure S1B). EcoliVGLUT3 particles embedded in vitreous ice are imaged with good contrast at a relative high defocus value (>3µm). In spite of that, they all appear as single dots lacking any structural features (Figure S1C). It is obvious that images of such particles are difficult to be aligned with each other. The ABC transporter is a ~130kDa heterodimer, which by itself is rather small for single particle cryoEM studies. We used EcoliVGLUT3-Fab and ABC transporter-Fab complexes to demonstrate the Fab selection process.
Selection of a suitable Fab
With the technological advances in generating recombinant monoclonal Fabs, such as phage-display technology, it is possible to produce multiple Fabs against almost any protein (Fellouse et al., 2007). However, it is unlikely that each and every Fab is a good facilitator for single particle cryoEM study of its target protein. Some Fabs may bind with a high affinity to a flexible domain of the protein and thus do not form a rigid complex with its target. In the past, Fabs were seldom used as fiducial markers for image alignment in single particle cryoEM because of such potential flexibility of Fab decoration (Harris et al., 2011; Jiang et al., 2004). Because we rely on the added structural feature of a tightly bound Fab to enable accurate image alignment, a suitable Fab must not only bind with high affinity but also form a rigid complex with the target protein. Therefore, the first and the most critical step is the selection of suitable candidates from those already available for the target protein. When viewed by negative stain EM, particles of the protein-Fab complex must show certain level of homogeneity, as this would be expected from rigid complexes.
HIV-1 IN
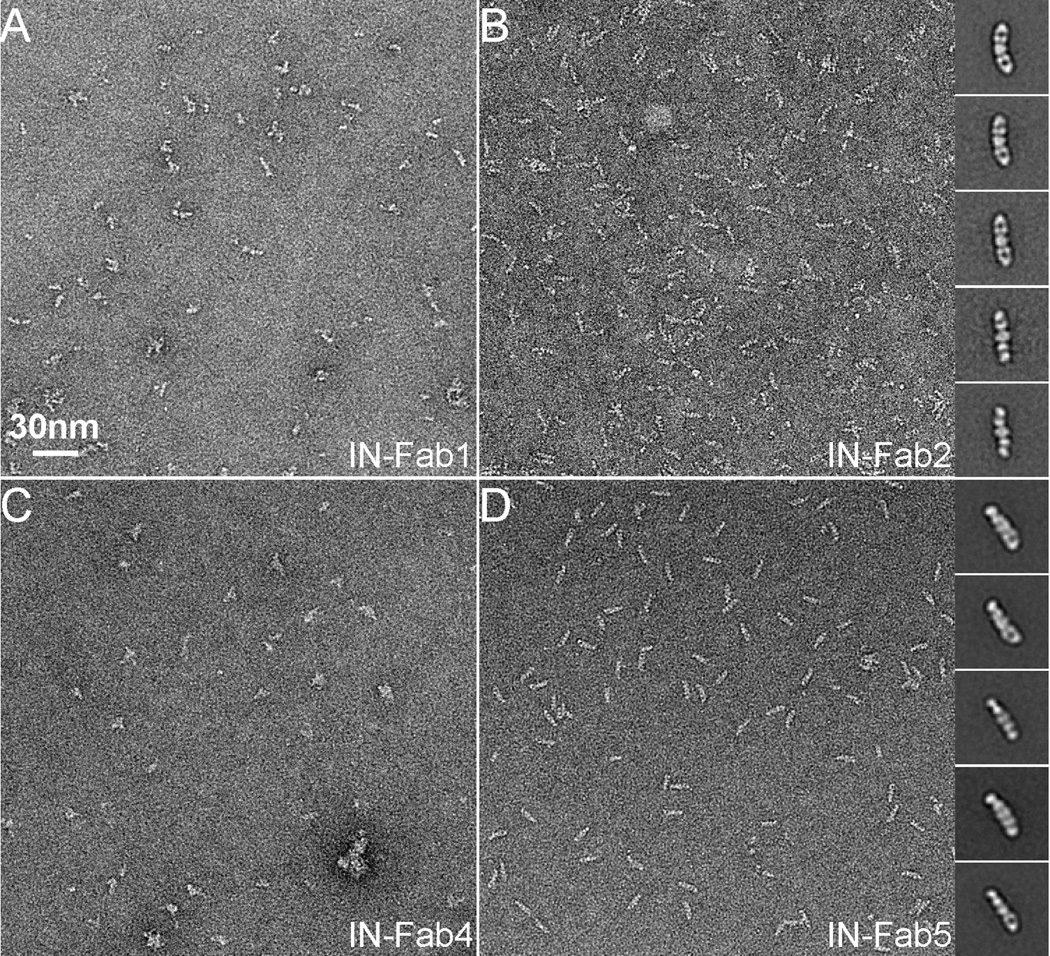
We indentified five different Fabs against the catalytic core of HIV-1 IN by phage-display experiments. IN and each Fab were mixed together and subjected to size exclusion chromatography. Only four of the five Fabs eluted with IN in higher molecular weight fractions. Although this indicated the formations of IN-Fab complexes, it was difficult to conclude whether the Fabs were bound to a dimeric or a tetrameric form of IN. Brief examination by negative stain EM showed that only two of them (Fab2 and Fab5) form a stable complex with the IN and appeared homogeneous in shape and size (Figure 2). We manually selected ~1,500 particles from IN-Fab2 and ~2,500 from IN-Fab5 for further image analysis. Selected particles were boxed out from the raw micrographs and were subjected to multiple rounds of MRA and classification. Inserted in Figure 2B and D are 2D class averages of the IN-Fab2 and IN-Fab5 complexes. By comparing these class averages with those from a Fab alone (Figure 1B), we easily identified densities corresponding to two Fabs bound to IN from opposite sides. The visibility of the Fab with its characteristic shape in 2D class averages suggests that both IN-Fab2 and IN-Fab5 are stable and reasonably rigid complexes.
Figure 2. Selection of a suitable Fab to enable single particle cryoEM studies of HIV-1 IN.
(A–D): EM micrographs of HIV-1 IN in complex with four different Fabs. It is obvious that IN-Fab2 and IN-Fab5 are more homogeneous than the other two IN-Fab complexes. Inserted in (B) and (D) are representative 2D class averages of IN-Fab2 and IN-Fab5 complexes. In all 2D class averages, there are two Fabs bound to one IN dimer.
PCSK9
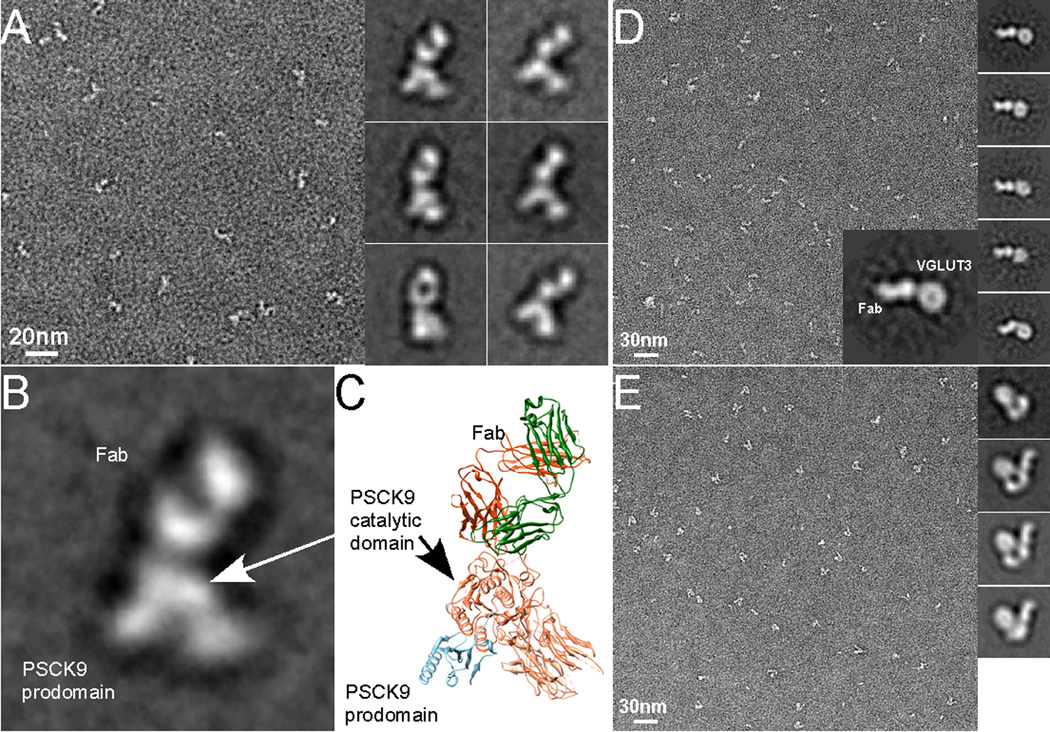
A neutralizing Fab, J16, generated by hybridoma technology (Liang et al., 2011) forms a stable complex with PCSK9. In contrast to the particles of PCSK9 alone where impurities were often observed, particles of the PCSK9-J16 complex were highly homogeneous, both in size and shape (Figure 3A). The standard MRA and classification procedure revealed a number of different class averages of the complex (right panel of Figure 3A), which were confirmed later as corresponding to different orientations of the same complex. They all have recognizable features. The shape of the domain in the top is similar to the image of an isolated Fab that has a hole in the middle as shown in Figure 1B. The domain in the bottom resembles the atomic structure of PCSK9. As a simple comparison, we placed the crystal structures of PCSK9 (pdb code: 2P4E) (Cunningham et al., 2007) and a representative Fab (pdb code: 1M71) together and correlated their orientations as revealed in a class average. Such qualitative similarity suggests that the Fab binds to the catalytic domain of PCSK9 (Figure 3B and C).
Figure 3. Negative stain EM analysis of protein-Fab complexes.
(A) An EM micrograph of negatively stained PCSK9-J16 complex and its representative 2D class averages. (B) On the left is an enlarged view of a 2D class average of PCSK9-J16 complex. On the right is a ribbon diagram in which a PCSK9 (pdb code: 2P4E) and a typical Fab were placed together with their orientations closely matching that of the 2D class average on the left. Domains of PCSK9 and Fab are indicated. (C) An EM micrograph of negatively stained EcoliVGLUT3- BC1 complex and its representative 2D class averages. The large insertion is one of the 2D class averages, in which domains corresponding to Fab BC1 and EcoliVGLUT3 are indicated. (D) An EM micrograph of the ABC transporter in complex with a Fab. On the right are representative 2D class averages. The first 2D class average is from ABC transporter alone and the rest are those of the complex.
EcoliVGLUT3
Fabs against EcoliVGLUT3 were identified by phage display experiments using a human naïve B cell phage library (Duriseti et al.). Only one Fab, BC1, was shown to bind EcoliVGLUT3 through size exclusion chromatography. The EcoliVGLUT3-BC1 complex was analyzed by negative stain EM to assess homogeneity of the complex. Images of negatively stained EcoliVGLUT3-BC1 complex were recorded (Figure 3D) and 1,196 particles were manually selected for standard 2D image analysis. In a typical class average of the complex (right panel and insertion of Figure 3D), EcoliVGLUT3 is readily recognized. The Fab is also prominent with its characteristic shape that matches the dimensions of an isolated Fab. Since there is only one Fab in the complex, the round shaped EcoliVGLUT3 is most likely a monomer.
ABC transporter
We identified a number of Fabs against the ABC transporter by phage display experiments, using the same phage library used for EcoliVGLUT3. Among the Fabs that were shown to bind the ABC transporter, three were used in this study. These ABC transporter – Fab complexes were analyzed by negative stain EM. One of the three complexes is shown in Figure 3E. The ABC transporter has a relative large soluble domain and its molecular weight is considerably larger than EcoliVGLUT3. Nevertheless, the bound Fab can still be seen clearly in all 2D class averages.
In all the examples presented here, the characteristic shape of the bound Fab can be identified in 2D class averages. This observation indicates that in each of these cases, the Fab forms a stable and rigid complex with its target protein. An unstably bounded Fab would result in the smeary density in 2D class averages without well-defined typical Fab features. As a selection criterion, Fabs that show clear structural features in 2D class averages are good candidates to facilitate image alignment and structure determination of their target proteins by single particle cryoEM.
Negative stain 3D reconstruction of protein-Fab complex
Having identified suitable Fabs, we then tested if we could use negative stain EM to calculate reliable 3D reconstructions of protein-Fab complexes. Here, we judge the correctness of a 3D reconstruction by the shapes of a chosen target protein and the bound Fab. Out of all mentioned samples, only HIV-1 IN and PCSK9 have had their crystal structures determined and are available in the protein data bank (PDB). We, therefore, only determined 3D reconstructions of these two proteins in complex with their corresponding Fabs, and used crystal structures of the uncomplexed proteins and representative Fabs to evaluate the results. We used a procedure that combines random conical tilt (RCT) procedure with projection matching of images from untilted micrographs only (henceforth referred to as 0° images). RCT is a well-established single particle EM procedure that produces a reliable 3D reconstruction without an initial model (Radermacher et al., 1987; Shaikh et al., 2008). But 3D reconstructions calculated from this procedure suffer from flattening and missing cone artifacts (Cheng et al., 2006). Flattening is often caused by adsorbing proteins to the supporting carbon supporting film during the process of negative staining (Cheng et al., 2006), and it is more visible in tilted image than in 0° image. Therefore, we used a RCT reconstruction as an initial model for projection matching to align all 0° images. We excluded all tilted images in this step. With a sufficiently large number of different views, this approach produces a 3D reconstruction with less flattening artifacts.
PCSK9-J16 complex
We collected 50 tilt-pair micrographs of the PCSK9-Fab complex at 60°/0° tilt and selected 1,422 particles from this set (Figure S2A and B). Additionally, we recorded 22 more micrographs from untilted specimen and selected a total of 2,454 particles. In the first step, the untilted particles from the tilt-pair images were classified into 20 classes. We selected a number of classes with the highest number of particles and computed RCT reconstructions from each of them. Certain features were easily recognized as common to all 3D reconstructions obtained directly from the RCT, i.e. two domains linked together in the shape of a foot. But these RCT 3D reconstructions lack clear structural features perpendicular to the carbon support film (Figure S2C) likely due to flattening induced by the negative staining procedure (Cheng et al., 2006) as well as to the missing cone effect (Radermacher et al., 1987). Nevertheless, their overlaps in space suggest that these 3D reconstructions and their class averages correspond to the same object adsorbed to the carbon film in different orientations. In the next step, we calculated a 3D reconstruction of the PCSK9-J16 complex by projection matching, as described above. The final 3D reconstruction (Figure 4, movie S1) has a resolution of 26 Å, as determined by the 0.5 criteria of Fourier Shell Correlation (FSC) curve. Distinctive structural features of both PCSK9 and Fab are clearly visible in the 3D reconstruction. The J16 appears with the characteristic shape of a Fab, i.e. a bi-lobed shape in one view and a “hole” in the center in another view. The three domains of PCSK9 are also well resolved.
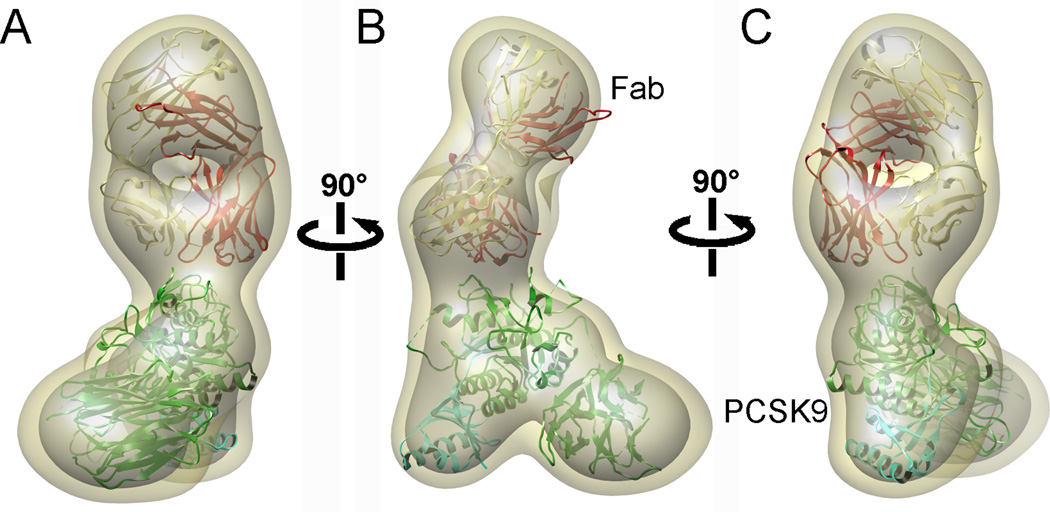
Figure 4. Negative stain 3D reconstruction of PCSK9-Fab complex at a resolution of 26 Å.
(A–C): Three different views of the 3D reconstruction rotated 90° around a vertical axis. The atomic structure of PCSK9 and Fab were docked as rigid bodies into the 3D reconstruction. The density of the 3D reconstruction is displayed at two different isosurface levels (high in grey and low in khaki). See also Figure S2 and Movie S1.
We assembled an atomic model of the PCSK9-J16 complex by docking the crystal structures of PCSK9 and Fab separately as rigid bodies into the 3D reconstructions (Fig. 4). In this model, as suggested by the 2D class average (Figure 3B and C), the Fab binds to the subtilisin-like catalytic domain of PCKS9, overlapping with residues comprising the previously identified LDL-R binding site (Kwon et al., 2008; Zhang et al., 2007). A separate study has shown that J16 has an antagonistic effect on the interaction between PCSK9 and the LDL-R (Liang et al., 2011), which leads to the degradation of LDL-R in the lysosome (Horton et al., 2007). Our model therefore, reveals how this antagonizing effect by the Fab J16 occurs by directly blocking the interaction between PCSK9 and LDL-R.
HIV-1 IN-Fab5 complex
A 3D reconstruction of HIV-1 IN-Fab5 complex was also determined by the same procedure described above. In this 3D reconstruction, densities corresponding to two Fabs are easily recognized. They bind to IN from opposite sides. Atomic structures of a representative Fab (pdb code: 1M71) and the dimeric IN catalytic core (pdb code: 1EXQ) fit well into the density map. The density between the two Fabs can only accommodate a dimer, but not a tetramer, indicating that the Fabs selectively bind to the IN dimer. Considering that the crystal structure of IN catalytic core has a pseudo 2-fold axis and that the initial 3D reconstruction appears as a 2-fold symmetrical object, we applied a 2-fold symmetry and the final 3D reconstruction was calculated at a resolution of 25 Å (Fig. 5).
Figure 5. Single particle negative stain EM study of HIV-1 IN-Fab complex.
(A–C): Three different views of the 3D reconstruction (resolution 25 Å) rotated 45° around a horizontal axis. The density is displayed at two different isosurface levels (high in grey and low in khaki). The atomic structures of a Fab and an HIV-1 IN catalytic core were docked in the density map as rigid bodies.
The resolution of the maps calculated from negative stain single particle EM is, however, not sufficient by itself to distinguish the heavy and light chain of the Fab. In both cases, docking of the atomic structures of the Fabs in two different orientations yield a small difference in the cross correlations between the docked structure and the density map (0.75 vs. 0.71 for the Fab in PCSK9-J16 complex, and 0.768 vs. 0.761 for the Fab in IN-Fab5 complex, calculated using UCSF Chimera (Pettersen et al., 2004)). We chose the orientation with a higher cross correlation value. This turned out to be the correct Fab orientation for J16, confirmed by comparison with the crystal structure of PCSK9-J16 (PDB code: 3SQO) determined in a separate study (Liang et al., 2011). As for the Fab5, the difference of cross correlations between docking the Fab in two different orientations increased to 0.76 vs. 0.7 in the single particle cryoEM 3D reconstruction of IN-Fab5 at a significantly better resolution.
In both examples presented here, despite limited resolution, the shapes of the individual components, i.e. Fab, PCSK9 and IN catalytic core dimer, match well with the corresponding densities in the 3D reconstruction. Such unambiguous correspondence provides a good validation for the quality and the correctness of these 3D reconstructions.
CryoEM 3D reconstruction of protein-Fab complex
The goal of this study is to demonstrate that Fabs enable the determination of reliable 3D reconstructions of frozen hydrated small proteins by single particle cryoEM, a task that is otherwise impossible. We therefore determined a 3D reconstruction of ice-embedded IN-Fab5 complex. We have chosen to use IN-Fab5 for this demonstration, because it possesses a 2-fold symmetry that is an advantage over PCSK9-J16. By virtue of that symmetry, the total number of images required to reconstruct a 3D density maps to a given resolution is significantly reduced.
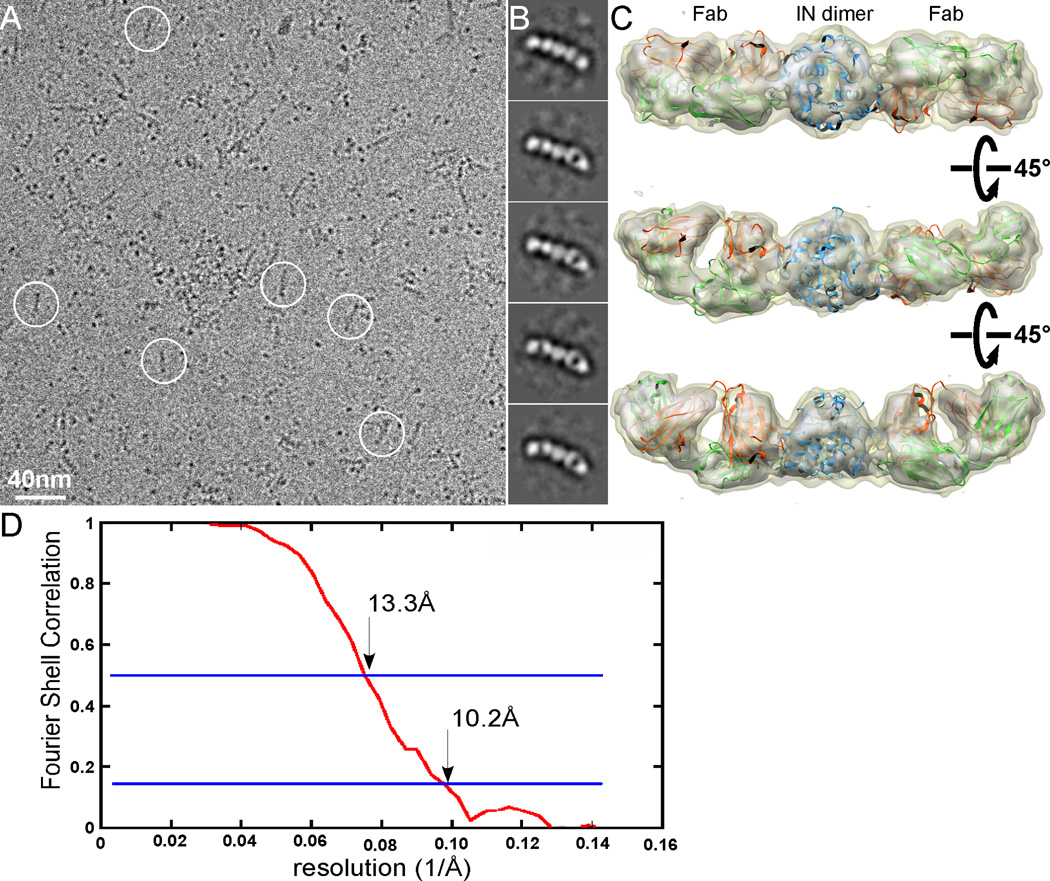
The molecular weight of the full length HIV-1 IN dimer is ~65kDa. Particles of vitreous ice-embedded IN dimer, being similar in size to EcoliVGLUT3 (Figure S1C), are too small to be visualized except as featureless dots. However, the IN-Fab5 complex has a total molecular weight of ~165kDa and a recognizable elongated shape. Figure 6A shows a raw low-dose image of the frozen hydrated IN-Fab5 complex embedded in a thin layer of vitrified ice. Many particles in this image have shapes similar to those seen in the images of the negatively stained sample, although there are also many particles that appear as single dots. These dots are either IN dimer alone, or complexes oriented with their long axis perpendicular to the ice surface. Even in the presence of such potential heterogeneity, it is easy to select out the particles that have an elongated shape, which are the IN-Fab5 complex in different side views. A total of 14,829 particles were selected, corrected for contrast transfer function (CTF), and subjected to multiple rounds of standard MRA and classification. Representative class averages are shown in Figure 6B.
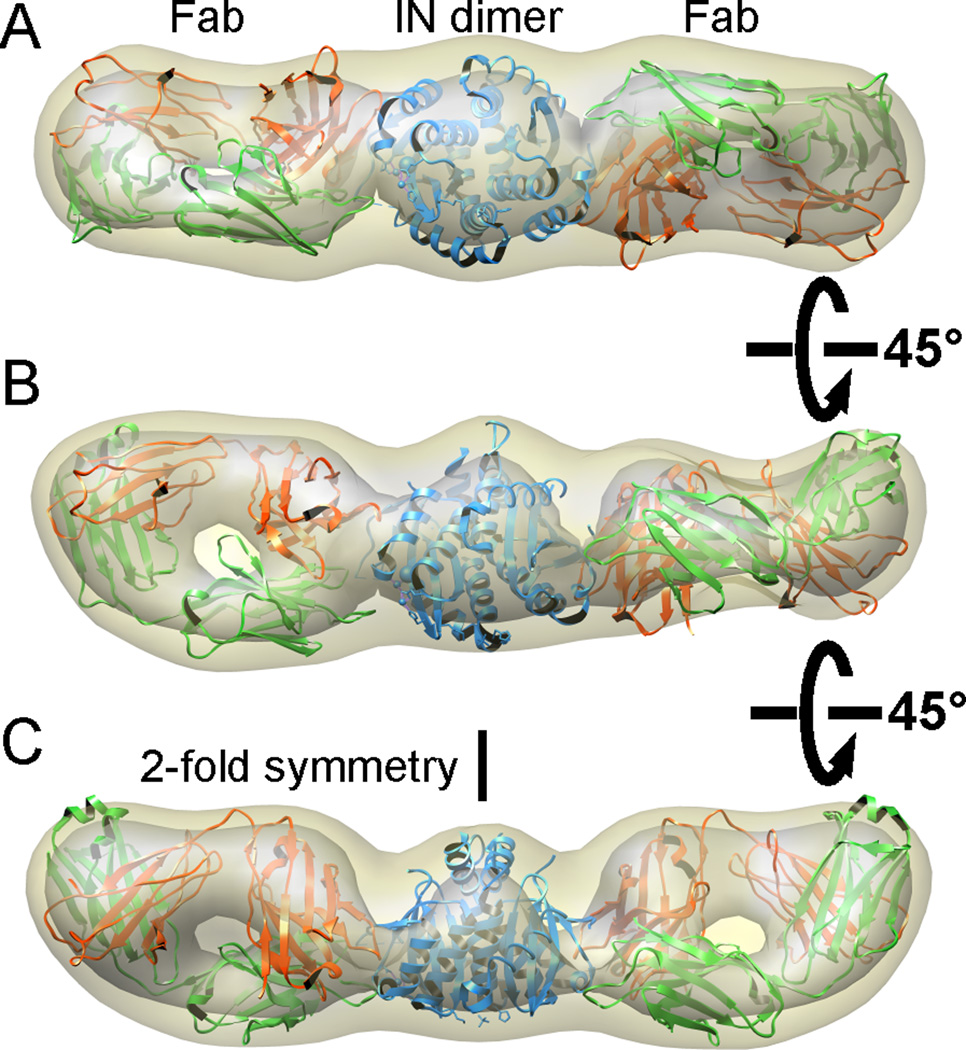
Figure 6. Single particle cryoEM of HIV-1 IN-Fab complex.
(A): A typical image of frozen hydrated IN-Fab complex embedded in vitreous ice. (B) Representative class averages of IN-Fab complex. Fabs can be clearly recognized in the raw image as well as in class averages. (C) Three different views of the 3D reconstruction oriented around a horizontal axis. The density of the 3D reconstruction is displayed at two different isosurface levels (high in grey and low in khaki). The atomic structures of a generic Fab (pdb code: 1M71) and an HIV-1 IN catalytic core were docked in the density map as rigid bodies. (D) The FSC curve of the 3D reconstruction, with the resolutions that correspond to FSC=0.5 and 0.143 marked. See also Figures S3 and S4, and Movie S2.
We used the negative stain 3D reconstruction (Fig. 5) as an initial reference model to align individual frozen hydrated IN-Fab5 particles. After multiple cycles of iterative refinement, a final 3D reconstruction of IN-Fab5 was calculated (Figure 6C, movie S2). The resolution was estimated as 13.3 Å by the FSC = 0.5 criteria or 10.2 Å by the FSC = 0.143 criteria (Figure 6D). The structural features of both Fab and IN are much clearer than those of the negative stain 3D reconstruction. Docking the atomic structures of a representative Fab (pdb code: 1M71) and the HIV-1 catalytic core (pdb code: 1EXQ) yields excellent fittings. The cross correlation coefficient between the 3D reconstruction and the density generated from the assembled atomic model to the same resolution is 0.82, calculated using UCSF Chimera (Pettersen et al., 2004). A visual comparison of our 3D reconstruction to the map calculated from the docked atomic structures truncated at various resolutions suggests that the resolution of our reconstruction is, indeed, as estimated from FSC curve (Figure S3). At such a resolution, docking of the Fab atomic structure into the cryoEM density map in two different orientations yields a larger difference in cross-correlations (0.76 vs. 0.7) than what obtained from the similar docking to the negative stain 3D reconstruction (Figure 5). From the experience of docking the Fab J16 (Figure 4), this difference seems to be sufficient to distinguish the light and heavy chains of the Fab.
The density corresponding to the HIV-1 IN dimer (Figure 6C and Figure S4) could only accommodate the catalytic core, but not the full length IN. Neither the C- nor the N-terminal domains are visible in the 3D reconstruction, indicating that these domains are relative flexible in solution. This observation is consistent with the model of full length IN, in which the N- and C-terminal domains are separated from the catalytic core (Wang et al., 2001) and the linkers between them are flexible (Chen et al., 2000).
DISCUSSION
In spite of its many achievements during recent years, the technique of single particle cryoEM has not been widely used to study proteins smaller than 100kDa, because it is generally difficult to align images of small particles embedded in vitreous ice accurately and even harder to validate the 3D reconstruction. For large macromolecular assemblies, each single particle image contains sufficient information for accurate alignment to near-atomic resolution (Armache et al., 2010; Chen et al., 2009; Wolf et al., 2010; Yu et al., 2008; Zhang et al., 2010). However, small particles often do not have easily recognizable structural features and appear as a single dot in images of vitrified samples (Figure S1C). These images simply do not contain sufficient structural information for accurate image alignment (Henderson, 1995), even when they can be visualized with high contrast. Thus, the limitation inherent to aligning such images to each other is a major obstacle for high-resolution structural analysis of small proteins by single particle cryoEM. As pointed out many years ago by Henderson (Henderson, 1995), overcoming this limitation requires some kind crystals or geometrically ordered aggregates such as a large assembly with a icosahedral symmetry. Following this suggestion, many efforts have been invested in developing novel approaches to enable structure determination of small proteins by single particle cryoEM. One of the methods proposed was to fuse a small protein to the nucleocapsid of hepatitis B virus (HBV) particles so that the highly symmetrical HBV particle facilitates the alignment of the fused small particles (Kratz et al., 1999). However, such an approach has very limited applicability, and is not suitable for studying small protein complexes. Also, the linker between the large assembly and the small target protein is often flexible and limits the achievable resolution.
The method we presented here uses a Fab to overcome these limitations by increasing the effective particle size and providing a recognizable structural feature required for accurate image alignment. The molecular weights of PCSK9 and IN dimer by themselves are each less than 70 kDa, but when bound to one or two Fabs, the complexes are more than 100 kDa. More importantly, clearly recognizable Fab feature in the complexes allow accurate image alignment. Also, when there is no previous knowledge about the overall architecture of a small protein, it is difficult to generate a reliable 3D initial model and, given the low resolution, there are no good means to validate the correctness of the final 3D reconstruction. However, when a Fab forms a complex with the protein, its presence in the 3D density map strongly supports the correctness of the reconstruction. In addition, there are other practical benefits of forming a protein-Fab complex for single particle cryoEM studies. For instance, sample heterogeneity could be overcome by identifying and selecting correct particles facilitated by the presence of Fabs in the complex.
Our 3D reconstruction of the frozen hydrated IN-Fab5 complex, with a molecular weight of less than 160kDa, has a resolution of close to 10 Å. This demonstrates the feasibility of generating reliable 3D reconstructions of small proteins by single particle cryoEM. We did not pursue a higher resolution reconstruction of this particular complex by collecting and averaging more images because the HIV-1 IN dimer is not a physiologically active form of the integrase. However, the quality of the current map, obtained only with a limited amount of data, suggests that it should be possible to improve the resolution further.
One concern of using a Fab as a fiducial marker is the potential flexibility between the Fab and its target, which would impair image alignment. In the past, Fabs were not used as fiducial markers for image alignment in single particle cryoEM because of such flexibility (Harris et al., 2011; Jiang et al., 2004). There are two types of Fab epitope: linear and conformational (Van Regenmortel, 1992). The flexibility could result from Fab binding to a linear epitope, which tends to be more flexible than a conformational epitope, or binding to a conformational epitope but located in a flexible domain. Previous cryoEM studies have shown that the densities of Fabs were not well defined in the 3D reconstruction when Fabs were bound to linear epitopes (Stewart et al., 1997). However, Fabs bound to conformational epitopes forming rigid complexes displayed well-defined Fab densities in the 3D reconstruction (Conway et al., 2003). Our current studies also demonstrated that once a suitable Fab is identified to form rigid complex with its target protein it can be used as fiducial marker. For the same concern, we prefer to use Fabs generated directly against the target proteins, but not the Fabs recognizing a genetically introduced protein tag, because a linker between a tag and a target protein is often flexible.
Another concern is the potential internal flexibility within a Fab between its variable and constant domains. Fabs are thought to be flexible because there is a wide range of variation in angles between variable and constant domains, the so-called “elbow” angle, among the known crystal structures of Fabs. It was found, however, that the “elbow” angle variation of the same Fab crystallized in different conditions is in general less than 15° (Sotriffer et al., 2000). And it has been pointed out that some Fabs may be less flexible in the elbow region than others and the “elbow” angle fluctuations of Fabs in solution is not fully understood (Stanfield et al., 2006). Some earlier studies also support that Fabs may be sufficiently rigid to be visualized in single particle cryoEM. For examples, 3D reconstruction of Fab decorated icosahedral hepatitis B capsids showed well-defined densities of tightly bound Fabs at a resolution of near 10 Å and produced excellent fitting of Fab atomic structures (Conway et al., 1998; Conway et al., 2003; Kandiah et al., 2011). Furthermore, in our current study, the two 3D reconstructions of the PCSK9-J16 and IN-Fab5 complexes also produced well-defined densities of both variable and constant domains of the bound Fabs (Figure 4 and Figure 6C). It is possible that a small fluctuation in the elbow region may not be detectable at the resolutions achieved in the current and previous studies. Nevertheless, these results suggest that the Fabs used in these studies were sufficiently rigid to produce 3D reconstructions at near 10 Å resolutions, and strongly support that a Fab forming a rigid complex with its target protein can serve as a fiducial marker to facilitate image alignment and as internal control to validate the correctness of the final 3D reconstruction.
To make our method generally applicable, various Fab candidates need to be readily generated against a wide range of target proteins. This can be achieved by method based on highly diverse phage-display Fab libraries (Fellouse et al., 2007). The phage display method is of particular interest because the selected Fabs often recognize conformational epitopes in their target proteins (Duriseti et al., 2010; Kim et al., 2011). Thus, this method increases the chances of identifying a Fab that forms a rigid complex with its target protein. As demonstrated here, a suitable Fab can easily be identified by simple inspection of 2D images of the complex in negative stain and their class averages, in which a well resolved Fab would indicate the formation of a stable and rigid complex.
Similar as shown before (Conway et al., 2003), single particle cryoEM can also be used for epitope mapping of small proteins without high symmetry. In the case of PCSK9, the density of the Fab in the 3D reconstruction clearly indicates where the antibody binds so that one can firmly narrow down the location of the epitope to a single domain (Figure 4). Given the high correlation between the map and the fitted atomic structures, one can suggest which residues are more likely to be involved in the interaction. In view of the several studies that indicate that the antibody fragment has antagonistic effects on the binding of PCSK9 to the LDL-R (Horton et al., 2007; Kwon et al., 2008; Liang et al., 2011; Zhang et al., 2007), these observations are relevant in the context of the search for drugs with potential use in the treatment of hypercholesterolemia.
In summary, we have established a new approach that enables single particle cryoEM studies of proteins or protein complexes smaller than 100 kDa. In conjunction with other developments in the field of cryoEM, this method has the potential of determining correct 3D reconstructions of small proteins, including integral membrane proteins, as demonstrated here by frozen hydrated HIV-1 IN-Fab complex.
EXPERIMENTAL PROCEDURES
Sample preparation
PCSK9 and Fab J16 were expressed and purified as previously described (Liang et al., 2011). Purified PCSK9 and Fab were mixed in equimolar ratio and incubated on ice for 1 hour, followed by size exclusion chromatography. The fraction corresponding to the PCSK9-Fab complex was used to prepare negative stain EM grids immediately. HIV-1 IN was expressed and purified as previous described (Alian et al., 2009). IN-specific Fabs were a generous gift from Dr. Kossiakoff (University of Chicago) and were expressed and purified as described elsewhere (Fellouse et al., 2007). IN-Fab complexes were formed by mixing equimolar concentrations of both proteins, incubated on ice for 1 hour and subsequently purified using size exclusion chromatography. The fraction corresponding to the complex was used for cryoEM studies. An E. coli homologue of the mammalian vesicular glutamate transporters (EcoliVGLUT3) with a C-terminal His-tag was expressed in BL21 Gold competent cells. EcoliVGLUT3 was solubilized by 20 mM α–dodecyl maltoside (DDM) and purified by Ni-NTA affinity column, followed by size exclusion chromatography. The heterodimeric ABC transporter complex was expressed and purified as described elsewhere (Zutz et al., 2010). Fabs against both EcoliVGLUT3 and ABC transporter were generated by phage display experiments using a human naïve B-cell library (Duriseti et al., 2010). EcoliVGLUT3-Fab was prepared by size exclusion chromatography and ABC transporter-Fab complexes were prepared by pull-down.
Electron microscopy and image processing
EM grids of negatively stained samples were prepared as previously described (Ohi et al., 2004). Negatively stained EM grids were observed either on a Tecnai T20 microscope or a Tecnai T12 microscope (FEI Company, USA), both operated at 120kV. Images were recorded at a nominal magnification of 50kX (T20) or 52kX (T12) using a 4K × 4K CCD camera (UltraScan 4000, Gatan Inc., USA), corresponding to a pixel size of 2.1Å/pixel (T20) on the specimen. Tilt pair images for RCT 3D reconstruction were recorded at 60°/0°, using the RCT data acquisition function of the UCSFTomo package (Zheng et al., 2007). Particles were selected manually. Individual particles were windowed out from the raw images and were subjected to 10 cycles of MRA and K-means classification using SPIDER (Frank et al., 1996). For RCT 3D reconstruction, the untilted particles were subjected to the same procedure of 10 cycles of MRA and classification. Initial 3D reconstructions were calculated from tilted particles, whose corresponding untilted images belonged to the same classes, using the back-projection, back-projection refinement and angular refinement procedures in SPIDER. In the final step, all CTF-corrected images of untilted particles were grouped together as a single image stack and were aligned to a 3D reference model using FREALIGN (Grigorieff, 2007). The initial 3D reference model was the 3D reconstruction obtained directly from RCT. The resolution of each reconstruction was estimated using the FSC = 0.5 criterion. This process was concluded when no further resolution improvement was detected.
CryoEM grids of the IN-Fab5 complex were prepared by standard plunge-freezing procedure. Quantifoil grids were glow-discharged for 10s. 2ul IN-Fab complex was loaded to glow-discharged grid and plunge-frozen using a Vitrobot Mark III (FEI Company, USA) with 0mm offset, room temperature, 100% humidity, and 6.5 sec blotting. Grids of frozen-hydrated IN-Fab5 complexes were imaged using a Tecnai TF20 electron microscope equipped with a field emission source (FEI Company, USA) and operated at 200kV. An objective aperture of 70µm (resolution cut-off at 2 Å) was used. Images were collected at a nominal magnification of 80 kX using a TemF816 8K × 8K CMOS camera (TVIPS, Germany), corresponding to a pixel size of 0.9 Å/pixel on the specimen. All images were binned by a factor of 2 for further processing. Defocus values were determined for each micrograph using CTFFIND (Mindell and Grigorieff, 2003), and ranged from −5µm to −2µm. Only side-view particles, which were easily recognized, were selected. 3D reconstructions were calculated and refined using GeFREALIGN (Li et al., 2010). A cylindrical mask was applied to the 3D reconstruction before calculating the FSC curve. The resolution of the final 3D reconstruction was estimated using multiple criteria, namely FSC=0.5/0.143 (Rosenthal and Henderson, 2003).
The map was sharpened by a resolution dependent amplitude-scaling factor determined as the ratio between rotational averaged power spectra of the reference versus the experimental map (Penczek et al., 1999; Saad et al., 2001). In the case of IN-Fab complex, the reference map was calculated from the docked atomic structures of IN-Fab complex. The formula used to scale the amplitude is
where Frec(H) is the Fourier amplitude of the original density map at frequency H, and
is the scaling coefficient, ∑H±ΔH/2 indicates an integration of power spectra within the Fourier shell of thickness ΔH. The density map is filtered to 10 Å with a cosine edge low-pass filter.
Docking of crystal structures
Atomic structures of PCSK9, HIV-1 IN core and Fabs, were docked separately into the final 3D reconstructions by using UCSF Chimera (Pettersen et al., 2004). The “fit-in-map” option in Chimera was used to optimize the docking of atomic structures into 3D density maps. It maximizes the cross correlation coefficient between the cryoEM density map and the fitted atomic structure. All molecular graphics images were produced in Chimera.
Data deposition
Density maps of single particle 3D reconstructions have been deposited in the Electron Microscopy Data Bank (EMDB) with access code: EMD-5294 (IN-Fab5) and EMD-5295 (PCSK9-J16).
Highlights.
A novel approach of enabling single particle cryoEM of small proteins by Fabs
A rigidly bound Fab is a fiducial marker to facilitate accurate image alignment
A well resolved Fab density is an internal control of the 3D reconstruction
A 3D reconstruction of a 65kDa protein in complex with Fabs by single particle cryoEM
Supplementary Material
ACKNOWLEDGEMENTS
This work is partly supported by grants from NIH (5R01GM098672 and 1S10RR026814-01 (Y.C.), P50GM082250 - HARC Center (A. Frankel) and 1P50 GM073210 - Membrane Protein Expression Center (R.M.S.)), UCSF Program for Breakthrough Biomedical Research (Opportunity Award in Basic Science and New Technology Award (Y.C.)), and The German Research Foundation (SFB 807 Transport and Communication across Biological Membranes and TA157/7 (R.T.)). DSB is supported by a NIGMS-IMSD Fellowship. The authors thank Jaume Pons, Arvind Rajpal, Dave Shelton and Hong Liang for their advises and insightful discussion in generating PCSK9 and Fab J16 samples.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information contains additional supplementary figures and movies.
We declare no conflict of interest.
REFERENCES
- Aebi U, ten Heggeler B, Onorato L, Kistler J, Showe MK. New method for localizing proteins in periodic structures: Fab fragment labeling combined with image processing of electron micrographs. Proc Natl Acad Sci U S A. 1977;74:5514–5518. doi: 10.1073/pnas.74.12.5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alian A, Griner SL, Chiang V, Tsiang M, Jones G, Birkus G, Geleziunas R, Leavitt AD, Stroud RM. Catalytically-active complex of HIV-1 integrase with a viral DNA substrate binds anti-integrase drugs. Proc Natl Acad Sci U S A. 2009;106:8192–8197. doi: 10.1073/pnas.0811919106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armache JP, Jarasch A, Anger AM, Villa E, Becker T, Bhushan S, Jossinet F, Habeck M, Dindar G, Franckenberg S, et al. Localization of eukaryote-specific ribosomal proteins in a 5.5-A cryo-EM map of the 80S eukaryotic ribosome. Proc Natl Acad Sci U S A. 2010;107:19754–19759. doi: 10.1073/pnas.1010005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JC, Piper DE, Cao Q, Liu D, King C, Wang W, Tang J, Liu Q, Higbee J, Xia Z, et al. A proprotein convertase subtilisin/kexin type 9 neutralizing antibody reduces serum cholesterol in mice and nonhuman primates. Proc Natl Acad Sci U S A. 2009;106:9820–9825. doi: 10.1073/pnas.0903849106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JC, Krucinski J, Miercke LJ, Finer-Moore JS, Tang AH, Leavitt AD, Stroud RM. Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding. Proc Natl Acad Sci U S A. 2000;97:8233–8238. doi: 10.1073/pnas.150220297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JZ, Settembre EC, Aoki ST, Zhang X, Bellamy AR, Dormitzer PR, Harrison SC, Grigorieff N. Molecular interactions in rotavirus assembly and uncoating seen by high-resolution cryo-EM. Proc Natl Acad Sci U S A. 2009;106:10644–10648. doi: 10.1073/pnas.0904024106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Wolf E, Larvie M, Zak O, Aisen P, Grigorieff N, Harrison SC, Walz T. Single particle reconstructions of the transferrin-transferrin receptor complex obtained with different specimen preparation techniques. J Mol Biol. 2006;355:1048–1065. doi: 10.1016/j.jmb.2005.11.021. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Zak O, Aisen P, Harrison SC, Walz T. Structure of the human transferrin receptor-transferrin complex. Cell. 2004;116:565–576. doi: 10.1016/s0092-8674(04)00130-8. [DOI] [PubMed] [Google Scholar]
- Cong Y, Baker ML, Jakana J, Woolford D, Miller EJ, Reissmann S, Kumar RN, Redding-Johanson AM, Batth TS, Mukhopadhyay A, et al. 4.0-A resolution cryo-EM structure of the mammalian chaperonin TRiC/CCT reveals its unique subunit arrangement. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.0913774107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway JF, Cheng N, Zlotnick A, Stahl SJ, Wingfield PT, Belnap DM, Kanngiesser U, Noah M, Steven AC. Hepatitis B virus capsid: localization of the putative immunodominant loop (residues 78 to 83) on the capsid surface, and implications for the distinction between c and e-antigens. J Mol Biol. 1998;279:1111–1121. doi: 10.1006/jmbi.1998.1845. [DOI] [PubMed] [Google Scholar]
- Conway JF, Watts NR, Belnap DM, Cheng N, Stahl SJ, Wingfield PT, Steven AC. Characterization of a conformational epitope on hepatitis B virus core antigen and quasiequivalent variations in antibody binding. J Virol. 2003;77:6466–6473. doi: 10.1128/JVI.77.11.6466-6473.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costet P, Krempf M, Cariou B. PCSK9 and LDL cholesterol: unravelling the target to design the bullet. Trends Biochem Sci. 2008;33:426–434. doi: 10.1016/j.tibs.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Cunningham D, Danley DE, Geoghegan KF, Griffor MC, Hawkins JL, Subashi TA, Varghese AH, Ammirati MJ, Culp JS, Hoth LR, et al. Structural and biophysical studies of PCSK9 and its mutants linked to familial hypercholesterolemia. Nat Struct Mol Biol. 2007;14:413–419. doi: 10.1038/nsmb1235. [DOI] [PubMed] [Google Scholar]
- Duriseti S, Goetz DH, Hostetter DR, LeBeau AM, Wei Y, Craik CS. Antagonistic anti-urokinase plasminogen activator receptor (uPAR) antibodies significantly inhibit uPAR-mediated cellular signaling and migration. J Biol Chem. 2010;285:26878–26888. doi: 10.1074/jbc.M109.077677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellouse FA, Esaki K, Birtalan S, Raptis D, Cancasci VJ, Koide A, Jhurani P, Vasser M, Wiesmann C, Kossiakoff AA, et al. High-throughput generation of synthetic antibodies from highly functional minimalist phage-displayed libraries. J Mol Biol. 2007;373:924–940. doi: 10.1016/j.jmb.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Glaeser RM, Typke D, Tiemeijer PC, Pulokas J, Cheng A. Precise beam-tilt alignment and collimation are required to minimize the phase error associated with coma in high-resolution cryo-EM. J Struct Biol. 2011;174:1–10. doi: 10.1016/j.jsb.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorieff N. FREALIGN: high-resolution refinement of single particle structures. J Struct Biol. 2007;157:117–125. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Henderson R. The potential and limitations of neutrons, electrons and X-rays for atomic resolution microscopy of unstained biological molecules. Q Rev Biophys. 1995;28:171–193. doi: 10.1017/s003358350000305x. [DOI] [PubMed] [Google Scholar]
- Horton JD, Cohen JC, Hobbs HH. Molecular biology of PCSK9: its role in LDL metabolism. Trends Biochem Sci. 2007;32:71–77. doi: 10.1016/j.tibs.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Cohen JC, Hobbs HH. PCSK9: a convertase that coordinates LDL catabolism. J Lipid Res. 2009;50 Suppl:S172–S177. doi: 10.1194/jlr.R800091-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen GJ, Kornberg RD. Single-particle selection and alignment with heavy atom cluster-antibody conjugates. Proc Natl Acad Sci U S A. 1998;95:9262–9267. doi: 10.1073/pnas.95.16.9262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang QX, Wang DN, MacKinnon R. Electron microscopic analysis of KvAP voltage-dependent K+ channels in an open conformation. Nature. 2004;430:806–810. doi: 10.1038/nature02735. [DOI] [PubMed] [Google Scholar]
- Kandiah E, Watts NR, Cheng N, Cardone G, Stahl SJ, Heller T, Liang TJ, Wingfield PT, Steven AC. Cryo-EM study of Hepatitis B virus core antigen capsids decorated with antibodies from a human patient. J Struct Biol. 2011 doi: 10.1016/j.jsb.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Stroud RM, Craik CS. Rapid identification of recombinant Fabs that bind to membrane proteins. Methods. 2011 doi: 10.1016/j.ymeth.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratz PA, Bottcher B, Nassal M. Native display of complete foreign protein domains on the surface of hepatitis B virus capsids. Proc Natl Acad Sci U S A. 1999;96:1915–1920. doi: 10.1073/pnas.96.5.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan L, Li X, Naraharisetty HL, Hare S, Cherepanov P, Engelman A. Structure-based modeling of the functional HIV-1 intasome and its inhibition. Proc Natl Acad Sci U S A. 2010;107:15910–15915. doi: 10.1073/pnas.1002346107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukolj G, Skalka AM. Enhanced and coordinated processing of synapsed viral DNA ends by retroviral integrases in vitro. Genes Dev. 1995;9:2556–2567. doi: 10.1101/gad.9.20.2556. [DOI] [PubMed] [Google Scholar]
- Kwon HJ, Lagace TA, McNutt MC, Horton JD, Deisenhofer J. Molecular basis for LDL receptor recognition by PCSK9. Proc Natl Acad Sci U S A. 2008;105:1820–1825. doi: 10.1073/pnas.0712064105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederkremer GZ, Cheng Y, Petre BM, Vogan E, Springer S, Schekman R, Walz T, Kirchhausen T. Structure of the Sec23p/24p and Sec13p/31p complexes of COPII. Proc Natl Acad Sci U S A. 2001;98:10704–10709. doi: 10.1073/pnas.191359398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Grigorieff N, Cheng Y. GPU-enabled FREALIGN: accelerating single particle 3D reconstruction and refinement in Fourier space on graphics processors. J Struct Biol. 2010;172:407–412. doi: 10.1016/j.jsb.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Chaparro-Riggers J, Strop P, Geng T, Sutton JE, Tsai D, Bai L, Abdiche Y, Dilley J, Yu J, et al. PCSK9 Antagonism Reduces LDL-cholesterol in Statin-treated Hypercholesterolemic Non-human Primates. Journal of Pharmacology and Experimental Therapeutics. 2011 [Google Scholar]
- Maignan S, Guilloteau JP, Zhou-Liu Q, Clement-Mella C, Mikol V. Crystal structures of the catalytic domain of HIV-1 integrase free and complexed with its metal cofactor: high level of similarity of the active site with other viral integrases. J Mol Biol. 1998;282:359–368. doi: 10.1006/jmbi.1998.2002. [DOI] [PubMed] [Google Scholar]
- Marasco WA, Sui J. The growth and potential of human antiviral monoclonal antibody therapeutics. Nat Biotechnol. 2007;25:1421–1434. doi: 10.1038/nbt1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- Murata K, Liu X, Danev R, Jakana J, Schmid MF, King J, Nagayama K, Chiu W. Zernike phase contrast cryo-electron microscopy and tomography for structure determination at nanometer and subnanometer resolutions. Structure. 2010;18:903–912. doi: 10.1016/j.str.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Cheng Y, Ramm E, Sheng M, Walz T. Structure and different conformational states of native AMPA receptor complexes. Nature. 2005;433:545–549. doi: 10.1038/nature03328. [DOI] [PubMed] [Google Scholar]
- Newton K, Matsumoto ML, Wertz IE, Kirkpatrick DS, Lill JR, Tan J, Dugger D, Gordon N, Sidhu SS, Fellouse FA, et al. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell. 2008;134:668–678. doi: 10.1016/j.cell.2008.07.039. [DOI] [PubMed] [Google Scholar]
- Ohi M, Li Y, Cheng Y, Walz T. Negative Staining and Image Classification - Powerful Tools in Modern Electron Microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osenkowski P, Li H, Ye W, Li D, Aeschbach L, Fraering PC, Wolfe MS, Selkoe DJ. Cryoelectron microscopy structure of purified gamma-secretase at 12 A resolution. J Mol Biol. 2009;385:642–652. doi: 10.1016/j.jmb.2008.10.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penczek P, Ban N, Grassucci RA, Agrawal RK, Frank J. Haloarcula marismortui 50S subunit-complementarity of electron microscopy and X-Ray crystallographic information. J Struct Biol. 1999;128:44–50. doi: 10.1006/jsbi.1999.4157. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Radermacher M, Wagenknecht T, Verschoor A, Frank J. Three-dimensional reconstruction from a single-exposure, random conical tilt series applied to the 50S ribosomal subunit of Escherichia coli. J Microsc. 1987;146:113–136. doi: 10.1111/j.1365-2818.1987.tb01333.x. [DOI] [PubMed] [Google Scholar]
- Rasmussen SG, Choi HJ, Fung JJ, Pardon E, Casarosa P, Chae PS, Devree BT, Rosenbaum DM, Thian FS, Kobilka TS, et al. Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature. 2011;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G, Gao K, Bushman FD, Yeager M. Single-particle image reconstruction of a tetramer of HIV integrase bound to DNA. J Mol Biol. 2007;366:286–294. doi: 10.1016/j.jmb.2006.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- Roth J. The silver anniversary of gold: 25 years of the colloidal gold marker system for immunocytochemistry and histochemistry. Histochem Cell Biol. 1996;106:1–8. doi: 10.1007/BF02473197. [DOI] [PubMed] [Google Scholar]
- Saad A, Ludtke SJ, Jakana J, Rixon FJ, Tsuruta H, Chiu W. Fourier amplitude decay of electron cryomicroscopic images of single particles and effects on structure determination. J Struct Biol. 2001;133:32–42. doi: 10.1006/jsbi.2001.4330. [DOI] [PubMed] [Google Scholar]
- Savarino A. A historical sketch of the discovery and development of HIV-1 integrase inhibitors. Expert Opin Investig Drugs. 2006;15:1507–1522. doi: 10.1517/13543784.15.12.1507. [DOI] [PubMed] [Google Scholar]
- Shaikh TR, Gao H, Baxter WT, Asturias FJ, Boisset N, Leith A, Frank J. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat Protoc. 2008;3:1941–1974. doi: 10.1038/nprot.2008.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotriffer CA, Rode BM, Varga JM, Liedl KR. Elbow flexibility and ligand-induced domain rearrangements in antibody Fab NC6.8: large effects of a small hapten. Biophys.J. 2000;79:614–628. doi: 10.1016/S0006-3495(00)76320-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanfield RL, Zemla A, Wilson IA, Rupp B. Antibody elbow angles are influenced by their light chain class. J Mol Biol. 2006;357:1566–1574. doi: 10.1016/j.jmb.2006.01.023. [DOI] [PubMed] [Google Scholar]
- Steinberg D, Witztum JL. Inhibition of PCSK9: a powerful weapon for achieving ideal LDL cholesterol levels. Proc Natl Acad Sci U S A. 2009;106:9546–9547. doi: 10.1073/pnas.0904560106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PL, Chiu CY, Huang S, Muir T, Zhao Y, Chait B, Mathias P, Nemerow GR. Cryo-EM visualization of an exposed RGD epitope on adenovirus that escapes antibody neutralization. EMBO J. 1997;16:1189–1198. doi: 10.1093/emboj/16.6.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Regenmortel MHV. Molecular dissection of protein antigens. Vol 1. Boca Raton: CRC Press; 1992. [Google Scholar]
- Wang JY, Ling H, Yang W, Craigie R. Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein. EMBO J. 2001;20:7333–7343. doi: 10.1093/emboj/20.24.7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf M, Garcea RL, Grigorieff N, Harrison SC. Subunit interactions in bovine papillomavirus. Proc Natl Acad Sci U S A. 2010;107:6298–6303. doi: 10.1073/pnas.0914604107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye JD, Tereshko V, Frederiksen JK, Koide A, Fellouse FA, Sidhu SS, Koide S, Kossiakoff AA, Piccirilli JA. Synthetic antibodies for specific recognition and crystallization of structured RNA. Proc Natl Acad Sci U S A. 2008;105:82–87. doi: 10.1073/pnas.0709082105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Jin L, Zhou ZH. 3.88 Å structure of cytoplasmic polyhedrosis virus by cryo-electron microscopy. Nature. 2008;453:415–419. doi: 10.1038/nature06893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DW, Lagace TA, Garuti R, Zhao Z, McDonald M, Horton JD, Cohen JC, Hobbs HH. Binding of proprotein convertase subtilisin/kexin type 9 to epidermal growth factor-like repeat A of low density lipoprotein receptor decreases receptor recycling and increases degradation. J Biol Chem. 2007;282:18602–18612. doi: 10.1074/jbc.M702027200. [DOI] [PubMed] [Google Scholar]
- Zhang X, Jin L, Fang Q, Hui WH, Zhou ZH. 3.3 A cryo-EM structure of a nonenveloped virus reveals a priming mechanism for cell entry. Cell. 2010;141:472–482. doi: 10.1016/j.cell.2010.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Settembre E, Xu C, Dormitzer PR, Bellamy R, Harrison SC, Grigorieff N. Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proc Natl Acad Sci U S A. 2008;105:1867–1872. doi: 10.1073/pnas.0711623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Kollman JM, Braunfeld MB, Sedat JW, Agard DA. Automated acquisition of electron microscopic random conical tilt sets. J Struct Biol. 2007;157:148–155. doi: 10.1016/j.jsb.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zutz A, Hoffmann J, Hellmich UA, Glaubitz C, Ludwig B, Brutschy B, Tampe R. Asymmetric ATP hydrolysis cycle of the heterodimeric multidrug ABC transport complex TmrAB from Thermus thermophilus. J Biol Chem. 2010;286:7104–7115. doi: 10.1074/jbc.M110.201178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.