Summary
Aureochrome1, a signaling photoreceptor from a eukaryotic photosynthetic stramenopile, confers blue-light-regulated DNA binding on the organism. Its topology, in which a C-terminal LOV sensor domain is linked to an N-terminal DNA-binding bZIP effector domain, contrasts with the reverse sensor-effector topology in most other known LOV-photoreceptors. How then is signal transmitted in Aureochrome1? The dark- and light-state crystal structures of Aureochrome1 LOV domain (AuLOV) show that its helical N- and C-terminal flanking regions are packed against the external surface of the core β-sheet, opposite to the FMN chromophore on the internal surface. Light-induced conformational changes occur in the quaternary structure of the AuLOV dimer and in Phe298 of the Hβ strand in the core. The properties of AuLOV extend the applicability of LOV domains as versatile design modules that permit fusion to effector domains via either the N- or C-termini, to confer blue-light sensitivity.
Introduction
The plant kingdom thrives on light, which controls almost all aspects of the plant life cycle from growth to maturation. Phototropism is one of the most well-recognized and well-studied light-dependent phenotypes, and is mediated by a serine-threonine kinase known as phototropin (Briggs and Christie, 2002; Huala et al., 1997). Phototropin contains two FMN-binding LOV (light-oxygen-voltage) domains, which constitute a subgroup of the PAS (Per-Arnt-Sim) superfamily (Gu et al., 2000; Möglich et al., 2009a) and confer sensitivity of the serine-threonine kinase activity to blue light. Upon absorption of a photon, LOV domains initiate a unique photochemical reaction in which a metastable covalent bond is formed between a conserved cysteine residue and atom C4a of the FMN, buried in the core of the LOV domain (Salomon et al., 2000; Salomon et al., 2001; Swartz et al., 2001). LOV blue light sensor domains are found covalently linked to various effector domains that display important biological activities such as signal transduction, enzymatic activity or DNA binding (Nash et al., 2011). These normally light-inert activities are thereby placed under the control of blue light. Phototropins are, however, absent in certain plants living in an aquatic environment such as the photosynthetic stramenopiles. The discovery in the marine alga Vaucheria frigida by Takahashi et al. of a novel form of LOV protein known as Aureochrome1 (Fig. 1A) has extended our understanding of photoperception, photointegration and photomorphogenesis mediated by LOV domains (Takahashi et al., 2007). Aureochromes are blue-light-responsive transcription factors in which an N-terminal bZIP (basic region / leucine zipper) DNA binding domain, the effector domain, is covalently linked to a C-terminal LOV sensor domain. Two copies of Aureochromes denoted Aureo1 and 2 have been found in Vaucheria sp. but only Aureo1 is capable of light-dependent, specific DNA binding to a TGACGT sequence, typical of S- or D-type bZIP transcription factors (Jacoby et al., 2002). Of the 11 residues associated with flavin binding and formation of the covalent adduct state (Crosson et al., 2003), all 11 are conserved in Aureo1 and 9 in Aureo2 (Takahashi et al., 2007). Extensive phylogenetic analysis based on LOV domain diversity (Ishikawa et al., 2009) suggests the independent evolution of Aureo1 and Aureo2 LOV even before the LOV1 and LOV2 domains in phototropin diverged.
Figure 1.
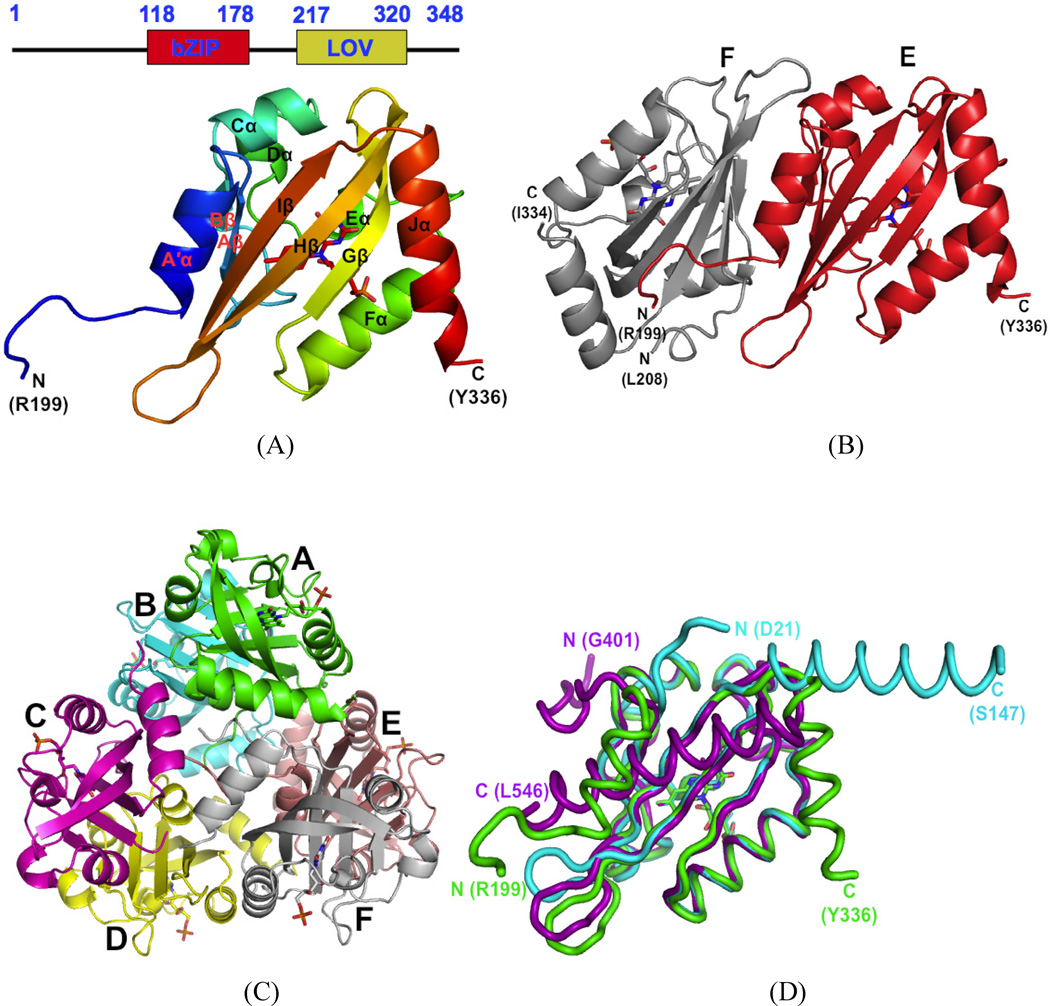
Crystal structure of AuLOV. A) Domain structure of Aureochrome1 LOV and the best-resolved E subunit from AuLOV is shown in detail; B) The EF dimer of AuLOV, showing the A′α helix from the E chain sandwiched between the monomers; C) The 6 molecules A–F in the asymmetric unit of AuLOV are arranged as a trimer of antiparallel dimers; D) Superimposed monomers from the crystal structures of AuLOV (green), YtvA (cyan) and Oat Phot1 LOV2 (purple) show distinct differences in the arrangement of their N- and C-terminal helices but otherwise very similar core structures. See also Figure S1.
The crystal structures of isolated LOV domains in their dark and light states (Crosson and Moffat, 2001, 2002) show that light-induced structural changes are limited in magnitude in crystal lattices and are largely restricted to FMN and its immediate environment. This contrasts with spectroscopic findings in solution where more substantial changes are noted (Corchnoy et al., 2003; Salomon et al., 2001). An important factor is the exact construct under study; structural changes between the dark and light states may well differ among truncated and full-length constructs. High-resolution crystal structures of extended LOV constructs from Avena sativa (Halavaty and Moffat, 2007) and Bacillus subtilis (Möglich and Moffat, 2007) identified modest light-dependent structural changes in the linker Jα helix that forms the C-terminus of the LOV domain in most photoreceptors (Harper et al., 2003; Harper et al., 2004). In solution, the Jα helix unfolds upon illumination in both natural and engineered Avena sativa LOV-based photoreceptors (Harper et al., 2004; Strickland et al., 2008). It is believed that this unfolding transmits signal from the sensor to the effector domain. Most other known LOV structures containing LOV-linker-effector domains, including a recent full-length structure of EL222 (Nash et al., 2011), possess an N-terminal LOV domain followed by a C-terminal Jα helix. This topology directly contrasts with the reverse effector-sensor topology found in Aureochromes, in which the N-terminal effector domain is linked to a C-terminal LOV domain. A few other natural photoreceptors with Aureochrome-like domain topology exist including HTH-LOV and GGDEF-LOV-EAL constructs which merit further characterization (supplementary material in Losi and Gärtner, 2011), The standalone, FAD-binding LOV photoreceptor VVD from the fungal protein Vivid (Zoltowski et al., 2007; Vaidya et al., 2011) is a well-studied exception in which signal originating in the core of the LOV domain is propagated via the N-terminal cap consisting of a short β-strand and an α-helix. VVD tunes the blue-light response in Neurospora by attenuating activation of the White Collar complex (Zoltowski et al., 2007) but apparently does not contain a separate N-terminal effector domain within the same protein, unlike Aureochromes. This raises the question of exactly how signal is transmitted from the sensor to the effector domain in Aureo1.
Blue-light-responsive LOV domains serve as one of the primary design modules in the emerging field of optogenetics. Optogenetics aims to modulate cellular functions via genetically-encoded natural and artificial photoreceptors (Deisseroth, 2011; Deisseroth et al., 2006). The approach originated in control of processes in the nervous system by naturally-occurring signaling photoreceptors such as channelrhodopsin but the term has been extended beyond natural photoreceptors by ‘biologically-inspired design’ of engineered photoreceptors with desired properties (Möglich and Moffat, 2010). LOV domains have been successfully used to confer sensitivity to blue light on normally light-inert effector domains as diverse as the Trp repressor (Strickland et al., 2008), a histidine kinase (Möglich et al., 2009b) and the small GTPase Rac1 (Wu et al., 2009), by domain fusion between a C-terminal LOV domain and an N-terminal effector domain. Successful design of engineered photoreceptors depends on issues such as the nature of the linker by which fusion is achieved and the ability to generate a large population of a stable signaling state. Since the domain topology of Aureo1 differs from that of all other known LOV domains, Aureo1 offers new design strategies for optogenetics.
To explore the unusual topology and signaling mechanism exhibited by Aureochrome1, we cloned, expressed, purified, crystallized and determined the structure of Aureochrome1 LOV from V. frigida in both the dark and light states. This construct contains residues 176–337, and is designed to include flanking regions in both termini. To evaluate the potential of Aureo1 LOV as an optogenetic design module, we performed site-directed mutagenesis to identify variants with different lifetimes of the signaling state. We also characterized important biophysical properties such as oligomeric state in solution, light-induced secondary, tertiary and quaternary structural changes, and fluorescence properties.
Results and Discussion
Crystal structure of Aureo1 LOV in the dark state
Of the four LOV constructs of V. frigida Aureochrome1 (Fig. 1A) spanning residues 176–348, 176–337, 183–337 and 187–337, we succeeded in crystallizing the construct 176–337 in its dark-adapted state. We denote this construct AuLOV, and determined its dark state structure at 2.75Å resolution. We obtained the initial model by molecular replacement using the dark state structure of the phot1 LOV1 domain (Fedorov et al., 2003) from C. reinhardtii (PDB code 1N9L) as the search template, and further refined it to a final R-factor and free R-factor of 0.192 and 0.256, respectively (Table 1). Electron density for N-terminal residues 176–198 and residue 337 at the C-terminus are not visible in the structure (Fig. 1A). The asymmetric unit (ASU) in the P43 space group contains six monomers denoted A to F arranged as a trimer of antiparallel dimers (Fig. 1B), in which the monomers are related by non-crystallographic 2-fold axes (Fig. 1C and Fig. S1A.). Interface area calculations using the PISA server (Krissinel and Henrick, 2007) reveals most probable dimeric associations between AB, CD and EF with interface areas of 1194, 822 and 1399 Å2 respectively (Fig. S1B). The dimer interfaces exhibit an extensive network of salt bridges and H-bonds. The positioning in AuLOV of the conserved Cys254 relative to the FMN chromophore is very similar to those in other LOV proteins. The cavity of the chromophore binding pocket is largely lined with hydrophobic residues, although Gln317, Gln258, Asn286 and Asn296 stabilize the isoalloxazine ring via hydrogen bonds. Arg255 and Arg271 directly interact with the phosphate group of the ribityl side chain.
Table 1.
Data collection and structure refinement of AuLOV in dark/light states.
| Dark | Light | ||
|---|---|---|---|
| Data collection | |||
| Cell dimensions (Å) | a=b=73.987, c=177.468 | a=b=73.98, c=176.18 | |
| Solvent content | 40% | ||
| Resolution/highest shell (Å) | 50-2.75 (2.82-2.75) | 50-2.9 (2.95-2.9) | |
| Completeness | 99.8% (100%) | 99.5% (99%) | |
| Rmerge | 0.066 (0.064) | 0.074 (0.069) | |
| # of unique reflections | 24770 (1203) | 21007 (1028) | |
| Average redundancy | 7.3(7.5) | 6.4 (6.0) | |
| I/σ(I) | 34.1 (1.98) | 24.0 (1.64) | |
| Refinement | |||
| Resolution range (Å) | 33-2.75 (2.86-2.75) | 34-2.9 (3.03-2.9) | |
| R-factor Rfree |
0.192 (0.28) 0.256 (0.37) |
0.201 (0.27) 0.264 (0.36) |
|
| # of reflections | 24612 | 20832 | |
| Completeness | 99.6% | 99.4% | |
| TLS groups | A, B, C, D, E, F chains | ||
| r.m.s.d. Bond Length (Å)/angles (°) |
0.004/0.763 | 0.003/0.703 | |
| ASU: Protein Chromophore Waters Total # of atoms |
6 AuLOV (aa 176–337) 6 (FMN) 68 6392 |
6 AuLOV (aa 176–337) 6 (FMN) 66 6370 |
|
| Missing segments | A: 176–201, 337; B: 176–206, 335–337; C: 176–215, 336– 337; D: 176–205, 336–337; E: 176–198 (176–201 in light structure), 337; F: 176–207, 335–337 |
||
| PDB accession code | 3UE6 | 3ULF | |
The core domain of AuLOV adopts a topology typical of the PAS superfamily (Gu et al., 2000; Möglich, Ayers and Moffat, 2009a), in which a five-stranded, anti-parallel β-sheet whose strands are in the topological order 2-1-5-4-3 is distended to accommodate the chromophore in a cavity at the inner face of the β-sheet. A tunnel, originating at the core of the β-sheet, is a feature common to PAS/LOV structures (Fig. S1C). It further extends to the exterior surface of LOV in between loops and α–helical regions. At the N- and C-termini, two largely helical extensions of the core are packed against the outer face of the β-sheet. These two helices, however, are not close enough to form the helical bundles observed in the structurally-related GAF domains of bacteriophytochromes (Yang et al., 2008). In most structures in the LOV family, the structurally conserved core is flanked by a C-terminal Jα helix. This also holds in AuLOV, though here the Jα helix is partially folded back on the outer surface of the β-sheet (Fig. 1D) in contrast to Oat LOV2 (Halavaty and Moffat, 2007) where it is fully docked on this surface, or in YtvA LOV (Möglich and Moffat, 2007) where it is completely undocked. As in other LOV domains such as Oat LOV2 and YtvA LOV, the inner surface of the β-sheet in AuLOV is predominantly hydrophobic (Fig. S1D). In VVD the N-terminal extension containing the Bβ strand and the Aα helix (Zoltowski et al., 2007) (Vaidya et al., 2011), folds back on the β-core. The N-terminal region of AuLOV, which we denote A′α, forms a three-turn helix folded back across the outer surface of the β-sheet in two monomers (A and E) of the AB and EF dimers. The N-termini in the CD dimer are not as well-resolved as in the AB and EF dimers. At the dimer interface, most of the interacting hydrophobic and polar residues on the outer surface of the β-sheet are related by non-crystallographic 2-fold symmetry. However, this local symmetry does not extend to the N-terminal helices. For example, the N-terminal A′α helix of monomers A and E is sandwiched between the β-sheets of monomers AB and EF, respectively. The N-termini of monomers A and B, and of E and F, appear to clash in the crystal lattice. Therefore, the A′α helices of monomers B and F extends into the solvent channel instead of packing against its own β-sheet as in monomers A and E.
Light-induced conformational changes
The structure of the light state was determined and refined against the light dataset at 2.9Å resolution using the dark structure as a starting model, with a final R-factor and free R-factors of 0.201 and 0.264, respectively (Table 1). The 2Fo-Fc map of the chromophore binding site (Fig. 2A) confirms the formation of the covalent Cys254-FMN adduct and doming of the isoalloxazine ring at C4 of FMN. We also observe light-induced rotamer changes in Phe298 of all six monomers in the asymmetric unit. Phe298 is strategically located in the Hβ-strand opposite to Cys254, across the FMN plane. Leu284 in the adjacent Gβ-strand also undergoes concerted conformational changes to accommodate motion of Phe298.
Figure 2.
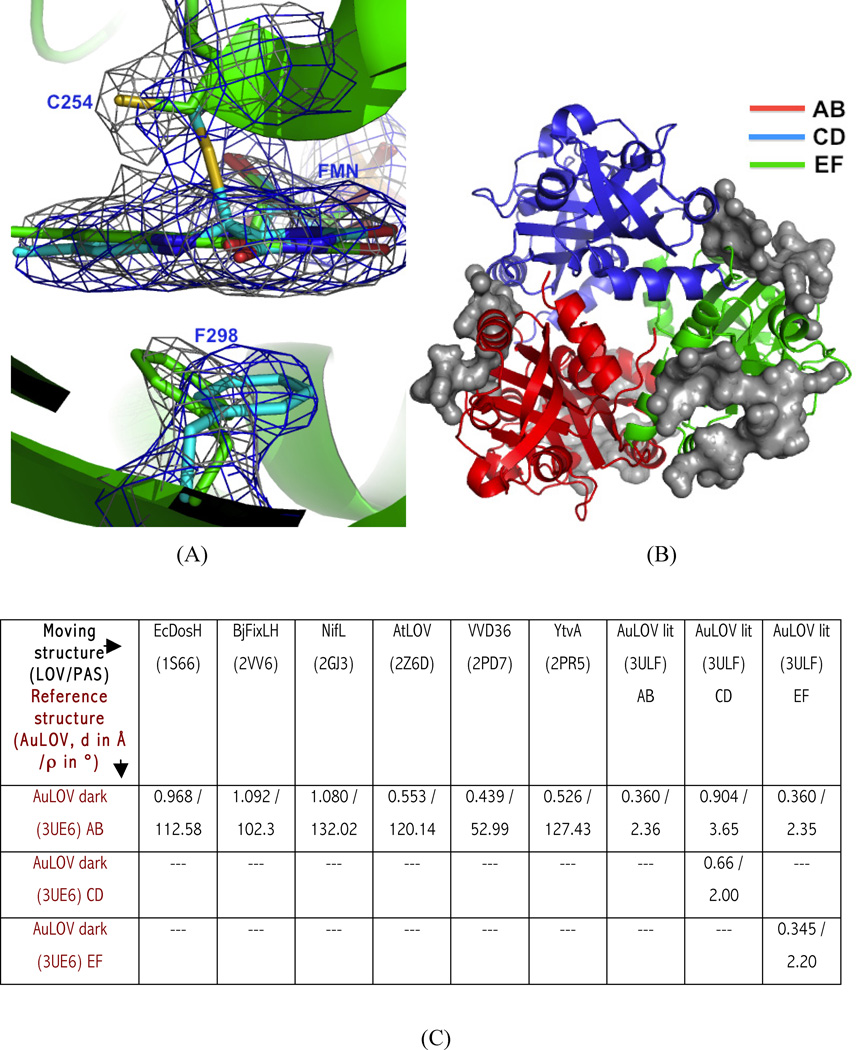
Dark and light state structures of AuLOV. (A) Overlay of 2Fo-Fc map in the dark (grey) and light structures (blue); both maps contoured at 1.4 σ. The most substantial light-induced change in the vicinity of FMN is found at Phe298, located underneath the FMN plane; it moves in the opposite direction upon light-irradiation; (B) Residues in the asymmetric unit involved in intermolecular contacts are highlighted (as grey surface). Crystal contacts with the other ASUs are observed only in the AB (red) and EF (green) dimers. The CD dimer (blue) is least constrained by the crystal lattice and makes contact only with the AB and EF dimers; (C) The monomer orientation of AuLOV in dark state is compared with other LOV/PAS-containing dimers, using rigid body screw-axis-rotation analysis. Screw rotation parameters [rotation (ρ) in degrees and translation (d) in angstroms] are given for each pairwise comparison with respect to AuLOV. See also Figures S2 and S3.
Although electron density for the AB and EF dimers is well-defined in both the dark and light structures, electron density for the CD dimer is significantly disordered in the light structure. Crystal packing analysis (Fig. 2B) reveals quite distinct molecular environments for the AB, CD and EF dimers. While the AB and EF dimers make most of the crystal contacts, the CD dimer only interacts with dimers AB and EF within the same asymmetric unit, but does not interact with other molecules related by crystallographic symmetry. Moreover, the CD dimer exhibits overall B-factor values that are larger than those of the AB and EF dimers (Fig. S2). In other words, the CD dimer is less restricted by intermolecular contacts, thereby permitting larger-amplitude tertiary or quaternary structural changes that evidently result in a less-ordered electron density map in the light structure.
To quantify light-induced quaternary structural changes, we performed screw-axis rotation-translation analysis (Ayers and Moffat, 2008); (Fig. 2C, where screw rotation is denoted as ρ and translation as d), to conduct a quantitative pairwise comparison. We found the largest relative displacement between the two monomers in the CD dimer; this represents the largest light-induced quaternary changes observed in any LOV crystal structure. For comparison, we also applied a similar analysis to other dimeric LOV and PAS structures (Fig. 2C) whose light-dependent changwe in quaternary structure depends on light. However, we reemphasize that PAS/LOV monomer orientation is dependent on whether the LOV domain is isolated or attached to other domain(s) (Ayers and Moffat, 2008).
Cycling variants in AuLOV
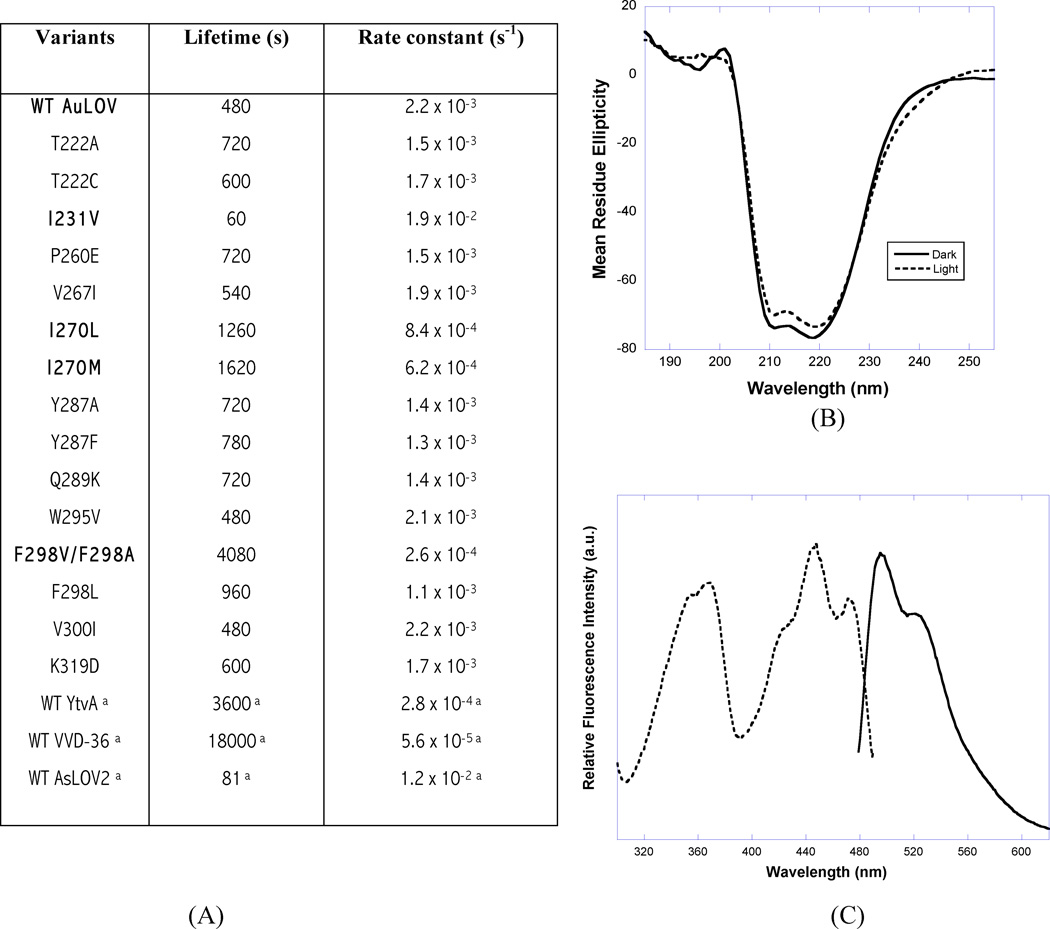
An important parameter for a light-responsive design module is the lifetime of the signaling state, here assumed to be the light state. LOV domains are reversible photoswitches; their reversible photochemistry is observed both in solution and in the crystal (Fig. S3). Naturally-occurring LOV proteins generally fall into two groups on the basis of this lifetime (or equivalently, their rate of dark reversion), although the range spanned by each group is wide. Phototropins, EL222 and Arabidopsis PhyB (Chen et al., 2004; Nash et al., 2011; Zoltowski et al., 2009) are examples of the fast-cycling group whose light state lifetime is on the order of seconds; in contrast FKF1, ZTL, LKP2, YtvA, LOVK, and VVD (Imaizumi et al., 2003; Losi et al., 2003; Nelson et al., 2000; Purcell et al., 2007; Somers et al., 2000; Zoltowski et al., 2009) etc. are examples of the slow-cycling group whose light state lifetime ranges from hours to days. AuLOV in solution has a lifetime of 480s. Since design modules with short or long signaling states may be desired in different optogenetic applications depending on the specific effector function to be targeted, we explored residues in AuLOV that modulate the light state lifetime using a strategy similar to that reported for the LOV photoreceptor Vivid (Zoltowski et al., 2009).
We produced 16 AuLOV variants by site-directed mutagenesis in which candidate sites were based on a multiple sequence alignment of a range of LOV proteins including Aureo1, Aureo2 and VVD36 (Zoltowski et al., 2009) (Fig. S4). Kinetic data on dark reversion are summarized in Fig. 3A. We identified two residues in AuLOV, Ile270 and Phe298 (highlighted in Fig. 3A), mutations at which show significantly increased light state lifetime relative to WT. In Aureo1, 11 residues contribute to FMN-binding, of which 9 are conserved in Aureo2. Presence of leucine and valine respectively at the remaining two residues at 270 and 298 (Aureo1 numbering), are presumably responsible for the absence of FMN-binding in Aureo2 (Takahashi et al., 2007; the location of I270 in AuLOV is shown in Fig. S5). However, the single site Aureo1 variants (I270L, I270M, F298A, F298L and F298V) retained FMN-binding and photoactivity although they did alter the light state lifetime relative to WT with a 2.5-fold increase in both I270L and I270M, a ~9-fold increase in both F298V and F298A, but no significant change in F298L. In YtvA and VVD-36, the corresponding residues to Phe298 are Leu106 and Leu163, respectively (Fig. S4), though the effect of substituting leucine with phenylalanine at this position has not been studied in these systems. We attribute the range of lifetimes of Phe298 variants to steric contributions from the side chain of this residue, located ~3.5Å from the FMN plane. As noted above, Phe298 displays distinct side chain conformations in the dark and light states (Fig. 2A) and may play an important role in signal transduction. The I231V AuLOV variant (location shown in Fig. S5) equivalent to I74V-I85V variant in VVD-36 (Zoltowski et al., 2009) (residue 220 is valine in WT AuLOV) decreased the lifetime by 8-fold. This side chain stabilizes the active site Cys in both proteins. The V300I variant in AuLOV corresponds to ‘on’ state variants in VVD-36 with greatly increased lifetime (Zoltowski et al., 2009). However, the lifetime of V300I is unaltered in AuLOV, perhaps because V300 does not make any van der Waals contact with the chromophore, unlike its counterpart Met165 in VVD-36.
Figure 3.
Physico-chemical properties of AuLOV. (A) Parameters for dark-state recovery kinetics in AuLOV variants. a represents literature value from (Zoltowski et al., 2009). (B) AuLOV CD spectroscopy recorded at the far-UV region (190–260 nm); (C) Representative fluorescence excitation spectra (dashed line) were recorded at an emission wavelength of 495 nm, and emission spectra (solid line) by excitation at 450 nm. See also Figures S4, S5 and S6.
Biophysical characterization
The crystal structure of AuLOV is clearly based on packing of dimers. To determine whether AuLOV also forms dimers in solution, we conducted sedimentation velocity experiments. At concentrations of 18.6 and 7.15 µM, a single species (~96%) was observed with a molecular mass of 45.7 ± 5.2 kDa, consistent with the molecular mass predicted for an AuLOV dimer of 41.2 kDa. This suggests that AuLOV is dimeric in solution and places an upper limit on the dimer-monomer dissociation equilibrium constant at around 4 µM.
To probe secondary structural changes upon illuminating AuLOV in solution, we performed CD spectroscopy and found a clear difference between the dark and light states (Fig. 3B) that were both fully reversible and reproducible through several photocycles. We observed a ~6% decrease in α-helix content upon illumination, somewhat lower than the value of ~10–13% in other LOV proteins (Losi et al., 2005; Möglich and Moffat, 2007). The near-UV CD difference spectrum between the light and dark states (Fig. S6A) resembles that of AsLOV (Corchnoy et al., 2003) and the strong 290 nm peak confirms formation of the C254-FMN adduct upon illumination.
It is possible to extend optogenetic approaches to imaging, in which the spatial location of the target and its activity are deliberately unperturbed. Recent studies successfully designed blue- and red-light-based fluorescent sensors (Chapman et al., 2008; Filonov et al., 2011) for in vivo imaging, in which these sensors addressed certain limitations of the widely-used, GFP-based fluorescent proteins (Tsien, 2009). Robust and small LOV-based sensors are effective even under anaerobic conditions and photochemically-inactive variants can have a high quantum yield for fluorescence (Chapman et al., 2008; Drepper et al., 2007; Tsien, 2009). This suggests that they may be developed for imaging purposes (Chapman et al., 2008; Losi and Gärtner, 2011). However, their dimeric nature, high background fluorescence arising from the wide cellular distribution of flavins and the possibility of cellular damage by the production of singlet oxygen species are concerns. The FMN chromophore in WT AuLOV and all its variants is naturally fluorescent with maximum emission at 495 nm when excited at 450 nm. Upon light irradiation, the fluorescence intensity decreases sharply (Fig. S6B). The decrease in fluorescence intensity is fully reversible as evidenced by the I231V variant, which exhibited fast dark recovery as monitored by either absorbance or fluorescence (Fig. S6B). To examine the potential benefit of AuLOV as an in vivo reporter, we produced the photoinactive C254A variant in five Aureo1 LOV constructs (176–348, 187–335, AuLOV (176–335), I231V-AuLOV and Y287F-AuLOV) and determined their fluorescence quantum yields relative to free FMN (ΦF = 0.26) (Drepper et al., 2007). The excitation and emission spectra of Aureo1 LOV is shown in Fig. 3C. The value of ΦF for all Aureo1 LOV variants falls in the range of 0.21–0.244, with the maximum value found for the I231V-AuLOV variant. This value is comparable to that of other LOV-based fluorescent proteins (0.17, 0.39, 0.32 for BsYtvA, PpSB2 and iLOV respectively (Chapman et al., 2008; Drepper et al., 2007; Losi et al., 2005)) and closely matches that of the blue form of GFP with a ΦF of 0.24 (Patterson et al., 2001).
Aureochrome1 LOV as an optogenetic design module
Applications of optogenetics are being extended beyond naturally occurring photoreceptors to designed photoreceptors (Möglich and Moffat, 2010) in which the activity of a desired effector domain is made sensitive to light by fusion to a sensor domain. The design and engineering of useful, biologically-inspired, artificial photoreceptors is based on signaling mechanisms characterized in naturally-occurring photoreceptors.
The oligomeric state of candidate sensor domains is important to successful design, but the mechanism of light sensitivity differs in different protein examples. For example, Jα unfolding upon illumination is crucial to signaling in phototropin kinase (Harper et al., 2004), LOV-TAP (Strickland et al., 2008) and GTPase-RacI (Wu et al., 2009). AsLOV is monomeric and its C-terminal Jα helix is docked on the surface of the core β-sheet (Halavaty and Moffat, 2007). Combining this natural property with alternate, mutually exclusive helical conformations, the DNA binding activity of Trp repressor has been made light-sensitive (Strickland et al., 2008). Similarly, by taking advantage of the monomeric nature of the prototypical PAS domain, photoactive yellow protein, the DNA binding activity of GCN4 has also been made light-sensitive. However, the mechanism is different: here, light promotes dimerization (Morgan et al., 2010). A similar phenomenon is observed in the natural photoreceptor EL222 LOV-HTH, where the fully-folded HTH effector domain binds to the dimerization interface of LOV and renders monomeric LOV-HTH unable to bind DNA in the dark state (Nash et al., 2011). Light decouples this sensor-effector complex, exposes the dimer interface of both the LOV and HTH domains, promotes formation of dimeric HTH and thus DNA binding. A further example is VVD, where light lowers the affinity of the N-terminal cap for the monomeric core of LOV, produces a hinge motion between them and generates a light-activated dimer (Vaidya et al., 2011). Finally, in the designed YF1 construct, the histidine kinase activity of FixL is made light-sensitive by fusing it to the LOV domain of YtvA (Möglich et al., 2009b). In YF1, the oligomerization state of the LOV domain is exploited in a different way. The naturally dimeric YtvA LOV is proposed to transmit signal by exerting torque through a C-terminal, coiled-coil Jα linker (Möglich et al., 2010) to the N-terminal DHp domain of the FixL kinase, thereby positioning the phosphoacceptor histidine of one kinase monomer at the ATP-binding, catalytic site of the other.
Our results on Aureo1 are consistent with a more general aspect of signaling by LOV domains, namely, the key feature of signaling is light-dependent structural changes in the β-sheet that weaken the binding of any elements packed on its external surface (Möglich et al., 2009a; Nash et al., 2011; Vaidya et al., 2011). These elements comprise the C-terminal Jα helix in all LOV domains including Aureo1, the N-cap in VVD and the A′α helix in Aureo1. We note also that weakened binding of these elements exposes a new area on the external surface of the β-sheet, and this area may in turn bind a new element. Thus, the topology of linkage between the LOV and effector domains is of less consequence. A general signaling mechanism holds for LOV domains, and indeed may extend more generally to PAS and GAF domains that share the same structure of the core.
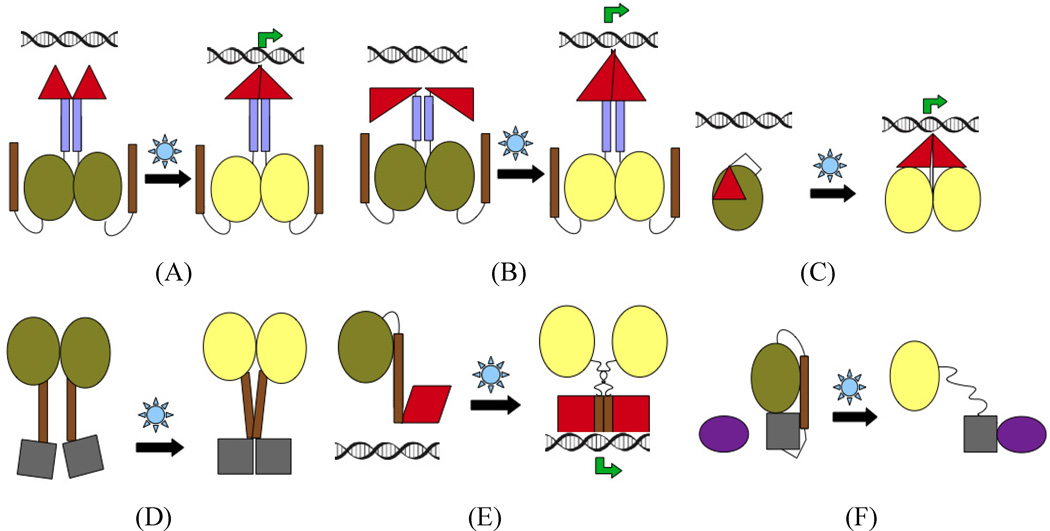
The amphipathic pattern of residues in Jα (Harper et al., 2004) is believed to play an important role in the affinity of the β-sheet for elements packed on its external surface, and thus in transmitting signaling from the LOV core to the spatially-distant effector domain. In the A′α and Jα helices of AuLOV, this amphipathic pattern is preserved though it is not as pronounced as in YtvA or AsLOV (Fig. S7). Moreover, the coiled-coil propensity of the A′α and Jα helices in AuLOV is very low in comparison to the Jα helices in AsLOV or YtvA LOV. However, we propose that in full-length Aureochrome1, the strongly coiled-coil nature of the N-terminal bZIP domain influences the conformation of the A′α helix and is essential for signal transmission in Aureo1. Whether or not both A′α and Jα in AuLOV are absolutely necessary for signal transduction is a matter for further investigation. Thus, not only is the nature of the linker connecting sensor and effector domains important in LOV-based designs, but the properties of the effector domain itself may play a role. Light-induced quaternary structural changes (Fig. 4A) within the effector domain or sequence-dependent hinge-type motions (Fig. 4B) favoring the coiled-coil pattern in the effector might play a role in Aureo1 signaling. Finally, the A′α and Jα helices may interact directly (though they do not do so in the AuLOV crystal structure). We also cannot rule out the possibility of signaling using the C-terminal Jα helix of AuLOV in synthetic photoreceptors.
Figure 4.
Proposed signaling strategies in AuLOV (panels A, B) and comparison with other biologically-inspired designs of synthetic photoreceptors, e.g. GCN4-PYP fusion (Morgan et al., 2010) (C), YtvA-FixL design (Möglich et al., 2009b) (D), LOV-TAP (Strickland et al., 2008) (E), GTPase Rac1 (Wu et al., 2009) (F) and. Top panel: Signalling through N terminus; bottom panel: Signaling through C-terminus. Color codes: olive – LOV at dark-state, yellow – LOV at light-state, violet – N-ter helix, brown – C-ter helix, red – DNA binding domain, grey – other output domains, dark violet – secondary output domain. See also Figure S7.
Based on all the design strategies discussed above, we present a cartoon representation of light-dependent signal transduction in Fig. 4.
Aureo1 LOV, with its unusual effector-sensor topology, A′α/Jα linker properties, dimeric nature and fluorescence properties, may offer a unique, genetically-encoded optogenetic design module that could either control the activity of an effector domain or, in photoinactive variants, be applicable to fluorescence-based imaging.
Experimental Procedures
Cloning, Expression and Purification of AuLOV
The coding region corresponding to residues 176–348 of V. frigida Aureo1 was synthesized with codons optimized for E. coli expression system (Geneart) and subcloned in pET28c (Novagen, Madison, MI) at NdeI and SacI (New England Biolabs, Beverly, MA) restriction sites. The PCR products of three shorter constructs (corresponding to residues 176–337, 183–337 and 187–337) were inserted into the pET28c vector between NheI and SacI sites. E. coli BL21(DE3) cells transformed with these constructs were initially grown at 37°C. After IPTG induction, cell culture continued to grow at 18°C for additional 16–20 h. Cell pellets were re-suspended in buffer containing 20 mM Tris-HCl, pH 8.0, 50 mM NaCl, 10 mM imidazole, 10% glycerol in the presence of protease inhibitor (Roche, Indianapolis, IN), and lysed by sonication on ice. The supernatant was incubated for ~2 h with excess FMN to ensure efficient chromophore incorporation, and was further incubated with Co2+-enriched Talon metal affinity resin (Clontech, Mountain View, CA) for 1 h at 4°C. The protein-resin mixture was loaded on to a 10 ml disposable polypropylene column (Thermo Scientific, USA) followed by extensive washing. His6-tagged protein was eluted using buffer containing 20 mM Tris-HCl, pH 8.0, 50 mM NaCl, 300 mM imidazole, 10% glycerol. The His-tag was subsequently removed using Thrombin CleanCleave resin (Sigma-Aldrich). The eluted, tag-free protein was concentrated (Millipore, Billerica, MA), and further purified by size-exclusion chromatography using a Sephacryl S-100 gel filtration column (Amersham Pharmacia). All growth and purification steps were carried out either in the dark or in the presence of dim red light. Protein concentrations were determined from absorbance at 450 nm and an extinction coefficient of 12,500 M−1 cm−1.
Mutagenesis and Purification of AuLOV variants
All AuLOV variants were generated using standard site-directed mutagenesis protocols. Variants were purified using the same protocol as for WT constructs, except that steps for His6-tag removal and S-100 gel filtration were omitted. Imidazole and excess FMN were removed using a PD-10 desalting column. The authenticity of all variants was confirmed by DNA sequencing.
UV-Visible Spectroscopy and dark-recovery kinetics
UV-Vis spectra of WT and mutant AuLOV were recorded at room temperature from 900 to 230 nm with a Shimadzu UV-1650 PC spectrophotometer. Absorption spectra were recorded in the dark state and after 5min illumination with fiber-optic white light (136 mW) at 60–600s time intervals, until no further change at 447 nm was observed. To measure dark recovery kinetics, values of OD447 as a function of time were fitted to a single exponential: OD447 = const(1−e−kt), using Kaleidagraph.
Crystallization and Data Collection
Crystals of AuLOV grew at 20°C via vapor diffusion at a final concentration of 15.5 mg/ml in crystallization conditions containing 100 mM phosphate-citrate buffer (pH 4.2), 20% (w/v) PEG 8000, and 200 mM NaCl. Yellow, diamond-shaped crystals of up to 0.2×0.2×0.4 mm appeared within three weeks. Crystals in the dark state were frozen under dim red light. Crystals of the light state were obtained by illuminating the dark state for 2 minutes using white fiber-optic light (136 mW power) and then quickly freezing in liquid-N2.
Both dark and light monochromatic oscillation datasets were collected at BioCARS beamline 14-BMC at the Advanced Photon Source, Argonne National Laboratory. During data collection of the light state, crystals were continuously illuminated using white fiber-optic light. All images were indexed, integrated and scaled using HKL2000 (Otwinowski and Minor, 1997).
Structure determination
The crystal structure of AuLOV was determined in space group P43 by molecular replacement using PHASER (McCoy et al., 2005) in CCP4 (Collaborative Computational Project, 1994), with the Phot1 LOV1 domain from C. reinhardtii (PDB code 1N9L) as the search template. The N- and C-terminal segments were initially built modeled on improved electron densities following density modification using Resolve (Terwilliger, 2000). The crystal structure containing six AuLOV monomers in the asymmetric unit was further built using Coot (Emsley et al., 2010) and refined by Phenix (Afonine et al., 2005) and Refmac5 (Winn et al., 2001). All structure figures were generated using Pymol (Delano Scientific). Crystal contacts were analyzed using Areaimol (Lee and F M Richards, 1971) in CCP4 and PISA (Krissinel and Henrick, 2007). Pairwise screw-axis-rotation analysis (Ayers and Moffat, 2008) of all dimer structures were carried out using lsqkab (Kabsch, 1976) in CCP4.
Analytical ultracentrifugation
Sedimentation velocity studies were performed in a Beckman Optima XL-A analytical ultracentrifuge equipped with an AN-60 Ti rotor (Beckman Coulter) using absorption optics at a wavelength of 280 and 450 nm. Experiments used a 1.2-mm two-channel Epon centerpiece at a rotor speed of 40,000 rpm for 18 hr at 20°C. Absorbance was monitored at time intervals of 360–480 s and a step size of 0.003 cm. Multiple scans at different time points were fitted to a continuous size distribution (c(s)) model and an integrated distribution function using SEDFIT version 11.3. The partial specific volume of proteins and buffer density were calculated from standard tables using SEDNTERP.
Circular dichroism spectroscopy
CD spectroscopy was carried out in phosphate buffer, pH 7.5, using an Aviv CD spectrometer with 0.1 cm path-length. Buffer-only reference spectra were subtracted. Spectra were recorded in the dark, or 5–10min after illumination. Best fit was obtained using CDSSTR program (OLIS Globalworks).
Fluorescence spectroscopy and determination of quantum yields
Fluorescence emission spectra for WT AuLOV and variants were recorded upon excitation at 450 nm; excitation spectra were taken with emission at 520 nm at 25°C, using a Horiba FluoroMax-3. Spectra of the light state were taken after 5 minutes illumination on the sample using fiber-optic white light. The quantum yield was calculated based on photo-inactive C254A mutants using FMN as standard.
Highlights.
Crystal structures in dark and light states determined for Aureochrome1 LOV
Significant light induced change observed in F298 and quaternary structure
Potential photocycle mutant variants, altering the light-state lifetime, identified
Possible mechanism of signaling in topologically unique Aureo1 speculated
Supplementary Material
Acknowledgements
We thank Dr. Andreas Möglich for providing the plasmid containing Aureochrome1 sequence (176–348) and for helpful suggestions. We thank Drs. Vukica Srajer and Elena Solomaha for assistance in microspectrophotometry and biophysical experiments; and Drs. Masakazu Sugishima and Elena Davydova for useful discussions. DM thanks the BioCARS staff for assistance in X-ray data collection, and speakers of the CCP4 summer school 2011 for providing valuable suggestions during structure determination. This study was supported by NIH Grant GM036452 to KM. BioCARS is funded by NIH grant RR007707 to KM; the APS is supported by the US Dept. of Energy.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflict of interest.
References
- Afonine P, Grosse-Kunstleve R, Adams P. phenix.refine. CCP4 Newsl. 2005;42 Contribution 8. [Google Scholar]
- Ayers R, Moffat K. Changes in quaternary structure in the signalling mechanisms of PAS domains. Biochemistry. 2008;47:12078–12086. doi: 10.1021/bi801254c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs WR, Christie JM. Phototropins 1 and 2: versatile plant blue-light receptors. Trends. Plant. Sci. 2002;7:204–210. doi: 10.1016/s1360-1385(02)02245-8. [DOI] [PubMed] [Google Scholar]
- Chapman S, Faulkner C, Kaiserli E, Garcia-Mata C, Savenkov E, Roberts A, Oparka K, Christie J. The photoreversible fluorescent protein iLOV outperforms GFP as a reporter of plant virus infection. Proc. Natl. Acad. Sci. USA. 2008;105:20038–20043. doi: 10.1073/pnas.0807551105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Chory J, Fankhauser C. Light signal transduction in higher plants. Annu. Rev. Genet. 2004;38:87–117. doi: 10.1146/annurev.genet.38.072902.092259. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, N. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. sect D. 1994 doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Corchnoy SB, Swartz TE, Lewis JW, Szundi I, Briggs WR, Bogomolni RA. Intramolecular proton transfers and structural changes during the photocycle of the LOV2 domain of phototropin 1. J. Biol. Chem. 2003;278:724–731. doi: 10.1074/jbc.M209119200. [DOI] [PubMed] [Google Scholar]
- Crosson S, Moffat K. Structure of a flavin-binding plant photoreceptor domain: Insights into light-mediated signal transduction. Proc. Natl. Acad. Sci. USA. 2001;98:2995–3000. doi: 10.1073/pnas.051520298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosson S, Moffat K. Photoexcited structure of a plant photoreceptor domain reveals a light-driven molecular switch. Plant Cell. 2002;14:1067–1075. doi: 10.1105/tpc.010475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosson S, Rajagopal S, Moffat K. The LOV domain family: photoresponsive signalling modules coupled to diverse output domains. Biochemistry. 2003;42:2–10. doi: 10.1021/bi026978l. [DOI] [PubMed] [Google Scholar]
- Deisseroth K. Optogenetics. Nat. Methods. 2011;8:26–29. doi: 10.1038/nmeth.f.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisseroth K, Feng G, Majewska AK, Miesenbock G, Ting A, Schnitzer MJ. Next-generation optical technologies for illuminating genetically targeted brain circuits. J. Neurosci. 2006;26:10380–10386. doi: 10.1523/JNEUROSCI.3863-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drepper T, Eggert T, Circolone F, Heck A, Krauss U, Guteri J-K, Wendorff M, Losi A, Gartner W, Jaeger K-E. Reporter proteins for in vivo fluorescence without oxygen. Nat. Biotechnol. 2007;25:443–445. doi: 10.1038/nbt1293. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Cryst. 2010;D66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov R, Schlichting I, Hartmann E, Domratcheva T, Fuhrmann M, Hegemann P. Crystal structures and molecular mechanism of a light-induced signalling switch: The phot-LOV1 domain from Chlamydomonas reinhardtii. Biophys. J. 2003;84:2474–2482. doi: 10.1016/S0006-3495(03)75052-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filonov G, Piatkevich K, Ting L-M, Zhang J, Kim K, Verkhusa V. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nat. Biotechnol. 2011;29:757–761. doi: 10.1038/nbt.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Hogenesch J, Bradfield C. The PAS superfamily: sensors of environmental and developmental signals. Annu. Rev. Pharmacol. Toxicol. 2000;40:519–561. doi: 10.1146/annurev.pharmtox.40.1.519. [DOI] [PubMed] [Google Scholar]
- Halavaty AS, Moffat K. N- and C- terminal flanking regions modulate light-induced signal transduction in the LOV2 domain of the blue light sensor Phototropin 1 from Avena sativa. Biochemistry. 2007;46:14001–14009. doi: 10.1021/bi701543e. [DOI] [PubMed] [Google Scholar]
- Harper SM, Christie JM, Gardner KH. Disruption of the LOV-J helix interaction activates phototropin kinase activity. Biochemistry. 2004;43:16184–16192. doi: 10.1021/bi048092i. [DOI] [PubMed] [Google Scholar]
- Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin light switch. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- Huala E, Oeller PW, Liscum E, Han IS, Larsen E, Briggs WR. Arabidopsis NPH1: a protein kinase with a putative redox-sensing domain. Science. 1997;278:2120–2123. doi: 10.1126/science.278.5346.2120. [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Tran H, Swartz T, Briggs W, Kay S. FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature. 2003;426:302–306. doi: 10.1038/nature02090. [DOI] [PubMed] [Google Scholar]
- Ishikawa M, Takahashi F, Nozaki H, Nagasato C, Motomura T, Kataoka H. Distribution and phylogeny of the blue light receptors aureochromes in eukaryotes. Planta. 2009;230:543–552. doi: 10.1007/s00425-009-0967-6. [DOI] [PubMed] [Google Scholar]
- Jacoby M, Weisshaar B, Droge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F. bZIP transcription factors in Arabidopsis. Trends. Plant. Sci. 2002;7:106–111. doi: 10.1016/s1360-1385(01)02223-3. [DOI] [PubMed] [Google Scholar]
- Kabsch W. A solution for the best rotation to relate two sets of vectors. Acta. Cryst. A. 1976;32:922–923. [Google Scholar]
- Krissinel E, Henrick K. Interface of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Lee B, Richards FM. The interpretation of protein structures: estimation of static accesibility. J. Mol. Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. J. Mol. Biol. 55, 379–400 (1971) [DOI] [PubMed] [Google Scholar]
- Losi A, Gärtner W. Old chromophores, new photoactivation paradigms, trendy applications: Flavins in blue light-sensing photoreceptors. Photochem. Photobiol. 2011;87:491–510. doi: 10.1111/j.1751-1097.2011.00913.x. [DOI] [PubMed] [Google Scholar]
- Losi A, Ghiraldelli E, Jansen S, Gärtner W. Mutational effects on protein structural changes and interdomain interactions in the blue-light sensing LOV protein YtvA. Photochem. Photobiol. 2005;81:1145–1152. doi: 10.1562/2005-05-25-RA-541. [DOI] [PubMed] [Google Scholar]
- Losi A, Quest B, Gärtner W. Listening to the blue: the time resolved thermodynamics of the bacterial blue-light receptor YtvA and its isolated LOV domain. Photochem. Photobiol. Sci. 2003;2:759–766. doi: 10.1039/b301782f. [DOI] [PubMed] [Google Scholar]
- McCoy A, Grosse-Kunstleve R, Storoni L, Read R. Likelihood-enhanced fast translation function. Acta Crystallog. sect. D. 2005;53:2126–2132. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- Möglich A, Ayers R, Moffat K. Structure and signaling mechanism of Per-ARNT-Sim domains. Structure. 2009a;17:1282–1294. doi: 10.1016/j.str.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möglich A, Ayers RA, Moffat K. Design and signalling mechanism of light-regulated histidine kinases. J. Mol. Biol. 2009b;385:1433–1444. doi: 10.1016/j.jmb.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möglich A, Ayers RA, Moffat K. Addition at the molecular level: signal integration in designed Per-ARNT-Sim receptor proteins. J. Mol. Biol. 2010;400:477–486. doi: 10.1016/j.jmb.2010.05.019. [DOI] [PubMed] [Google Scholar]
- Möglich A, Moffat K. Structural basis for light-dependent signalling in the dimeric LOV domain of the photosensor YtvA. J. Mol. Biol. 2007;373:112–126. doi: 10.1016/j.jmb.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möglich A, Moffat K. Engineered photoreceptors as novel optogenetic tools. Photochem. Photobiol. Sci. 2010;9:1286–1300. doi: 10.1039/c0pp00167h. [DOI] [PubMed] [Google Scholar]
- Morgan SA, Wahid SA, Woolley GA. Structure-based design of a photocontrolled DNA binding protein. J. Mol. Biol. 2010;399:94–112. doi: 10.1016/j.jmb.2010.03.053. [DOI] [PubMed] [Google Scholar]
- Nash A, McNulty R, Shillito M, Swartz T, Bogomolni R, Luecke H, Gardner K. Structural basis of photosensitivity in a bacterial light-oxygen-voltage/helix-turn-helix (LOV-HTH) DNA-binding protein. Proc. Natl. Acad. Sci. USA. 2011;108:9449–9454. doi: 10.1073/pnas.1100262108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D, Lasswell J, Rogg L, Cohen M, Bartel B. FKF1, a clock controlled gene that regulates the transition to flowering in Arabidopsis. Cell. 2000;101:331–340. doi: 10.1016/s0092-8674(00)80842-9. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meth. Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Patterson G, Day R, Piston D. Fluorescent protein spectra. J. Cell. Sci. 2001;114:837–838. doi: 10.1242/jcs.114.5.837. [DOI] [PubMed] [Google Scholar]
- Purcell E, Siegal-Gaskins D, Rawling D, Fiebig A, Crosson S. A photosensory two-component system regulates bacterial cell attachment. Proc. Natl. Acad. Sci. USA. 2007;104:18241–18246. doi: 10.1073/pnas.0705887104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon M, Christie JM, Knieb E, Lempert U, Briggs WR. Photochemical and mutational analysis of the FMN binding domains of the plant blue light receptor, phototropin. Biochemistry. 2000;39:9401–9410. doi: 10.1021/bi000585+. [DOI] [PubMed] [Google Scholar]
- Salomon M, Eisenreich W, Durr H, Schleicher E, Knieb E, Massey V, Rudiger W, Muller F, Bacher A, Richter G. An optomechanical transducer in the blue light receptor phototropin from Avena sativa. Proc. Natl. Acad. Sci. USA. 2001;98:12357–12361. doi: 10.1073/pnas.221455298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers D, Schultz T, Milnamow M, Kay S. ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell. 2000;101:319–329. doi: 10.1016/s0092-8674(00)80841-7. [DOI] [PubMed] [Google Scholar]
- Strickland D, Moffat K, Sosnick TR. Light-activated DNA binding in a designed allosteric protein. Proc. Natl. Acad. Sci. USA. 2008;105:10709–10714. doi: 10.1073/pnas.0709610105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz TE, Corchnoy SB, Christie JM, Lewis JW, Szundi I, Briggs WR, Bogomolni RA. The photocycle of a flavin-binding domain of the blue light photoreceptor phototropin. J. Biol. Chem. 2001;276:36493–36500. doi: 10.1074/jbc.M103114200. [DOI] [PubMed] [Google Scholar]
- Takahashi F, Yamagata D, Ishikawa M, Fukamatsu Y, Ogura Y, Kasahara M, Kiyosue T, Kikuyama M, Wada M, Kataoka H. AUREOCHROME, a photoreceptor required for photomorphogenesis in stramenopiles. Proc. Natl. Acad. Sci. USA. 2007;104:19625–19630. doi: 10.1073/pnas.0707692104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T. Maximum likelihood density modification. Acta Crystallogr. sect. D. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien R. Nobel Lecture: Green fluorescent protein - Constructing and exploiting the fluorescent protein paintbox. Angew. Chem. Int. Ed. 2009;48:5612–5626. doi: 10.1002/anie.200901916. [DOI] [PubMed] [Google Scholar]
- Vaidya A, Chen C-H, Dunlap J, Loros J, Crane B. Structure of a light-activated LOV protein dimer that regulates transcription. Sci. Signal. 2011;4:ra50. doi: 10.1126/scisignal.2001945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn M, Isupov M, Murshudov G. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, Hahn KM. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461:104–108. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Kuk J, Moffat K. Crystal structure of Pseudomonas aeruginosa bacteriophytochrome: Photoconversion and signal transduction. Proc. Natl. Acad. Sci., USA. 2008;105:14715–14720. doi: 10.1073/pnas.0806718105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoltowski B, Schwerdtfeger C, Widom J, Loros J, Bilwes A, Dunlap J, Crane B. Conformational switching in the fungal light sensor Vivid. Science. 2007;316:1054–1057. doi: 10.1126/science.1137128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoltowski B, Vaccaro B, Crane B. Mechanism-based tuning of a LOV domain photoreceptor. Nat. Chem. Biol. 2009;5:827–834. doi: 10.1038/nchembio.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.