Abstract
Mutations in the untranslated regulatory regions of genes may result in abnormal gene expression or transcriptional regulation. In this study, we characterize the ornithine transcarbamylase (OTC) mRNA isoforms of the X-linked OTC gene involved in the urea formation in the liver. Our data revealed that two major transcripts (OTC-t1 and OTC-t2) are more highly expressed than any of the other isoforms in all the tissues analyzed, though a longer transcript (OTC-t3) was also isolated and characterized from the brain sample. The OTC-t2 sequence fully matches the OTC mRNA reference sequence (NM_000531.5). All three isoforms use a canonical AAUAAA hexamer that is predicted to fold into a hairpin secondary structure which might be exposed to the cleavage and polyadenylation specificity factor. In addition, we observed that the OTC-t1 and OTC-t2 transcripts display heterogeneity at the cleavage sites in a tissue-dependent manner. Taken together, our data demonstrate that several mRNA isoforms are transcribed from the OTC gene, thereby indicating a wide degree of variability in post-transcriptional regulation.
Introduction
Impairment of ornithine transcarbamylase (OTC; EC: 2.1.3.3) function causes the most frequent birth defect in the urea cycle, namely ornithine transcarbamylase deficiency (OTCD, MIM 311250), which occurs at an estimated frequency of 1 in 14,000 births (Brusilow and Maestri, 1996). The gene that encodes the OTC protein is mapped to an X-linked, 73 kb genomic sequence that is comprised of ten small exons (Ricciuti et al., 1976; Hata et al., 1988). Routinely, a diagnosis of OTCD is initially made through biochemical measurements. Affected individuals have high levels of ammonia in their plasma and increased urinary excretion of orotic acid, which results from the difficulty in incorporating carbamylphosphate into the cycle. Subsequent molecular confirmation of the disease-associated mutation in the OTC gene is essential, but routine sequencing methods fail to identify large deletions in heterozygous females or duplications in both men and women (Suess et al., 1992; Quental et al., 2009; Shchelochkov et al., 2009; Balasubramaniam et al., 2010).
Mutations located within introns that modify the normal splicing pattern (Matsuda and Tanase, 1997; Climent and Rubio, 2002; Ogino et al., 2007), as well as mutations in regulatory elements, and in the 5′ and 3′ untranslated regulatory regions (UTRs) are thought to be responsible for the disease phenotype and may explain a portion of the clinical cases without an identified mutation that is linked to the phenotype. Recently, the 5′ UTR of the gene was studied, and a disruption of the enhancer-promoter binding site was found to be associated with clinical presentation (Luksan et al., 2010). This finding has reinforced the hypothesis that some uncharacterized OTCD cases may present with mutations in regulatory regions. Although alterations at the 5′ UTR may affect gene expression, mutations at the 3′ UTR in any gene may associate with impaired mRNA processing, which can lead to deregulation of gene expression, nuclear export, mRNA stability, and translational efficiency (Mazumder et al., 2003). Among all disease-associated mutations, some are associated with an impairment in the process of mRNA maturation in diseases such as thalassemia (Higgs et al., 1983; Ma et al., 2001), thrombophilia (Gehring et al., 2001), and amyotrophic lateral sclerosis (Lin et al., 1998).
To date, no data have been documented regarding the characterization of the OTC-3′ UTR, and this knowledge is critically relevant in planning future diagnostic approaches that may embrace both UTRs in combination with coding and splice-site sequences. In this study, we characterized the 3′ UTR region of OTC transcripts in two tissues where the gene is highly expressed (the liver and small intestine) and in two other tissues with lower expression (the brain and muscle). We isolated two abundant and a less expressed mRNA isoform that use canonical AAUAAA sequences as polyA signal. In addition, we discover tissue-dependent heterogeneity at the cleavage sites that reveals the variability associated with OTC mRNA processing.
Materials and Methods
RT-PCR analyses
Total RNA from the liver, small intestine, brain, and skeletal muscle was obtained from the FirstChoice Human Total RNA Survey Panel (Ambion). Each tissue-sample (lot 1004068) contains 10 μg of total RNA from three distinct donors as follows: liver and small intestine (two men and one woman), muscle and brain (two women and one man). First-strand cDNA synthesis was performed by using an RETROscript First Strand Synthesis Kit (Ambion) and 1 μg of mRNA, according to specifications of the manufacturer. Gene-specific cDNA amplification was performed by using the OTCF (5′ GGACACCCTGGCTAAAGAAGCATCC 3′; located at position 658–682 in the reference sequence NM_000531.5) and OTCR (5′ TTGGAGTAGCTGCCTGAAGGTGC 3′; position 853–875) primer pair to evaluate the OTC expression in each tissue. The GAPDH housekeeping gene was also amplified as an internal cDNA control, using the following primers: GAPDHF (5′ CCAGCCGAGCCACATCG 3′) and GAPDHR (5′ GGTCATGAGTCCTTCCACG 3′) (Pereira-Castro et al., 2011). The amplification conditions were defined as follows: an initial denaturation at 95°C for 15 min, 35 cycles of 94°C for 30 s, annealing at 62°C for 30 s and extension at 72°C for 90 s, and a final extension at 72°C for 10 min.
3 ′ Rapid amplification of cDNA ends analysis
The cDNA was synthesized using a SMARTer™ 3′ rapid amplification of cDNA ends (RACE) Amplification Kit (Clontech) and 1 μg of total RNA from the liver, small intestine, brain, or muscle (Ambion) after muscle according to the manufacturer's protocol. The 3′ RACE PCR reactions were performed using 12.5 μL of 2X QIAGEN PCR kit (QIAGEN, Hilden, Germany), 1 μL cDNA obtained from the 3′ RACE cDNA syntheses, 2.5 μL 10X Universal Primer Mix provided with the Clontech kit, 0.5 μL OTC-specific primer (10 μM; GSP1: 5′ ATGGCTGTCATGGTGTCCCTGCTGA 3′; position 1217–1241)), and 8.5 μL RNAse-free water for a final volume of 25 μL per reaction. The amplification conditions were set as recommended by the manufacturer.
The 3′ RACE PCR amplicons were resolved on a 1% agarose gel, gel extracted, and purified. Nested PCR was performed with 1 μL of the 3′ RACE-amplified products by using 1 μL nested forward OTC-specific primer (NGSP: 5′ CTCACCTCAGCTCCAGAAGCCTA 3′; position 1249–1271), 1 μL nested universal reverse primer provided by the manufacturer (Clontech), 12.5 μL 2xQIAGEN PCR kit, and 9.5 μL RNAse-free water for a final volume of 25 μL per reaction. The amplification was performed by using the manufacturer's recommended conditions. The PCR products were resolved on a 1% agarose gel, extracted, and purified.
Sequencing analyses
Sequencing reactions were performed with a Big Dye Terminator Cycle Sequencing Ready Reaction Kit version 3.1 (Applied Biosystems), according to the manufacturer's recommendations, and the reaction conditions were defined as follows: an initial denaturation at 96°C for 4 min, followed by 35 cycles of denaturation (94°C, 10 s), annealing (60°C, 10 s), and extension (60°C, 2 min). The primers used were NSGP1, for all the products amplified, and a downstream OTC primer (5′ CTTGTGAGAATGTGGTAGTGGGGAATCC 3′) to amplify the larger PCR product obtained in the brain. The sequencing products were run on an ABI PRISM 3130XL Genetic Analyzer (Applied Biosystems).
Computational analyses of mRNAs and expressed sequence tags
The 3′ expressed sequence tags (ESTs) available at the UniGene database (www.ncbi.nlm.nih.gov/unigene) (Wheeler et al., 2008) for human (Hs.117050) and non-human mammals (Mus musculus: Mm.2611; Rattus norvegicus: Rn.2391; Sus scrofa: Ssc.16155 and Canis lupus familiaris: Cfa.24035) were collected and analyzed. To analyze their 3′ends, all the ESTs were aligned by using the default settings in the MUSCLE 3.6 software (Edgar, 2004), which was running in the Geneious program v5.3 (Drummond et al., 2011).
The secondary structures were calculated by using the fragment containing the poly(A) site and the 50 flanking nucleotides (Sittler et al., 1995) by using the Mfold program (Zuker, 2003), which is available at www.bioinfo.rpi.edu/applications/mfold/.
Results
OTC expression by RT-PCR
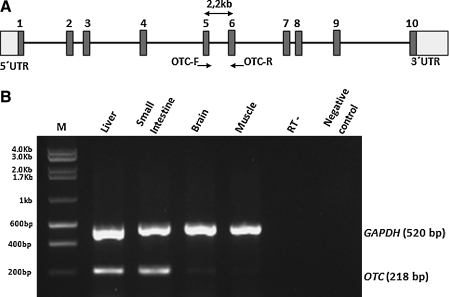
An RT-PCR strategy was applied by using a forward primer specific to OTC exon 5 and a reverse primer specific to exon 6. This primer set was predicted to yield a 2.2 kb fragment from genomic DNA, and a 218 bp fragment was expected for mRNA samples (Fig. 1A), thereby enabling us to distinguish true OTC transcripts from DNA contamination. This analysis, although semi-quantitative, revealed a higher expression of OTC in the liver and the small intestine (Neill et al., 2009) compared with the brain and muscle tissues (Fig. 1B).
FIG. 1.
Ornithine transcarbamylase (OTC) is highly expressed in the liver and the small intestine. (A) Schematic organization of the OTC gene and the localization of the primers that were used for the RT-PCR analyses. (B) Separation of the RT-PCR products on a 1% agarose gel. The housekeeping gene, GAPDH, was amplified as a control.
3 ′ RACE analyses of the OTC mRNA
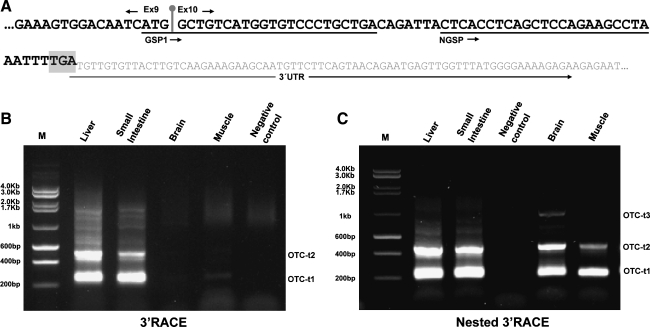
After we had established the presence of the OTC transcripts in the mRNA samples, we sought to characterize the OTC 3′ end in the four tissues. Thus, a 3′ RACE strategy was used with a forward gene-specific primer that matched both exon 9 and 10 (GSP1) and the poly-T primer that was supplied in the RACE kit (Fig. 2A). In the liver and the small intestine, we observed two highly expressed transcripts (herein designated as OTC-t1 and OTC-t2) (Fig. 2B). In addition, we observed the same transcripts in the brain and skeletal muscle though their expression level was lower. To assess these low amplified transcripts, nested PCR was performed to re-amplify the fragments that were obtained in the 3′ RACE PCR by using a NGSP located only 7 bp 3′ downstream from the end of GSP1 (Fig. 2A). Using this approach, the two transcripts just mentioned (OTC-t1 and OTC-t2) were successfully re-amplified in all tissues (Fig. 2C). In addition, a longer transcript was amplified in the brain sample, which we designated OTC-t3 (Fig. 2C).
FIG. 2.
The OTC gene is expressed in distinct mRNA isoforms. (A) Schematic localization of the GSP1 and nested forward OTC-specific primer (NGSP) primers in the OTC mRNA sequence. (B) 3′ rapid amplification of cDNA ends (RACE) products in the liver, small intestine, brain, and muscle. (C) PCR products obtained via nested PCR using the NGSP gene-specific primer.
Poly(A) signals and cleavage sites in the OTC mRNA isoforms
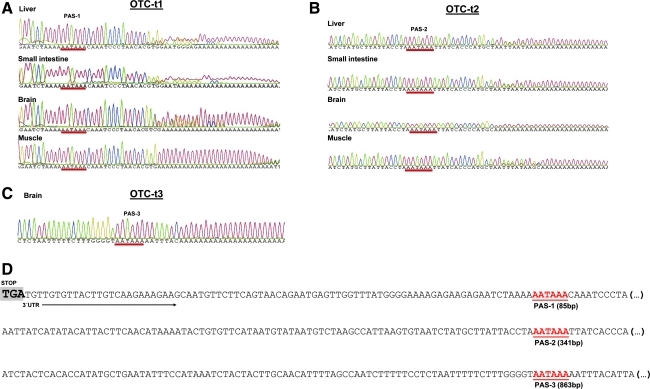
The fragments corresponding to OTC-t1 and OTC-t2 in all the tissues were sequenced along with the OTC-t3 transcript that was observed in the brain (Fig. 3). The smaller and more highly expressed transcript (OTC-t1) had the same sequence in all four tissues, though the sequence displayed heterogeneity at the cleavage site (i.e., the exact point of the initiation of the polyA tail) in the liver, small intestine, and brain. In muscle, however, a clear, distinct polyadenylation initiation site exists. Heterogeneity in cleavage sites has been reported to be a common event in mammalian mRNA sequences (Pauws et al., 2001; Tian et al., 2005), which also appears to be the case with OTC mRNA.
FIG. 3.
Distinct OTC mRNA isoforms revealed by 3′ RACE. (A–C) Sequencing results from the OTC transcripts corresponding to OTC-t1, OTC-t2, and OTC-t3. The predicted polyadenylation site for each transcript is underlined in red. (D) Position of the three polyadenylation sites identified in the OTC 3′ end.
The most common polyadenylation signal (PAS) is the canonical AAUAAA hexamer (reviewed in Lutz, 2008; Lutz and Moreira, 2011), which corresponds to more than half of the PASs observed in human and mouse sequences (Tian et al., 2005) within the expected range of 10 to 30–35 bases (reviewed in Lutz, 2008; Mandel et al., 2008; Lutz and Moreira, 2011) upstream of the polyA tail. Using the single muscle cleavage site as a reference, it is possible to assign a putative AAUAAA signal in the OTC-t1 transcript that is located 85 nucleotides downstream the OTC-stop codon and 17 nucleotides upstream of the OTC-t1 polyA tail (Fig. 3A, D). We hypothesize that this site corresponds to the PAS sequence used for the smaller and more highly expressed transcript, which was designated PAS-1.
In the case of the OTC-t2 transcript, the sequencing analyses revealed that the transcript matches the reference sequence for the OTC mRNA (NM_000531.5). In this transcript, a canonical PAS (PAS-2) is located 341 nucleotides downstream of the stop codon and 13 nucleotides upstream of the polyA tail (using the single brain cleavage site as a reference). Heterogeneity at the OTC-t2 cleavage site was observed in the liver, small intestine, and muscle sample but not in the brain (Fig. 3B), thus indicating that the variability observed in the OTC mRNA cleavage sites is tissue dependent.
Samples from the longer OTC-t3 transcript were obtained from the brain samples, though we cannot exclude the possibility that this mRNA isoform is also present in other tissues. However, if that is the case, then the level of OTC-t3 expression is insignificant compared with OTC-t1 and OTC-t2. Detailed inspection of this long transcript revealed a canonical AAUAAA hexamer (PAS-3) that is 863 nucleotides downstream of the stop codon and seven nucleotides from the cleavage site (Fig. 3C, D), which makes it a reliable PAS signal for this transcript.
Secondary structure of the OTC mRNA polyA signals
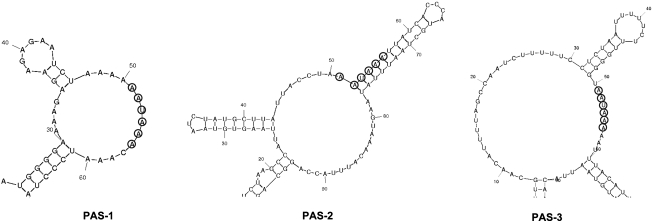
Previous data have established a functional role for the structural context of the signal sequence to maintain efficient recognition by the cleavage and polyadenylation specificity factor (CPSF) (Sittler et al., 1995; Graveley et al., 1996a, 1996b). Exposure of the AAUAAA sequence in a hairpin structure, where the polyA sequence is completely or partially embedded in the hairpin loop, would, therefore, facilitate its recognition by 3′ end processing factors (Sittler et al., 1995). To determine the secondary structure of the sequence neighboring PAS-1, PAS-2, and PAS-3, we employed a previously defined methodology (Sittler et al., 1995) that uses 50 nucleotides upstream and downstream of each AAUAAA site found in the OTC transcripts to predict structural elements. The resulting configurations (Fig. 4) revealed that the closest (PAS-1) and the most distal (PAS-3) signals to the termination codon of the OTC gene are part of a loop structure, whereas the PAS-2 signal is partially included in the stem that supports the hairpin structure as predicted, for example, in the case of the rat calcitonin gene (Sittler et al., 1995). The loop that includes the polyA site in all three structures is larger than the loops that have previously been observed in other vertebrate transcripts (i.e., the chicken β-tropomyosin-exon 9B, mouse neural cell adhesion molecule, and rat calcitonin gene) (Sittler et al., 1995), though its functional relevance, if any, is not known.
FIG. 4.
Predicted secondary structures of the canonical AAUAAA signals found in the OTC mRNA.
Discussion
Given that OTCD is the most common urea cycle disorder and that many cases are difficult to resolve by screening the OTC coding sequences, exon/intron-flanking sequences, or by deletion-detection methodologies, for example, haplotype analysis (Azevedo et al., 2003) and multiplex ligation-dependent probe amplification (Quental et al., 2009), the functional mapping of UTR sequences is critical. As recently demonstrated (Luksan et al., 2010), mutations in the OTC promoter region may have functional consequences preventing proper binding to the distal enhancer, in addition to mutations in the transcription factor binding sites that would impair gene expression. Therefore, it may be that a mutation in the 3′ UTR sequence could affect translational efficiency and/or stability of the mRNA precursor, thus leading to a disease-associated phenotype. Since the OTC gene is X-linked, the clinical outcome of a mutation in the 3′ UTR of the gene is expected to depend, as for any other disease-associated mutation, on the X-inactivation pattern in heterozygous women.
A previous in vitro study (Sheets et al., 1990) demonstrated the functional effect of all 18 possible single mutations on the AAUAAA canonical sequence, and data on the beta-globin (Higgs et al., 1983; Orkin et al., 1985; Rund et al., 1992; Ma et al., 2001), coagulation factor II (Gehring et al., 2001), glutamate transporter (Lin et al., 1998), and FOXP3 (Bennett et al., 2001) genes demonstrate the association between mutations affecting mRNA processing and diseases in some clinical cases.
To date, no studies have performed a detailed analysis of the OTC 3′UTR and the corresponding cis-regulatory elements that are necessary for mRNA processing. Our goal was to examine the mRNA isoforms that result from the 3′ RACE experiment to map these elements. We found three major mRNA isoforms in our sample set (liver, small intestine, brain, and muscle) that revealed an undocumented heterogeneity in OTC mRNA sequences. Two of the transcripts (OTC-t1 and OTC-t2) are expressed in all four of the tissues (Figs. 2 and 3). Although our methodology does not allow for direct quantitative inferences, our results revealed a tendency for high-level expression of the smaller transcript (OTC-t1). The second more abundant transcript (OTC-t2) matches the 3′ end of the reference OTC-mRNA sequence and is expressed in all four tissues. A third isoform was also purified and sequenced in the brain (OTC-t3), though it may also be expressed in other tissues at levels that are below the detection resolution of the technique used here (Fig. 3). We cannot discard the hypothesis that other OTC transcripts also exist though they represent less expressed isoforms.
All the three OTC transcripts that are discussed here are likely to use a canonical AAUAAA signal which is recognized by the CPSF, a 160 kDa protein that is involved in the processing of pre-mRNAs (reviewed in Lutz, 2008; Mandel et al., 2008). Several studies have demonstrated the relevance of secondary structures in the efficiency of mRNA processing (Sittler et al., 1995; Graveley et al., 1996a, 1996b, Klasens et al., 1999; Zarudnaya et al., 2003). In some viral and eukaryotic transcript examples, a hairpin structure exposes the AAUAAA signal for CPSF binding. We verified that the OTC AAUAAA-related sequences are either exposed in the hairpin loop (PAS-1 and PAS-3) or partially inserted into the stem that supports the loop (PAS-2) (Fig. 4). Although analysis of the sequence reveals a larger loop, the PAS structures strongly resemble those that have been previously documented (Sittler et al., 1995), which reinforces the possibility of being recognized and used by the CPSF.
In addition, as already demonstrated in other mRNA sequences (Pauws et al., 2001), we found heterogeneity at the cleavage sites in PAS-1 and PAS-2 that vary according to the tissue. For instance, for the OTC-t1, a single cleavage site was observed in the muscle sample, whereas more than one was detected in the liver, small intestine, and brain.
Alternative polyadenylation is thought to play a major role in gene regulation and may affect transcript stability, translation rate, and the localization of mRNA (reviewed in Lutz, 2008; Lutz and Moreira, 2011). In the case of the OTC gene, the outcome of each of these transcripts remains unclear. It is possible that each of them could be translated into an active protein, but such a conclusion cannot be made solely on the basis of our results.
The presence of two major transcripts in tissues other than the liver and the small intestine would allow for direct mRNA analyses without the need for invasive procedures. In agreement with the results presented here, previous data highlight the importance of using OTC cDNA from total RNA isolated from peripheral blood lymphocytes in molecular analyses (Giorgi et al., 2000; Bisanzi et al., 2002). This finding reinforces the idea that the OTC gene appears to have a low expression rate in a variety of tissues (Giorgi et al., 2000; Bisanzi et al., 2002), and its detection could require a second (or nested) PCR round as used here. Data from other mammals that were extracted from UNIGENE revealed that, as in humans, OTC transcripts are present in their livers and small intestines (Table 1). We also noticed that OTC is expressed in the muscle of mice and pigs and in the connective tissue and testes in humans, (Table 1) thus indicating that gene expression is not limited to the liver and small intestine. In addition, the 3′ ESTs from UNIGENE exhibited alternative polyadenylation in the mouse and rat Otc transcripts, as well as heterogeneity in the cleavage sites of mouse transcripts that result from the same polyA signal, which suggests the preservation of the process during evolution.
Table 1.
3′ Expressed Sequence Tags Data from UNIGENE Related to OTC Expression in Mammals
| Species | Number of EST sequences | Tissue expression |
|---|---|---|
| Homo sapiens (Hs.117050) | 28 | Connective tissue, liver, testis |
| Mus musculus (Mm.2611) | 129 | Intestine, liver, muscle |
| Rattus norvegicus (Rn.2391) | 26 | Liver, small intestine |
| Sus scrofa (Ssc.16155) | 29 | Intestine, liver, lung, muscle |
| Canis lupus familiaris (Cfa.24035) | 8 | Liver |
As our understanding about the OTC gene increases, more cases of disease-associated phenotypes are expected to be resolved, thereby reducing the rate of patients who lack molecular confirmation (Yamaguchi et al., 2006). In this sense, the functional impact of nucleotide substitutions that affect the 3′ end of OTC transcripts might now be better evaluated, as the boundaries of the sequence that corresponds to the exact 3′ UTR segment are known in humans.
Acknowledgments
This work was supported by the Fundação para a Ciencia e a Tecnologia (FCT) grants SFRH/BD/44264/2008 to I.P-C, and by POPH-QREN-Tipologia 4.2- Scientific employment promotion funded by the European Social Fund and by national funds from the Ministry of Science, Technology, and Higher Education (MCTES) to L.A., by research project PIC/IC/82794/2007 sponsored by FCT, COMPETE program and by FEDER funds. The Institute of Molecular Pathology and Immunology of the University of Porto (IPATIMUP) is an Associate Laboratory of the Portuguese MCTES and is partially supported by FCT.
Disclosure Statement
The authors declare that no conflicts of interest exist.
References
- Azevedo L. Stolnaja L. Tietzeova E. Hrebicek M. Hruba E. Vilarinho L., et al. New polymorphic sites within ornithine transcarbamylase gene: population genetics studies and implications for diagnosis. Mol Genet Metab. 2003;78:152–157. doi: 10.1016/s1096-7192(03)00019-2. [DOI] [PubMed] [Google Scholar]
- Balasubramaniam S. Rudduck C. Bennetts B. Peters G. Wilcken B. Ellaway C. Contiguous gene deletion syndrome in a female with ornithine transcarbamylase deficiency. Mol Genet Metab. 2010;99:34–41. doi: 10.1016/j.ymgme.2009.08.007. [DOI] [PubMed] [Google Scholar]
- Bennett C.L. Brunkow M.E. Ramsdell F. O'Briant K.C. Zhu Q. Fuleihan R.L., et al. A rare polyadenylation signal mutation of the FOXP3 gene (AAUAAA→AAUGAA) leads to the IPEX syndrome. Immunogenetics. 2001;53:435–439. doi: 10.1007/s002510100358. [DOI] [PubMed] [Google Scholar]
- Bisanzi S. Morrone A. Donati M.A. Pasquini E. Spada M. Strisciuglio P., et al. Genetic analysis in nine unrelated Italian patients affected by OTC deficiency: detection of novel mutations in the OTC gene. Mol Genet Metab. 2002;76:137–144. doi: 10.1016/s1096-7192(02)00028-8. [DOI] [PubMed] [Google Scholar]
- Brusilow S.W. Maestri N.E. Urea cycle disorders: diagnosis, pathophysiology, and therapy. Adv Pediatr. 1996;43:127–170. [PubMed] [Google Scholar]
- Climent C. Rubio V. Identification of seven novel missense mutations, two splice-site mutations, two microdeletions and a polymorphic amino acid substitution in the gene for ornithine transcarbamylase (OTC) in patients with OTC deficiency. Hum Mutat. 2002;19:185–186. doi: 10.1002/humu.9011. [DOI] [PubMed] [Google Scholar]
- Drummond A.J. Ashton B. Buxton S. Cheung M. Cooper A. Duran C., et al. Geneious v5.4. 2011. www.geneious.com www.geneious.com
- Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring N.H. Frede U. Neu-Yilik G. Hundsdoerfer P. Vetter B. Hentze M.W., et al. Increased efficiency of mRNA 3′ end formation: a new genetic mechanism contributing to hereditary thrombophilia. Nat Genet. 2001;28:389–392. doi: 10.1038/ng578. [DOI] [PubMed] [Google Scholar]
- Giorgi M. Morrone A. Donati M.A. Ciani F. Bardelli T. Biasucci G., et al. Lymphocyte mRNA analysis of the ornithine transcarbamylase gene in Italian OTCD male patients and manifesting carriers: identification of novel mutations. Hum Mutat. 2000;15:380–381. doi: 10.1002/(SICI)1098-1004(200004)15:4<380::AID-HUMU12>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Graveley B.R. Fleming E.S. Gilmartin G.M. Restoration of both structure and function to a defective poly(A) site by in vitro selection. J Biol Chem. 1996a;271:33654–33663. doi: 10.1074/jbc.271.52.33654. [DOI] [PubMed] [Google Scholar]
- Graveley B.R. Fleming E.S. Gilmartin G.M. RNA structure is a critical determinant of poly(A) site recognition by cleavage and polyadenylation specificity factor. Mol Cell Biol. 1996b;16:4942–4951. doi: 10.1128/mcb.16.9.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata A. Tsuzuki T. Shimada K. Takiguchi M. Mori M. Matsuda I. Structure of the human ornithine transcarbamylase gene. J Biochem. 1988;103:302–308. doi: 10.1093/oxfordjournals.jbchem.a122265. [DOI] [PubMed] [Google Scholar]
- Higgs D.R. Goodbourn S.E. Lamb J. Clegg J.B. Weatherall D.J. Proudfoot N.J. Alpha-thalassaemia caused by a polyadenylation signal mutation. Nature. 1983;306:398–400. doi: 10.1038/306398a0. [DOI] [PubMed] [Google Scholar]
- Klasens B.I. Thiesen M. Virtanen A. Berkhout B. The ability of the HIV-1 AAUAAA signal to bind polyadenylation factors is controlled by local RNA structure. Nucleic Acids Res. 1999;27:446–454. doi: 10.1093/nar/27.2.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.L. Bristol L.A. Jin L. Dykes-Hoberg M. Crawford T. Clawson L., et al. Aberrant RNA processing in a neurodegenerative disease: the cause for absent EAAT2, a glutamate transporter, in amyotrophic lateral sclerosis. Neuron. 1998;20:589–602. doi: 10.1016/s0896-6273(00)80997-6. [DOI] [PubMed] [Google Scholar]
- Luksan O. Jirsa M. Eberova J. Minks J. Treslova H. Bouckova M., et al. Disruption of OTC promoter-enhancer interaction in a patient with symptoms of ornithine carbamoyltransferase deficiency. Hum Mutat. 2010;31:E1294–E1303. doi: 10.1002/humu.21215. [DOI] [PubMed] [Google Scholar]
- Lutz C.S. Alternative polyadenylation: a twist on mRNA 3′ end formation. ACS Chem Biol. 2008;3:609–617. doi: 10.1021/cb800138w. [DOI] [PubMed] [Google Scholar]
- Lutz C.S. Moreira A. Alternative mRNA polyadenylation in eukaryotes: an effective regulator of gene expression. WIREs RNA. 2011;2:23–31. doi: 10.1002/wrna.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S.K. Lee A.C. Chan A.Y. Chan L.C. A novel AATAAA→CATAAA mutation at the polyadenylation site of the beta-globin gene. Br J Haematol. 2001;115:230–231. [PubMed] [Google Scholar]
- Mandel C.R. Bai Y. Tong L. Protein factors in pre-mRNA 3′-end processing. Cell Mol Life Sci. 2008;65:1099–1122. doi: 10.1007/s00018-007-7474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda I. Tanase S. The ornithine transcarbamylase (OTC) gene: mutations in 50 Japanese families with OTC deficiency. Am J Med Genet. 1997;71:378–383. [PubMed] [Google Scholar]
- Mazumder B. Seshadri V. Fox P.L. Translational control by the 3′-UTR: the ends specify the means. Trends Biochem Sci. 2003;28:91–98. doi: 10.1016/S0968-0004(03)00002-1. [DOI] [PubMed] [Google Scholar]
- Neill M.A. Aschner J. Barr F. Summar M.L. Quantitative RT-PCR comparison of the urea and nitric oxide cycle gene transcripts in adult human tissues. Mol Genet Metab. 2009;97:121–127. doi: 10.1016/j.ymgme.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino W. Takeshima Y. Nishiyama A. Okizuka Y. Yagi M. Tsuneishi S., et al. Mutation analysis of the ornithine transcarbamylase (OTC) gene in five Japanese OTC deficiency patients revealed two known and three novel mutations including a deep intronic mutation. Kobe J Med Sci. 2007;53:229–240. [PubMed] [Google Scholar]
- Orkin S.H. Cheng T.C. Antonarakis S.E. Kazazian H.H., Jr. Thalassemia due to a mutation in the cleavage-polyadenylation signal of the human beta-globin gene. EMBO J. 1985;4:453–456. doi: 10.1002/j.1460-2075.1985.tb03650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauws E. van Kampen A.H. van de Graaf S.A. de Vijlder J.J. Ris-Stalpers C. Heterogeneity in polyadenylation cleavage sites in mammalian mRNA sequences: implications for SAGE analysis. Nucleic Acids Res. 2001;29:1690–1694. doi: 10.1093/nar/29.8.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira-Castro I. Quental R. da Costa L.T. Amorim A. Azevedo L. Successful COG8 and PDF overlap is mediated by alterations in splicing and polyadenylation signals. Hum. Genet. 2011 doi: 10.1007/s00439-011-1075-9. [DOI] [PubMed] [Google Scholar]
- Quental R. Azevedo L. Rubio V. Diogo L. Amorim A. Molecular mechanisms underlying large genomic deletions in ornithine transcarbamylase (OTC) gene. Clin Genet. 2009;75:457–464. doi: 10.1111/j.1399-0004.2009.01172.x. [DOI] [PubMed] [Google Scholar]
- Ricciuti F.C. Gelehrter T.D. Rosenberg L.E. X-chromosome inactivation in human liver: confirmation of X-linkage of ornithine transcarbamylase. Am J Hum Genet. 1976;28:332–338. [PMC free article] [PubMed] [Google Scholar]
- Rund D. Dowling C. Najjar K. Rachmilewitz E.A. Kazazian H.H., Jr. Oppenheim A. Two mutations in the beta-globin polyadenylylation signal reveal extended transcripts and new RNA polyadenylylation sites. Proc Natl Acad Sci U S A. 1992;89:4324–4328. doi: 10.1073/pnas.89.10.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shchelochkov O.A. Li F.Y. Geraghty M.T. Gallagher R.C. Van Hove J.L. Lichter-Konecki U., et al. High-frequency detection of deletions and variable rearrangements at the ornithine transcarbamylase (OTC) locus by oligonucleotide array CGH. Mol Genet Metab. 2009;96:97–105. doi: 10.1016/j.ymgme.2008.11.167. [DOI] [PubMed] [Google Scholar]
- Sheets M.D. Ogg S.C. Wickens M.P. Point mutations in AAUAAA and the poly (A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res. 1990;18:5799–5805. doi: 10.1093/nar/18.19.5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittler A. Gallinaro H. Jacob M. The secondary structure of the adenovirus-2 L4 polyadenylation domain: evidence for a hairpin structure exposing the AAUAAA signal in its loop. J Mol Biol. 1995;248:525–540. doi: 10.1006/jmbi.1995.0240. [DOI] [PubMed] [Google Scholar]
- Suess P.J. Tsai M.Y. Holzknecht R.A. Horowitz M. Tuchman M. Screening for gene deletions and known mutations in 13 patients with ornithine transcarbamylase deficiency. Biochem Med Metab Biol. 1992;47:250–259. doi: 10.1016/0885-4505(92)90033-u. [DOI] [PubMed] [Google Scholar]
- Tian B. Hu J. Zhang H. Lutz C.S. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler D.L. Barrett T. Benson D.A. Bryant S.H. Canese K. Chetvernin V., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2008;36:D13–D21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S. Brailey L.L. Morizono H. Bale A.E. Tuchman M. Mutations and polymorphisms in the human ornithine transcarbamylase (OTC) gene. Hum Mutat. 2006;27:626–632. doi: 10.1002/humu.20339. [DOI] [PubMed] [Google Scholar]
- Zarudnaya M.I. Kolomiets I.M. Potyahaylo A.L. Hovorun D.M. Downstream elements of mammalian pre-mRNA polyadenylation signals: primary, secondary and higher-order structures. Nucleic Acids Res. 2003;31:1375–1386. doi: 10.1093/nar/gkg241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]