Abstract
Throughout the kingdoms of life, transfer RNA (tRNA) undergoes over 100 enzyme-catalyzed, methyl-based modifications. Although a majority of the methylations are conserved from bacteria to mammals, the functions of a number of these modifications are unknown. Many of the proteins responsible for tRNA methylation, named tRNA methyltransferases (Trms), have been characterized in Saccharomyces cerevisiae. In contrast, only a few human Trms have been characterized. A BLAST search for human homologs of each S. cerevisiae Trm revealed a total of 34 human proteins matching our search criteria for an S. cerevisiae Trm homolog candidate. We have compiled a database cataloging basic information about each human and yeast Trm. Every S. cerevisiae Trm has at least one human homolog, while several Trms have multiple candidates. A search of cancer cell versus normal cell mRNA expression studies submitted to Oncomine found that 30 of the homolog genes display a significant change in mRNA expression levels in at least one data set. While 6 of the 34 human homolog candidates have confirmed tRNA methylation activity, the other candidates remain uncharacterized. We believe that our database will serve as a resource for investigating the role of human Trms in cellular stress signaling.
Introduction
Transfer RNA (tRNA) is a noncoding RNA responsible for the accurate addition of an amino acid to the 3′ end of a translating protein. A tRNA is charged with a specific amino acid by an aminoacyl-tRNA synthetase after which the tRNA anticodon binds to the mRNA codon at the ribosome A site. tRNA has a distinct three-dimensional L-shape, often shown in two dimensions as a cloverleaf to display each of the five stem-loop arms (acceptor stem, variable arm, D arm, anticodon arm, and TψC arm) generated by self-dimerization. Based on the lengths of their D arm and variable arm, tRNA is divided into three different classes. Class I tRNA has a four base pair D stem and a four to five base variable loop. Class II tRNA has a three base pair D stem and a four to five base variable loop. Class III tRNA has a three base pair D stem and a large variable arm (13 to 21 bases). An additional distinguishing feature of tRNA is that over 100 known post-translational modifications have been reported (Dunin-Horkawicz et al., 2006).
The number of modified bases varies among individual tRNA types. The tRNAs of the budding yeast Saccharomyces cerevisiae have an average of 11 modifications per tRNA, while the average mammalian cytoplasmic tRNA has 13 to 14 modifications (Sprinzl et al., 1998). One of the most common tRNA modifications is methylation. Because tRNA methylation sites are conserved throughout most living organisms, their significance is implicit (Sprinzl et al., 1998; Dunin-Horkawicz et al., 2006). As many as 1% of the coding genomes of some bacterial species are dedicated to tRNA modification. Such a large commitment of biological resources by these bacteria to tRNA modification suggests that tRNA modification is critical to basic cellular function. Depending on their location, tRNA methylations have been connected to structure stability, frameshift prevention, and translation efficiency (Motorin and Helm, 2010). Methylations have the potential to be dynamic processes. A 2010 study by Chan et al. described a method for analyzing tRNA methylation patterns via mass spectrometry (MS) (Chan et al., 2010). This detection method allowed for detailed analysis of specific tRNA methylation changes in S. cerevisiae after exposure to the toxicants methylmethanosulfonate (MMS), sodium arsenite, hydrogen peroxide, and sodium hypochlorite. Each drug treatment caused a unique, dose-dependent series of tRNA methylation changes in the global pool. In addition, corresponding mutants deficient in specific tRNA methyltransferase (Trm) activities demonstrated decreased viability after exposure to one of the toxicants, therefore supporting the notion that enzyme-catalyzed tRNA methylation plays a dynamic role in the cellular response pathway.
Methylation reactions are catalyzed by enzymes known as methyltransferases. Many biomolecule types are potential targets of methylation, including proteins, DNA, and RNA. Most identified methyltransferases transfer the labile methyl group from S-adenosyl methionine (SAM or AdoMet) to its substrate, leaving S-adenosyl homocysteine as a byproduct (Fig. 1). This study focuses on Trms that have tRNA molecules as their substrate. In this study, we will summarize current information reported on each known S. cerevisiae Trm and we will identify and describe the closest human homologs to each S. cerevisiae Trm based on primary amino acid sequence.
FIG. 1.
The tRNA methyltransferase (Trm) reaction. (A–D) Each reaction diagram shows an example of a methylation reaction for adenine (A), cytosine (B), guanine (C), and uridine (D). In these examples, the methyl group is added to the 1-carbon position of the purine ring (A, C), the 5-carbon position of the pyrimidine ring (B) and the 2′-oxygen position of the ribose sugar (D). The added methyl is in bold. (E) The reactive methyl of S-adenosyl methionine (SAM) is removed by the methyltransferase and added to the substrate, leaving S-adenosyl homocysteine (AdoHcy) as a byproduct. tRNA, transfer RNA.
Trms methylate nucleotides at a target sequence in specific tRNAs. The nucleotide itself can be methylated on the purine or pyrimidine ring or ribose base, depending on the methylation (Fig. 1b–e). In S. cerevisiae, a total of 18 Trms comprising 15 holoenzymes are required for production of nearly all known methylated tRNA nucleotides. All Trms contain a methyltransferase domain, most often a class 1 domain characterized by a Rossmann fold domain consisting of a seven-strand β-sheet and a reverse two-stranded β-hairpin loop, flanked on both sides by an α-helix (Schubert et al., 2003). Despite their common function, methyltransferase domains vary widely in sequence, often only sharing a common GXGXG methyltransferase sequence (Kagan and Clarke, 1994). Some Trms have a distinct RNA binding domain, while others use unknown motifs to bind their target. S. cerevisiae deletion strains with a knock-out of each Trm gene have been generated. Of all the strains, only trm5Δ, trm6Δ, trm61Δ, and trm7Δ display a slow growth phenotype. A few others, for example, trm9Δ, only display an obvious phenotype while under dietary or chemical stress. Still other Trm deletions display a phenotype in combination with the removal of another Trm. For example, a decreased viability phenotype was observed for trm4Δ when deleted in combination with TRM8 or TRM82 (Alexandrov et al., 2005).
In our initial attempt to identify and characterize human Trms, we searched the Human Protein Resource Database (HPRD). HPRD does not currently maintain a Trm grouping; the closest category is RNA methyltransferases. HPRD categorizes 13 proteins as RNA methyltransferases. Of these 13, only 8 were Trms. Although most enzymes responsible for S. cerevisiae Trm activity have been characterized, very little concrete work has been undertaken to analyze their human counterparts. In this study, we report the creation of a database listing all of the predicted human Trm homologs. Each of the 18 known yeast Trms has at least one human homolog and there are 34 homologs in total. Our database aids in the compilation of relevant biochemical information about these homologs. We have also utilized the on-line tools HSLPred, Scansite, PhosphoSite Plus, and Oncomine to make initial predictions about subcellular localization for each protein, about potential phosphorylation sites, and about any potential connections between each protein's regulations in cancerous cells versus benign cells, respectively.
Materials and Methods
The human Trm database
A blastp search using the complete amino acid sequences of each known S. cerevisiae Trm was performed against all known human amino acid sequences. Matches with an E-value score <0.01 were considered human homolog candidates, with the match having the lowest score considered the primary homolog candidate. The NCBI nucleotide, gene, and protein entry for each candidate was cataloged, along with any projected alternate transcripts. The chromosomal location of each candidate was also cataloged. Candidates that did not appear to be expressed proteins in humans, based on analysis of their NCBI entry or sequence characteristics, were then eliminated as candidates. Our search results are summarized in Table 1. The molecular weight and isoelectric point of each candidate was calculated using the computational tools available on the ExPASy Proteomics Server, Swiss Institute of Bioinformatics (http://ca.expasy.org) (Gasteiger et al., 2005).
Table 1.
Saccharomyces cerevisiae Transfer RNA Methyltransferases, Their Products, and Homo sapiens Candidate Homologs
| Yeast Trm | tRNA methylation product | Human homolog candidate | Chromosome position | E-Value | Predicted/known localization |
|---|---|---|---|---|---|
| Trm1 | N2, N2-dimethylguanosine (m22G26) | TRM1 | 19p13.2 | 7.0E-97 | Cytoplasm (53%) |
| C1ORF25 (Trm1-like protein) | 1q25.2 | 3.0E-15 | Nucleus (53%) | ||
| Trm2 | 5-methyluridine (m5U54) | TRMT2A | 22q11.1–13; 22q11.21 | 1.0E-16 | Nucleus/cytoplasm (known) |
| TRMT2B1 | Xq22.1 | 1.00E-8 | Nucleus (53%) | ||
| Trm3 | 2′-O-methylguanosine (Gm18) | TARBP1 | 1q42.3 | 9.0E-38 | Nucleus (53%) |
| Trm4 | 5-methylcytosine (m5C, several positions) | NSUN2 | 5p15.31 | 3.0E-122 | Nucleolus (known) |
| NSUN1 | 12p13 | 2.0E-26 | Nucleus (95%) | ||
| NSUN3 | 3q11.1 | 1.0E-12 | Nucleus (68%) | ||
| NSUN4 | 1p34 | 2.0E-11 | Nucleus (80%) | ||
| NSUN5 | 7q11.23 | 9.0E-13 | Nucleus (80%) | ||
| NSUN6 | 10p12.31 | 7.0E-13 | Nucleus (68%) | ||
| Trm5 | 1-methylguanosine (m1G37) | TRMT5 | 14q23.1 | 2.0E-64 | Nucleus (53%) |
| Trm6/Gcd10p | 1-methyladenosine (m1A58) | TRM6 | 20p12.3 | 1.0E-14 | Nucleus (58%) |
| Trm7 | 2′-O-methyl(base), positions 32 and 34 | FTSJ1 | Xp11.23 | 2.0E-71 | Nucleolus (known) |
| FTSJ2 | 7p22 | 1.0E-26 | Nucleus (80%) | ||
| FTSJ3 | 17q23.3 | 2.0E-39 | Nucleus (80%) | ||
| Trm8 | (with Trm82) 7-methylguanosine (m7G46) | METTL1 | 12q13 | 4.0E-81 | Mitochondria (75%) |
| Trm9 | (with Trm112) 5-methylcarbonylmethyluridine or 5-methylcarbonylmethyl-2-thiouridine (mcm5U34, mcm5s2U34) | ABH8 | 11q22.3 | 1.0E-44 | Cytoplasm (GFP tagged) |
| KIAA1456 | 8p22 | 3.0E-35 | Nuclear (58%) | ||
| Trm10 | 1-methylguanosine (m1G9) | RG9MTD2 | 4q23 | 1.0E-31 | Nuclear (94%) |
| RG9MTD1 | 3q12.3 | 8.0E-12 | Cytoplasm (51%) | ||
| RG9MTD3 | 9p13.2 | 3.0E-15 | Nucleus (51%) | ||
| Trm11 | (with Trm112) 2-methylguanosine (m2G10) | TRMT11 | 6q11.1-q22.33 | 2.0E-71 | Nuclear (53%) |
| Trm12 | wybutosine (yW14, tRNAPhe) | TRMT12 | 8q24.13 | 6.0E-15 | Cytoplasm (53%) |
| Trm13 | 2′-O-methyl(base), position 4 | CCDC76 | 1pter-q31.3 | 1.0E-38 | Cytoplasm (75%) |
| Trm44 | 2′-O-methyluridine (Um44) | METTL19 | 4p16.1 | 3.0E-29 | Cytoplasm (53%) |
| Trm61/Gcd14p | 1-methyladenosine (m1A58) | TRM61A | 14q32.32–33 | 8.0E-47 | Mitochondria (51%) |
| TRM61B (hypothetical) | 2p23.2 | 1.0E-6 | Cytoplasm (51%) | ||
| Trm82 | See Trm8 | WDR4 | 21q22.3 | 8.0E-17 | Nucleus (98%) |
| Trm112 | See Trm9, Trm11 | TRMT112 | 11q13.1 | 4.0E-12 | Cytoplasm (51%) |
| Trm140 | 3-methylcytosine (m3C32) | METTL2B | 7q32.1 | 4.0E-49 | Cytoplasmic (51%) |
| METTL2A | 17q23.2 | 7.0E-49 | Cytoplasmic (51%) | ||
| METTL8 | 2q31.1 | 3.0E-45 | Cytoplasmic (51%) | ||
| METTL6 | 3p25.1 | 2.0E-44 | Cytoplasmic (51%) |
The amino acid sequences of each yeast Trm were used in a BLAST search against known human protein sequences. In case of multiple qualifying candidates, the protein with the lowest E-value is considered the primary candidate, while the others are considered the secondary candidates. The qualifying E-value threshold for homolog candidacy was 0.01. Protein localization predictions were performed by ESLPred. The localizations of the Trm6, Trm8, Trm9, Trm61, Trm112, and Trm82 homolog candidates were individually predicted since the program cannot predict the localization of protein complexes.
Trm, tRNA methyltransferase; tRNA, transfer RNA.
Protein localization prediction
The expected localization of each candidate was predicted using the online programs ESLPred and HSLPred (Bhasin and Raghava, 2004; Bhasin et al., 2005). All available prediction approaches (Composition based, Dipeptide composition based, physiochemistry based, PSI-BLAST based, and Hybrid based) were utilized for each amino acid sequence. The Hybrid-based prediction results and percent confidence were recorded, along with the consensus dissenting result from the aforementioned predictions.
Oncomine survey of Trm expression in cancer
Each gene listed in the Human Trm Database was surveyed using the Oncomine online database (www.oncomine.org). Each Trm homolog was searched under Cancer versus Normal-mRNA using the standard parameters (gene expression rank within the top 10% of change, expression change of greater than or equal to twofold, and threshold limit of 1e−4). The cancer type matching the parameters along with the number of analyses displaying the change and the total studies analyzed by Oncomine were recorded.
Trm alignments
The predicted methyltransferase domains of each of the potential human homologs were aligned to their S. cerevisiae Trm counterpart using the alignment program CLUSTALW2. Both the percent identity and percent highly conserved for each alignment were then recorded. The homologs to scTrm6, scTrm82, and scTrm112 were excluded from this analysis since scTrm6, scTrm82, and scTrm112 all lack methyltransferase activity.
Phosphorylation site predictions
The amino acid sequences of each S. cerevisiae and human Trm were analyzed for potential phosphorylation motifs using the online database Scansite (http://scansite.mit.edu/) at high stringency. The number of predicted sites and kinases potentially targeting these sites was recorded for each Trm. The recorded phosphorylation sites for each human Trm were also collected using the online database PhosphoSite Plus (Cell Signalling Technology, Danvers, MA; www.phosphosite.org). The listed sites were categorized by amino acid location and method of analysis used to confirm phosphorylation (site-specific [SS] or proteomic discovery mode MS).
Results
Potential tRNA methylation enzymes in humans
We designed a database cataloging each of the human Trm homologs that met our search criteria of a BLAST E-value <0.01 and the presence of a discernable methyltransferase motif. All yeast Trms generated at least one candidate (Table 1). A total of 34 proteins met our search criteria and 3 more proteins were just above the threshold (0.39–0.074). HTRMT61B had the highest E-value of all candidates with a score of 1*E−6, and hNSUN2 had the lowest E-value of all the candidates with a score of 3*E−122. While 9 of the 18 yeast Trms had only one homolog, the remaining 9 have multiple homolog candidates. With six, scTrm4 has the most homolog candidates of all the yeast Trms.
There appears to be no discernible pattern of chromosomal distribution among the human Trm candidates. All of the human chromosomes except for chromosomes Y, 13, 15, 16, and 18 have at least one Trm candidate gene. Chromosome 1 has the most candidate genes with four. We were also able to compile data on known or predicted transcript variants using the NCBI Gene entry of each candidate. Of the 34 candidates identified, 11 of the candidates have multiple transcript variants. Three initial homolog candidates (scTrm7 homolog hCG17134 and scTrm9 homolog hCG2042988 and scTrm112 homolog “similar to CG12975”) were eliminated from the list at this stage. Two of the these, hCG17134 and hCG2042988, appear to be transcript variants or mutations based on their near-complete identity to other homologs (hCG17134 with hFTSJ2 and hCG2042988 with hKIAA1456) and the vague chromosomal locations listed for their NCBI Gene entries. The third, “similar to CG12975” was an initial hit that has since been removed from the NCBI database after genome annotation processing.
After we compiled the amino acid sequence data for each homolog candidate, we used the database as a tool for initial functional analysis of each human Trm homolog. We focused on possible cell localizations, on potential links between homolog expression changes and human cancers, as well as on similarities between the methyltransferase domains of the homologs and their yeast counterparts. To make our predictions, we used several different online programs. To predict cellular localizations, we used the programs ESLPred and HSLPred. Analysis of each yeast or human protein for phosphorylation sites was determined by Scansite. For our initial analysis of homolog expression changes in cancer cells, we used the Oncomine online database (Table 2). For our comparisons of methyltransferase domain alignments, we used the alignment program CLUSTALW2. We then compared the percent similarity of the unknown candidates to the average similarity of the characterized homologs. All of the homologs display significant conservation between human Trms and yeast Trms, with conservation levels typically reaching 60% and, in the case of scTrm8/hMETTL1, as high as 82%. These high levels of conservation indicate that the methyltransferase domains of all of the homolog candidates are likely to be functional.
Table 2.
A Survey of mRNA Expression Change of Human TRMSin Cancer Cells
| |
Cancer type (significant Δ expression/total studies, up or down) |
||||
|---|---|---|---|---|---|
| Homolog | Breast | Colorectal | Bladder | Ovarian | Other |
| TRM1 | 0 | 2/12 up | 1/5 up | 1/9 up | 1/6 up |
| TRM1-like | 2/19 down | 0 | 0 | 0 | 4/33 up, 3/22 down |
| TRMT2A | 0 | 0 | 1/5 up | 0 | 2/25 up |
| TRMT2B | 0 | 1/10 up | 0 | 0 | 1/13 up |
| TARBP1 | 0 | 1/10 up | 0 | 1/11 up | 4/38 up |
| NSUN1 | 0 | 0 | 0 | 1/10 up | 8/63 up, 1/15 down |
| NSUN2 | 0 | 1/11 up | 0 | 0 | 3/13 up, 1/4 down |
| NSUN3 | 0 | 0 | 0 | 0 | 1/11 up |
| NSUN4 | 0 | 0 | 0 | 0 | 1/9 down |
| NSUN5 | 0 | 0 | 1/5 up | 1/5 up | 4/52 up |
| NSUN6 | 0 | 0 | 0 | 0 | 1/13 down |
| TRMT5 | 0 | 0 | 0 | 0 | 5/35 up, 3/16 down |
| TRMT6 | 0 | 2/11 up | 0 | 0 | 6/15 up, 1/14 down |
| FTSJ1 | 0 | 2/11 up | 0 | 0 | 6/50 up |
| FTSJ2 | 1/7 down | 0 | 1/5 up | 0 | 4/24 up, 2/9 down |
| FTSJ3 | 0 | 0 | 0 | 0 | 1/8 up |
| METTL1 | 0 | 3/11 up | 1/7 up | 0 | 3/39 up |
| ABH8 | No hits | ||||
| KIAA1456 | 1/11 down | 1/9 down | 0 | 0 | 1/11 up 1/16 down |
| RG9MTD1 | 0 | 1/8 up | 0 | 0 | 1/6 up |
| RG9MTD2 | No hits | ||||
| RG9MTD3 | No hits | ||||
| TRMT11 | 1/15 down | 0 | 0 | 0 | 0 |
| TRMT12 | 0 | 0 | 0 | 0 | 2/7 up, 1/13 down |
| CCDC76 | 0 | 0 | 0 | 0 | 2/25 up, 5/19 down |
| METTL19 | No hits | ||||
| TRMT61A | 0 | 0 | 0 | 0 | 1/6 down |
| TRMT61B | 0 | 0 | 0 | 0 | 1/9 up |
| WDR4 | 0 | 0 | 0 | 0 | 1/8 up |
| TRMT112 | 0 | 0 | 1/5 up | 0 | 1/9 up |
| METTL2B | 0 | 1/12 up | 0 | 0 | 1/11 up |
| METTL2A | 1/6 up | 0 | 0 | 2/5 up | 4/31 up |
| METTL8 | 1/11 up | 2/11 up | 0 | 0 | 4/25 up, 2/25 down |
| METTL6 | 0 | 0 | 0 | 0 | 1/8 up |
Each human Trm homolog gene was searched in Oncomine for significant change in mRNA expression. Trm homologs without clear gene assignments were not used in this analysis. Over 10 different cancer types displayed an expression change for at least one Trm.
Predicted cellular localizations and potential phosphorylation sites of Trm homologs
Using the localization prediction programs HSLPred and ESLPred, we predicted the localizations of each yeast Trm and human homolog candidate based on its amino acid sequence (Table 1). The predictions are limited to four categories (nucleus, cytoplasm, mitochondria, and extracellular/plasma membrane) and use prediction methods based on peptide and dipeptide composition, PSIBLAST searching, and biochemical properties along with a hybrid conclusion combining the results of all the methods. ESLPred and HSLPred also offer a percent confidence of the hybrid prediction ranging from 53% to 98% confidence. Since this program is not designed to make predictions about protein complexes, we excluded the Trms that primarily form complexes in vivo. We made an exception for scTrm9 and its homologs because scTrm9 appears capable of existing independently in a cellular context. For the human protein predictions, we primarily used HSLPred, but we also included the biochemical predictor data in ESLPred. According to ESLPred, 9 of the 11 yeast Trms analyzed are nuclear proteins, with the only exceptions being scTrm5 (mitochondrial) and scTrm9 (cytoplasmic). As expected, none of the yeast Trms was predicted to be extracellular. According to HSLPred, 18 of the homolog candidates are nuclear proteins, while 11 are cytoplasmic and three are mitochondrial. As expected, none of the homologs were predicted to be extracellular or plasma membrane proteins. However, only seven of the predictions scored above 75% confidence. This suggests that most of the homologs do not have an obvious localization indicator and could exist in multiple locations.
Using the phosphorylation site prediction tool Scansite, we categorized the phosphorylation sites identified with a high stringency search for each Trm (Table 3). Fifteen of the 18 S. cerevisiae Trms contain at least one predicted phosphorylation motif, while 31 of the 34 human sequences analyzed have at least one predicted phosphorylation motif. No obvious motifs appear to be conserved between yeast and human. Only one human Trm—hNSUN5—of the 34 homolog candidates displayed a changed motif prediction due to a stable transcript variant. HNSUN5 isoform 3 has an additional predicted 14-3-3 Mode 1 phosphorylation site (T441) that isoforms 1, 2, and 4 lacks. Otherwise, transcript variation does not appear to have an impact on phosphorylation motifs in the human Trms analyzed here.
Table 3.
Predicted and Detected Transfer RNA Methyltransferase Phosphorylation Sites
| Yeast Trm | Predicted-kinase/phosphorylation site | Human homolog candidate | Scansite-predicted phosphorylation site/kinase | Reported phosphorylation sites (MS, SS) |
|---|---|---|---|---|
| Trm1 | PKC delta (T376), PKC episilon (S359), Erk D-domain (L296, L422) | TRM1 | (T603/574)14-3-3 Mode 1, (T613/584) PKC delta | 8 (8 MS) |
| C1ORF25 (Trm1-like) | (Y635/479) Grb2 | 3 (3 MS) | ||
| Trm2 | PDGFR Kin/PLCg N-Term SH2 (Y419), Protein Kinase A(T269), PKC epsilon(S8), CDK5 Kinase (S233), Erk D-domain (L320, V322), PDK1 binding (E458, E13) | TRMT2A | DNA PK (S378), Erk1 Kinase (S602), Erk D-domain (L462, V141, V571) | 2 (2 MS) |
| TRMT2B1 | PKC zeta (S12), ATM Kinase (S251) | 0 | ||
| TRMT2B2 | PKC zeta (S12), ATM Kinase (S206) | 0 | ||
| TRMT2B3 | PKC zeta (S12), ATM Kinase (S251) | 0 | ||
| Trm3 | Lck Kinase (Y952), Fgr SH2 (Y462, Y952), Erk1 Binding (P1381), | TARBP1 | PLCg N-term. SH2 (Y457), Protein Kinase A (T738), AMP Kinase (S787), ATM Kinase (S1397), PDK1 Binding (S1234), Erk D domain (I665, V168) | 2 (2 MS) |
| Trm4 | PDK1 Binding (D578), Erk D domain (L63, L80) | NSUN2 | PKC alpha/beta/gamma (S685), PKC mu (S432), PKC zeta (S656), Erk D dom. (I236) | 12 (11 MS, 1 SS) |
| NSUN1 | 14-3-3 Mode 1 (S264), PKC alpha/beta/gamma (S562, T739), Casein Kin. 2 (T185, S150, S120, S111, T140), Casein kin. 1 (T140), PDK1 Binding (S588) | 18 (18 MS) | ||
| NSUN3 | (S81) PKC mu | 2 (2 MS) | ||
| NSUN4 | Cbl-Assoc. Protein C-SH3 (P144), GSK3 Kinase (S115), Erk D-dom. (L284) | 3 (3 MS) | ||
| NSUN5 | Isoform 1, 2, 4: Cbl-Assoc. Pro. C-SH3/p85SH3 mode 2 (P391). Isoform 3: 14-3-3 Mode 1 (T441), Cbl-Assoc. Pro. C-SH3/p85SH3 mode 2 (P391) | 8 (8 MS) | ||
| NSUN6 | Erk D-dom. (V359) | 1 (1 MS) | ||
| Trm5 | None | TRMT5 | 14-3-3 Mode 1 (T59), Src Kin. (Y177), Calmodulin dep. Kin. 2 (T494), Akt Kin. (T59), AMP Kin. (T494), ATM Kin. (T176), Erk D-dom. (I441) | 1 (1 MS) |
| Trm6/Gcd10p | p85 SH2 (Y237), Calmodulin DK 2 (S151), AMP Kinase (T125), Erk D domain (L115) | TRM6 | DNA PK (T107), Erk1 Binding (P247), Erk D-dom (I421), PIP3-binding PH (F47) | 5 (5 MS) |
| Trm7 | PKC mu (S222), Erk D domain (L84) | FTSJ1 | Abl SH2/Itk SH2/Nck SH2 (Y262), PKC alpha/beta/gamma (T279) | 3 (3 MS) |
| FTSJ2 | Erk D-dom. (L175) | 0 | ||
| FTSJ3 | 14-3-3 Mode 1 (S28), PKC epsilon(S534), Casein Kin. 1 (T351), Casein Kin 2 (S624, T622), PDK1 Binding (T693) | 17 (17 MS) | ||
| Trm8 | Akt Kinase (S59), Erk1 Binding (P183) | METTL1 | Erk1 Binding (P162), Erk D-dom. (L264). Act. Phos. Site at S27 (PKB alpha) | 1 (SS and MS) |
| Trm9 | None | ABH8 | Lck SH2/Fyn SH2/Src SH2 (Y379), Nck 2nd SH3 (P358), PKC alpha/beta/gamma (T332), PKC zeta (T28), DNA PK (S589) | 0 |
| KIAA1456 | 14-3-3 Mode 1/Akt Kinase (S214), Fgr Kinase/Lck Kinase/Insulin Receptor Kinase/Src Kinase (Y237), Cbl Associated proline C-SH3 (P286) | 0 | ||
| Trm10 | Cortactin SH3/Grb2 SH3 (P20) | RG9MTD2 | Casein Kinase (S306, S307) | 0 |
| RG9MTD1 | PKC epsilon (S73), Erk D-domain (L385) | 3 (3 MS) | ||
| RG9MTD3 | PKC epsilon (T243) | 0 | ||
| Trm11 | PKC epsilon (T101, T190), PKC delta (T190), PKC zeta (S100, T101), PDK1 Binding (D37) | TRMT11 | Shc SH2 (Y87), DNA PK (T120, S40), ATM Kinase (T120, S40) | 1 (1 MS) |
| Trm12 | Cbl-Assoc. protein C-SH3 (P169), pkc EPSILON (T300), ATM kin./DNA PK (S228), PDK1 Binding (S292, S257), Erk D domain (L362) | TRMT12 | None | 0 |
| Trm13 | Grb2 SH2 (Y357) | CCDC76 | None | 0 |
| Trm44 | Protein Kinase A (S351), Erk D domain (I198, I354) | METTL19 | 14-3-3 Mode 1 (T507) | 3 (3 MS) |
| Trm61/Gcd14p | PDK1 Binding (E185) | TRM61A | 14-3-3 Mode 1 (S244), Itk Kinase (Y67) | 4 (4 MS) |
| TRM61B (hypothetical) | PKC mu (T227), DNA PK (S421) | 0 | ||
| Trm82 | DNA PK/ATM Kin. (T187), Casein Kin. 1 (T163), PDK1 Binding (D279) | WDR4 | Protein Kinase A (S391), PKC Delta (S74), Casein Kinase 1 (S34) | 2 (2 MS) |
| Trm112 | None | TRMT112 | Erk D-Domain (V30) | 3 (3 MS) |
| Trm140 | Cdk2/PKC epsilon (S12), Casein kinase 2 (S162, S178), Casein kinase 1 (T260, S331) | METTL2B | ATM kinase (T326) | 2 (2 MS) |
| METTL2A | ATM kinase (T326) | 1 (1 MS) | ||
| METTL8 | None | 1 (1 MS) | ||
| METTL6 | PKC epsilon (T27), PKC zeta (S184), Fgr SH2, Fyn SH2 (Y236) | 0 |
The amino acid sequences of each Trm were analyzed for potential phosphorylation sites using the online prediction programs Scansite and PhosphoSite Plus. All ScanSite predictions shown were performed at high stringency. PhosphoSite Plus does not catalog S. cerevisiae proteins. For brevity, PhosphoSite Plus results were recorded as total sites and how many were determined by site-specific (SS) methods or proteomic mass spectroscopy (MS). All known human Trm transcript variants were scanned separately; predicted sites with identical amino acid sequences and kinase targets but different amino acid numbers between transcript variants were considered identical.
A variety of kinases are represented among the sites identified, from the SH2 family of kinases to the PKC family to DNA damage response kinases, offering a variety of potential clues to Trm regulation. The most represented kinases sites among the human homologs are Erk D-domain/MAP kinase (11/36) and ATM kinase (7/36). Erk kinases regulate cell proliferation, homeostasis, differentiation, and cell fate. It has been suggested that RNA-binding proteins play a role in regulating Erk kinases; this may offer a novel connection between Trms and Erk kinase regulation worthy of further investigation (Whelan et al., 2011). ATM kinase is one of the central figures in the cellular DNA damage response. At least two yeast Trms (scTrm2 and scTrm9) have been linked to DNA damage repair, so it stands to reason that human Trms may also play a role in DNA damage repair. One of the Trm2 homologs (hTRMT2B) has a high stringency ATM site, while one of the scTrm9 homologs (hABH8) has a medium stringency ATM site (data not shown).
We also used the protein modification database PhosphoSite Plus in our analysis of the human Trm homologs. Phosphosite Plus allowed us to record experimentally determined phosphorylation sites for each protein, as well as the method used to detect the site, whether by SS methods or proteomic mass spectroscopy. The results are listed in Table 3. Overall, 24 of the 34 human homologs have at least one reported phosphorylation site. The homolog with the most sites was scTrm4 homolog hNSUN1 with 18 sites, followed by scTrm7 homolog hFTSJ3 with 17 sites. The average number of phosphorylation sites rounds to three sites per homolog. The vast majority of reported phosphorylation sites were discovered in large-scale proteomic experiments; only hNSUN2 (S139) and hMETTL1 (S27) have reported phosphorylation sites determined with SS methods.
Most Trm homolog candidates display a significant expression change in cancer cells
Using the online database Oncomine, we searched for significant changes in mRNA expression by any of the homolog candidate (Oncomine™; Compendia Bioscience, Ann Arbor, MI; www.oncomine.org). Of the 30 homologs studied, 26 display more than a twofold increase or decrease in at least one cancer study (Table 2). A wide variety of cancers were represented in the matches: breast cancer, leukemia, ovarian cancer, colorectal cancer, lymphomas, and sarcomas. Although no homolog displayed a drastic change of expression in all collected studies of a cancer type, the wide variety of positive hits suggests that the human Trm homologs are important for general cellular function. These results offer a hint of a connection between tRNA methylation and cancer; however, there are studies that suggest a more direct connection between tRNA modification and cancer. For example, the Trm homolog hTRMT2A was originally identified as a biomarker for breast cancer. This will be discussed in detail in the Trm2 section. A 2009 study by Schaefer et al. found that the drug azacytadine inhibited the tRNA methylation activity of DNMT2. This inhibition decreased the metabolic activity of human cancer cell lines (Schaefer et al., 2009). For the remainder of this review, we shall focus on the individual yeast Trms and their human homolog candidates.
scTrm1
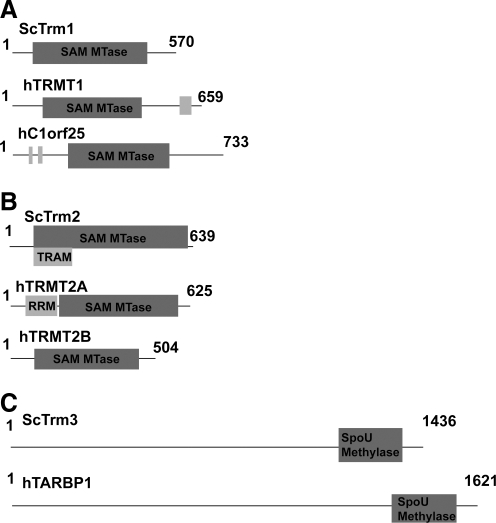
Current research has demonstrated that scTrm1 mediates both methylation events needed to generate m22G26 in 18 tRNA residues. The TRM1 gene was discovered in 1986 (Ellis et al., 1986). Ellis' study determined that in vitro scTrm1 was responsible for both methylations of guanine to N2,N2′-dimetguanine (m22G) at position 26 in tRNA. An alternate extended 5′ sequence was later found to direct scTrm1 to the mitochondria (Ellis et al., 1989). Beyond m22G significance as a modification found for both mitochondria and cellular tRNA, the biological purpose of the modification remains unclear.
The primary human homolog (reported and characterized in 2000) is named hTRM1 (Liu and Straby, 2000). The predicted methyltransferase domain of scTrm1 shares a 39.2% identity with hTRM1 and 61.9% of the amino acids are highly conserved (Fig. 2a). HTRM1 has three predicted transcript variants generating two isoforms according to its NCBI Gene entry. HTRM1 was discovered in a BLAST search with the recombinant protein methyltransferase activity confirmed in vitro using bacterial tRNA and mutant yeast tRNA, both lacking m22G26. Recombinant hTRM1 activity was confirmed in vivo in Escherichia coli cells expressing human tRNATyr, demonstrating that hTRM1 is responsible for both m2G26 and m22G26 tRNA methylations.
FIG. 2.
Line diagrams of scTrm1 (A), scTrm2 (B), scTrm3 (C), and their human homolog candidates. The known or predicted methyltransferase domains are in dark gray. Any known or predicted RNA-binding domains are in light gray. Domain assignments are from the protein entries in the NCBI database.
Found in 2001 during a genomic search for the Heriditary Prostate Cancer 1 locus, the secondary homolog candidate for scTrm1 in humans is hC1ORF25 (Sood et al., 2001). Because hC1ORF25 displays a 20% amino acid similarity to scTrm1, the gene was later designated “Trm1-like” (Fig. 2a). The predicted methyltransferase domain of scTrm1 shares a 23.2% identity with hTrm1-like and 44.9% of the amino acids are highly conserved. The protein characteristics of hC1ORF25 were not analyzed in the report by Sood et al., nor have they been studied since, providing no confirmation of methyltransferase activity. The mouse homolog of hTrm1-like, however, has been studied (Vauti et al., 2007). Mouse Trm1-like is expressed in a variety of tissues ranging from embryonic to adult, including numerous parts of the brain, spinal cord, esophagus, heart, and lung. Vauti's study also reported a Trm1-like knockout mouse. The knockout mice displayed a slight motor dysfunction phenotype; otherwise, the mice were viable and healthy.
scTrm2
ScTrm2 converts uridine to 5-methyl uridine (m5U) at position 54 of 42 tRNA residues in yeast (Dunin-Horkawicz et al., 2006). The trm2 mutation was first identified in S. cerevisiae in 1982; however, the gene itself was discovered in 1992 and initially named Rho-associated NuClease 1 (RNC1) (Chow et al., 1992). A later study corrected the RNC1 sequence and renamed it NUD1 (Van Vliet-Reedijk and Planta, 1993). NUD1 was later shown to play a role in DNA damage after overexpression in S. cerevisiae increased the cell survival time following exposure to gamma radiation (Sadekova and Chow, 1996). More specifically, scNud1 was connected to double strand break (DSB) repair via the homologous recombination pathway (Asefa et al., 1998). When expressed in HeLa cells, scNud1 seemed to stimulate the DSB pathway and interfere with other DNA repair pathways (Semionov et al., 1999).
In 2000, Nordlund et al. reported that the NUD1/RNC1 gene actually encoded scTrm2 and was not the exo/endonuclease protein previously described. The nuclease activity was likely from scRad52, reported in the earlier studies as a potential cofactor to Rnc1/Nud1. Nordlund's study also confirmed m5U methyltransferase activity in vitro and in vivo (Nordlund et al., 2000). It was later found that inactive scTrm2 is sufficient to stabilize mutant tRNASer(CGA), suggesting a second role in tRNA maturation (Johansson and Bystrom, 2002).
There are two human homolog candidates for scTrm2 arising from two separate genes: hTRMT2A and hTRMT2B. HTRMT2A is best known as a component of the breast cancer diagnostic tool Mammostrat. Mammostrat-positive patients are much more likely to have a cancer recurrence after Tamoxifen treatment, helping doctors formulate the optimal strategy for treatment (Bartlett et al., 2010). HTRMT2A was not identified as a Trm homolog until 2010. A breast cancer study linked the protein HTF9C to the risk of recurrence in HER2+ breast cancer patients (Hicks et al., 2010). Further analysis of the protein structure compelled the Hicks group to rename the protein hTRMT2A. In Hicks' study, immunohistochemistry analysis using a panel of antibodies linked cytosolic hTRMT2A staining to a likelihood of recurrence in HER2+ patients.
HTRMT2A has a projected RRM domain presumed responsible for RNA binding, followed by the projected SAM methyltransferase domain covering most of the protein (Fig. 2b). The predicted methyltransferase domain of hTRMT2A shares a 36% identity with scTrm2 and 61.3% of the amino acids are highly conserved. HTRMT2A has two transcript variants, both expressing the same gene product.
HTRMT2B remains unstudied beyond its confirmation as a human gene product. HTRMT2B has four predicted transcript variants producing three different predicted isoforms. In contrast to hTRMT2A, hTRMT2B does not have an apparent RNA-binding domain (Fig. 2b). The predicted methyltransferase domain of hTRMT2B shares a 28% identity with scTrm2 and 57.3% of the amino acids are highly conserved. Based on sequence similarity, particularly the presence of a known RNA binding domain, hTRMT2A is a human Trm homolog with the possibility of hTRMT2B acting as a different RNA methylase or is specific to a given tRNA species.
scTrm3
ScTrm3 converts guanosine at tRNA position 18 to 2-O′-ribomethylguanosine (Gm18) for 12 different tRNAs. ScTrm3 was originally identified as one of five eukaryotic homologs to the E. coli RNA methylase spoU (Cavaille et al., 1999). Deletion of the TRM3 gene caused a significant loss of G18 methylation, marking the gene as the third Trm found in S. cerevisiae. The deletion strain was viable under a variety of temperatures and media: 37°C, 30°C, and 19°C in rich, glucose-depleted, minimum, and nonfermentable media (Cavaille et al., 1999; Reynaud et al., 2001). The specific methylation activity was confirmed both in vitro—by analysis of recombinant scTrm3 protein in E. coli—and in vivo—by reintroduction of the TRM3 gene in the deletion strain.
The primary human homolog candidate TAR Binding Protein 1 (hTARBP1) was also identified by Caville et al. as a homolog candidate. HTARBP1 was originally described as TAR RNA-Binding Protein 185 (hTRP-185) in 1991. HTARBP1 is a 185 kDa protein that binds to the TAR regulatory element of HIV1, a stem-loop structure in the HIV long terminal repeat region required for gene activation by the protein tat (Wu et al., 1991). HTARBP1 was reported to displace RNA polymerase II from the TAR domain. HTARBP1 binding requires cofactors that were subsequently identified as hEF-1alpha, hPTB, and hSRB by Wu-Baer et al. (1996). The hTARBP1 methyltransferase domain was first identified in 2008 as a member of the SpoU-TrmD (SPOUT) methylase family (Wu et al., 2008). To date, the potential methyltransferase activity of hTARBP1 has not been tested.
The key similarities shared between scTrm3 and hTARBP1 are their large N-terminal regions and their SpoU methylase domains at their C-termini (Fig. 2c). SPOUT family methyltransferases are differentiated from Class 1 methyltransferases by their uncommon alpha/beta fold creating a topological knot (Kurowski et al., 2003; Tkaczuk et al., 2007). The hTARBP1 SPOUT domain shares 12% identity with the scTrm3 SPOUT domain and 34.7% of the amino acids are highly conserved. Although confirmation is required, it seems likely that hTARBP1 performs the Gm18 tRNA methylation.
scTrm4
ScTrm4 catalyzes the m5C methylation reaction at four different positions on tRNA: the major sites are at either 48 or 49 and the minor sites are 34 and 40. ScTrm4 was originally identified as scNCL1 (Nuclear protein 1). The protein was localized to the nucleolus and found to be nonessential (Wu et al., 1998). In 1999, the NCL1 gene was determined to be a Trm candidate in a sequence homology search against the E. coli rRNA methyltransferase SUN (Motorin and Grosjean, 1999; Tscherne et al., 1999). The Trm activity was confirmed both in vitro using scTrm4 recombinant protein and in vivo by gene replacement. Deletion of TRM4 alone has no effect on yeast growth; however, trm4 mutants have been found to be sensitive to MMS and H2O2 (Begley et al., 2004) (Chan et al., 2010). In addition, a trm4Δtrm8Δ double deletion creates a severe temperature-sensitive phenotype due to a drastic reduction in tRNAVal(AAC) levels (Alexandrov et al., 2006; Chernyakov et al., 2008). tRNA lacking both of these methylation activities degrades rapidly at nonpermissive growth temperatures.
Our search identified six proteins meeting our homology criteria for scTrm4, named hNSUN1-6. A similar previously performed search predicted hNSUN1-7 as potential m5C methyltransferases (Bujnicki et al., 2004). HNSUN2 is the closest match to scTrm4. HNSUN2 methyltransferase activity on tRNALeu(CAA) has been tested both in vitro and in vivo by transformation of this human gene into the trm4Δ yeast strain (Brzezicha et al., 2006). Like scTrm4, hNSUN2 requires the tRNA introns to be present for methylation (Strobel and Abelson, 1986; Brzezicha et al., 2006). Unlike scTrm4, hNSUN2 activity is specific to C34 in tRNALeu(CAA), with no evidence of C48 or C49 methylation, while C40 was not tested in the study. HNSUN2 has been identified as a nucleolar protein until anaphase (Frye and Watt, 2006; Sakita-Suto et al., 2007). HNSUN2 has been found in both immortalized cell lines and cancer lines (Frye et al., 2010). In particular, 32% of breast cancer lines studied in the report displayed an increased hNSUN2 copy number, leading to increased hNSUN2 mRNA and protein expression. The predicted methyltransferase domain of hNSUN2 shares a 38.9% identity with scTrm4 and 60.7% of the amino acids are highly conserved.
The other NSUN proteins are not well chronicled. HNSUN1 (aka hNOP2) depletion has been shown to reduce 27S pre-rRNA methylation (Hong et al., 1997). A site-directed mutagenesis study performed on hNSUN1 revealed that mutation of cysteine C424 caused a loss of hNSUN1 methylation activity (King et al., 1999). Published work on hNSUN3 is currently limited to a 2009 study on celiac disease, a genetic disease of the small intestine resulting in an immune overreaction to the wheat protein gliadin; however, this work did not establish a firm link to the disease (Trynka et al., 2009). The only published report on hNSUN4 is a mouse study in which the sperm head and tail associated protein (SHTAP) gene was found to be embedded within the hNSUN4 gene (Jamsai et al., 2010). There appears to be, however, no functional relationship between the two proteins. HNSUN5 consists of four known isoforms, differentiated by the lengths of their C-termini. It was first identified in a study of Williams-Beuren Syndrome, where deletion of the chromosome 7 long arm (where hNSUN5 is located) causes the neurodegenerative disorder. HNSUN6 has not been described in detail in any published work. The predicted methyltransferase domains of the hNSUN proteins share between 13.3% (hNSUN5) and 35.7% (hNSUN1) identity with scTrm4. Of the hNSUN proteins analyzed, hNSUN5 had the lowest amount of highly conserved amino acids (34.1%), while hNSUN1 had the highest amount of highly conserved amino acids 65.9% (hNSUN1) (Fig. 3).
FIG. 3.
Line diagrams of scTrm4 and the hNSUN family. The DNA Topoisomerase II and PUA domains are not believed to be involved with tRNA methylation. PUA, pseudouridine synthase and archeosine transglycolase domain.
Considering the sequence specificity of hNSUN2 methyltransferase activity and the phylogenic predictions previously made about the other members of the NSUN family, hNSUN3, hNSUN4, and hNSUN5 could be responsible for one tRNA m5C methylation each. However, it is also possible that at least one of the less conserved proteins (like hNSUN5) methylates a different RNA type entirely. Without rigorous methylase assay analysis and gene complementation analysis with the S. cerevisiae trm4Δ strain, it is impossible to predict which NSUN protein is responsible for which methylation event.
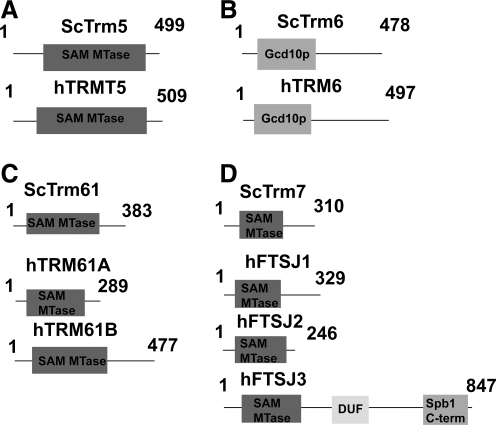
scTrm5
ScTrm5 is not only responsible for the m1G and m1I reaction at nucleotide 37 in several tRNAs but also catalyzes the first methylation toward wybutosine (yW) formation in tRNAPhe(GmAA). The trm5 mutant in S. cerevisiae was first discovered in 2001 by Bjork et al. and the TRM5 gene has been found in all kingdoms of life. Deletion of scTrm5 in yeast leads to a drastic decrease in growth, though the protein is not essential for cell survival (Bjork et al., 2001). Further analysis showed that scTrm5 is required for proper in-frame translation. Specifically, scTrm5 methylation at nucleotide 37 prevents +1 frameshifting but not −1 frameshifting (Urbonavicius et al., 2001, 2003).
The human homolog to scTrm5, named hTRM5, was first reported in 2004. HTRM5 was the third human tRNA modification enzyme identified (Brule et al., 2004). The predicted methyltransferase domain of hTRM5 shares a 47.6% identity with scTrm5 and 68.6% of the amino acids are highly conserved (Fig. 4a). Curiously, the original cDNA cloned in the study did not contain a start methionine. An additional exon was found by the group, but the possibility of alternative splicing exists. HTRM5 was found via a BLAST search and the putative protein was produced in E. coli. HTRM5 methylase activity was confirmed in vitro to produce m1G37 or m1I for several tRNAs, catalyzing the m1G37 reaction most efficiently for the class II tRNA1Leu. However, recombinant hTRM5 cannot methylate mitochondrial m1G37 in vitro. In addition, hTRM5 appears to be reliant on proper tRNA tertiary structure for optimal enzymatic activity. Specifically, altering the D-stem or D-loop structure of the tRNA substrate can cause a reduction in enzymatic activity. The biological role of hTRM5 remains to be determined.
FIG. 4.
Line diagrams of scTrm5 (A), scTrm6/61 (B, C), scTrm7 (D), and their human homolog candidates. ScTrm6 and scTrm61 are believed to form a heterotetramer. DUF, domain of unknown function.
scTrm6/61
ScTrm6 works in tandem with scTrm61 to methylate adenosine 58 (m1A58) on the TψC loop of initiator tRNAMet (tRNAiMet) as well as catalyzing m1A58 methylation on 16 other tRNAs. The genes now known as TRM6 and TRM61 were originally named GCD10 and GCD14, respectively. Gcd proteins regulate scGcn4, which, in turn, positively regulates a number of genes in S. cerevisiae responsible for amino acid biosynthesis. The gcd10 and gcd14 mutants were reported as two of a series of mutant strains capable of repressing scGcn4 activity, leading to the disruption of amino acid synthetic regulation (Harashima and Hinnebusch, 1986; Cuesta et al., 1998). A methyltransferase domain was found in scGcd14 and it was determined that gcd10 and gcd14 mutants cause slow growth caused by a defect in translation initiation (Calvo et al., 1999).
Anderson et al. (1998) found that scGcd10 played a role in tRNAiMet maturation but not in elongator methionine tRNA (tRNAeMet) maturation. Their study also revealed that tRNAiMet overexpression can mitigate the GCD10 deletion lethality at low temperatures. Confirmation by pulse chase analysis with tritiated uracil indicated that scGcd10 is required for tRNAiMet stability in vivo (Anderson et al., 1998). Using co-immunoprecipitation, this study also provided the first evidence that scGcd10 and scGcd14 associated with each other as a heterotetromer in vivo. Testing of scGcd14 for methyltransferase activity revealed that, indeed, scGcd14 was capable of methylating both tRNAiMet and tRNAeMet in vitro. Later work by Anderson et al. (2000) determined that both purified scGcd10 and scGcd14 are required to restore m1A58 to tRNA from gcd10Δ strains in vivo. Mutational analysis determined that scGcd10 is necessary for RNA binding, while scGcd14 is required for methyltransferase activity. Most recently, a search for deleted genes that produce a phenotype in combination with the temperature-sensitive trm6-504 mutant was performed (Ozanick et al., 2009). This search revealed that deletion of REX1, a gene encoding the exonuclease scRex1, in the Trm6-504 mutant strain caused a slow-growth phenotype due to reduced tRNAiMet 3′ end processing.
Our search for the scTrm6 and scTrm61 homologs identified hTRM6 and hTRM61, supporting findings previously made by Ozanick et al. in 2005 (Ozanick et al., 2005). Transformation of both hTrm6 and hTrm61 into Trm6-504 and Trm61-2 mutant yeast strains, respectively, were required for partial complementation of each strain's temperature-sensitive phenotype. Transformation with hTRM6 alone did not complement the growth defect in trm6 mutant yeast, implying that hTRM6 cannot interact with scTRM61 efficiently. In contrast, hTRM61 alone displayed a limited ability to complement the TRM61 mutant. The study also determined that hTrm6 and hTrm61 are both required for formation of m1A58 in vitro and in vivo. In 2010, Saikia et al. used microarray analysis to study the extent of m1A58 modification throughout five human cell lines (Saikia et al., 2010). Across the five cell lines studied, nearly a quarter of all tRNA species displayed m1A58 hypomodification. Many of the hypomodifications were common among all five cell lines, but initiator-tRNAMet maintained its m1A58 modification in all cell lines.
Our search revealed a homolog candidate related to hTrm61, called hTrmt61B, which has yet to be studied in detail. Htrmt61B (477 amino acids) is larger than hTrm61 (289 amino acids); sequence alignment of the two proteins indicates that the size difference is due to hTrmt61B having an extended 5′ region. The highest areas of similarity between the two proteins are between amino acids 49–152 in hTrm61 and amino acids 192–295 of hTrmt61B. Both proteins have a GXGXG sequence in this area, strongly indicating that this region is a common methyltransferase domain.
The predicted methyltransferase domain of hTrm61 shares a 42.6% identity with scTrm61 and 64.0% of the amino acids are highly conserved. In comparison, hTrmt61B shares a 17.3% identity with scTrm61 as well as having 44.9% and 45.2% highly conserved amino acids, respectively. hTrmt61B contains the GXGXG domain, making it a strong homolog candidate despite its increased size.
scTrm7
ScTrm7 is responsible for 2′-O ribose methylation at positions 32 and 34 for tRNALeu, Phe, Trp. ScTrm7 was originally found during a search for other 2′-O methyltransferases in budding yeast, based on the sequence of E. coli 2′-O methyltransferase FtsJ (Pintard et al., 2002). FtsJ was discovered in 1991 and methylates both Um2552 of 23S rRNA and several tRNA nucleotides at the 2′-O ribose sugar. Notably, deletion of ftsJ in E. coli caused severe growth defects (Ogura et al., 1991; Bugl et al., 2000; Caldas et al., 2000). Three S. cerevisiae homologs to FtsJ were found with each one methylating a different RNA species as the result of an evolutionary split: scTRM7 (tRNA), scMRM2 (21S rRNA), and scSPB1 (25S rRNA). Deletion of the TRM7 gene in yeast impairs protein production and increases sensitivity to the drug paramomycin (Pintard et al., 2002). ScTrm7 methyltransferase activity was confirmed both in vitro and in vivo at the ribose sugars at nucleotide 32 and 34. The analysis clearly implicated tRNAPhe and tRNATrp as scTrm7 substrates, while tRNALeu analysis was unclear. Pintard et al. also showed that this methyltransferase activity occurs after intron removal.
The E. coli FtsJ sequence was also used in a search for human 2′-O ribomethylases. Originally, one homolog was found (JM23, later renamed hFTSJ1) (Bugl et al., 2000); subsequently, two additional potential matches were found (FJH1, renamed hFTSJ2 and SPB1, renamed hFTSJ3) (Ching et al., 2002) and cataloged as members of the RrmJ family (Feder et al., 2003). By the parameters of our search, hFTSJ1 is the closest human homolog to scTrm7, while hFTSJ2 and hFTSJ3 are secondary candidates. HFTSJ1 and hFTSJ2 are close in amino acid length to scTrm7 (329 and 246 amino acids, respectively), while hFTSJ3 is 847 amino acids long (Fig. 4d). The predicted methyltransferase domain of scTrm7 shares a 59% identity with hFTSJ1, 35.7% identity with hFTSJ2, and 44.4% identity with hFTSJ3. ScTrm7 shares 79.5% similarity with hFTSJ1, 58% identity with hFTSJ2, and 65.3% similarity with hFTSJ3.
Northern blot analysis revealed that hFTSJ1 is expressed in both fetal and adult brains (Freude et al., 2004). Freude et al. (2004) studied hFTSJ1 in a genetic analysis of three families containing multiple males diagnosed with nonsyndromic X-linked mental retardation (NSXLMR). In the study, the hFTSJ1 gene was located on the X chromosome. A series of mutations in hFTSJ1 was found throughout the family. Some of these mutations caused a major reduction in hFTSJ1 mRNA compared to normal levels. HFTSJ1 mutations were also found in other familial genetic studies linking mutated hFTSJ1 not only to NSXLMR, but also to impaired cognition (Gong et al., 2008; Takano et al., 2008).
The only published study referencing hFTSJ2 or hFTSJ3 was undertaken in 2002. HFTSJ2 was localized to the nucleous in HeLa cells and using Northern blot analysis, hFTSJ2 was strongly detected in muscle, placenta, and heart tissues along with a variety of cancer cell lines (Ching et al., 2002). The group concluded that hFTSJ2 was possibly an rRNA methylase. Evolutionary analysis of hFTSJ3 matched the gene with scSPB1, marking it as a potential snoRNA methylase. No work has been undertaken confirming the methyltransferase activity of any of these homolog candidates.
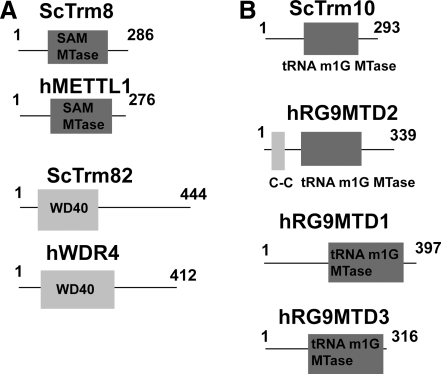
scTrm8/82
The scTrm8/82 complex is responsible for m7G46 methylation in 11 known tRNAs in S. cerevisiae. m7G46 is suspected to stabilize the tRNA tertiary structure. The scTrm8/82 proteins were isolated during an in vitro methyltransferase screen of a genomic set of purified glutathione-S-transferase (GST)-ORF fusion proteins (Alexandrov et al., 2002). Two of these ORF fusions, renamed scTrm8 and scTrm82, were able to generate m7G in vitro from tRNAPhe. Alexandrov et al. also tested trm8Δ and trm82Δ deletion strains for m7G methylation and found drastically reduced m7G levels in both strains in vivo and in vitro. Using affinity co-purification, the study also determined that scTrm8 and scTrm82 form an equimolar complex, and that this complex must form early in order to function properly as a methyltransferase unit. In a subsequent study, it was determined that while scTrm8 contains most of what is required for methyltransferase activity, scTrm82 is required to maintain the appropriate scTrm8 protein levels in the cell (Alexandrov et al., 2005).
The human homologs to scTrm8 and scTrm82 are hMETTL1 and hWDR4, respectively. As with their counterparts, hMETTL1 contains a putative methyltransferase domain, while hWDR4 only has four WD-40 repeats (Fig. 5a). The predicted methyltransferase domain of hMETTL1 shares a 66.4% identity with scTrm8 and 82.0% of the amino acids are highly conserved. HMETTL1 has two predicted isoforms from two transcript variants, while hWD4 has two transcript variants that only differ in their 3′ untranslated regions. HMETTL1 was originally suspected to be an RNA methylase of some sort (Bahr et al., 1999), while hWDR4 was originally found in a search for Down Syndrome genetic factors at chromosome 21 (Michaud et al., 2000). The WDR4 gene is located in the Down Syndrome chromosome deleted region (21q22.3), so WDR4 remains a potential candidate for Down Syndrome; nevertheless, further work is required to make any definite connection between the protein and the disease. Not only were these two Trm candidates found in a homology search by Alexandrov et al., but they were also tested for methyltransferase activity in vivo using the yeast deletion strains. Both hMETTL1 and hWDR4 were able to restore m7G46 in the tRNA of the deletion strains of their counterparts (Alexandrov et al., 2002). In 2005, Cartlidge et al. indicated that hMETTL1 is phosphorylated by protein kinase Bα (hPKBα) and hRSK at Serine 27 in HeLa cell extracts (Cartlidge et al., 2005). This phosphorylation event inactivates hMETTL1 but does not appear to affect hWDR4 binding to tRNA. While hRSK phosphorylation was considered to be an artifact by the researchers, hPKBα phosphorylation was mediated by insulin growth factor-1 (IGF1) in HEK-293 cells. In addition, site-directed mutants were made mimicking the phospho- (S-D/E) and dephospho- (S-A) forms of hMETTL1. While the S-A mutant was able to partially rescue trm8Δ cells, neither hMETTL1 S-D nor S-E was able to rescue this yeast Trm mutant. Cartlidge et al. speculated that this phosphorylation event helps to regulate protein levels since IGFs are commonly used to stimulate protein expression.
FIG. 5.
(A) Line diagrams of scTrm8, scTrm82, and their human homolog candidates. ScTrm8 and scTrm82 are believed to form a heterodimer. (B) Line diagrams of scTrm10 and its human homolog candidates.
scTrm9/scTrm112
ScTrm9 and scTrm112 form a complex responsible for the last methylation step needed to generate either mcm5U or mcm5s2U at position 34 of several tRNAs. In 2003, Kalhor and Clarke treated yeast cells with radiolabeled SAM to search for novel methylated cellular components. The Yml014w/f6 knockout strain displayed a significant loss of labeled methyl-RNA compared with the control, marking it as a candidate RNA methylase (Kalhor and Clarke, 2003). Further investigation revealed that scTrm9 was responsible for generating mcm5U and mcm5s2U at position 34 of tRNAGlu, Arg3 in vitro and in vivo. Kalhor and Clarke further determined that only scTrm9 was necessary for methylation. A trm9 deletion strain was generated (trm9Δ), and was found to have a temperature-sensitive growth defect in the presence of paramomycin (Kalhor and Clarke, 2003). Sctrm9 mutation or deletion was also found to confer resistance to zymocin, a K. lactis toxin that targets Trm9-modifcied tRNA (Lu et al., 2005; Jablonowski et al., 2006). A key factor for the survival of mutants resistant to zymocin was the loss of methylation at the wobble base of tRNAGlu. ScTrm9 has also been shown to play a role in cellular DNA damage response (Begley et al., 2007).
Initial findings suggested that scTrm9 acted alone; however, detailed analysis of scTrm9-based zymocin resistance showed otherwise. A study by Studte et al. (2008) found that zymocin resistance requires interaction between the C-terminus of scTrm9 and scTrm112. ScTrm9 was also found to compete with scMtq2 and scTrm11 for binding, further delineating the importance of scTrm9/112 interaction. For example, coexpression of scTrm9 and scTrm112 in E. coli generated more soluble scTrm9 (Mazauric et al., 2010). The scTrm9/Trm112 interaction had been previously reported in a large-scale protein study (Gavin et al., 2002), but gene deletion experiments were not attempted because trm112 deletion was considered lethal. When a trm112 deletion was made, it was shown that isolated tRNA lack mcm5U and mcm5s2. In addition, scTrm9 and scTrm112 coexpression in trm9Δ resulted in mcm5U (but not mcm5s2) generation (Mazauric et al., 2010). These results demonstrated that while scTrm9 is capable of tRNA methylation by itself in vitro, it works optimally in vivo with scTrm112.
Our search suggests that there are two potential human homolog candidates to scTrm9: Human AlkB homolog 8 (hALKBH8 or hABH8) and hKIAA1456 (also known as C8ORF79). HABH8 has only one known transcript, but hKIAA1456 has two transcript variants producing two isoforms. Surprisingly, hKIAA1456 isoform 2 does not qualify as a Trm homolog (E value=0.39). A CLUSTALW amino acid alignment comparison revealed that hKIAA1456 isoform 2 has significantly less homology with scTrm9 (39.8% conservation between isoform 2 and scTrm9 compared to 60.2% conservation between isoform 1 and scTrm9). Although hABH8 has the strongest match to scTrm9 with an E-value of 1.0e−44, its overall structure is quite different than that of scTrm9. HABH8 is much larger and contains an Fe(II) oxygenase domain/AlkB domain in addition to a methyltransferase domain, while scTrm9 does not (Fig. 6a). The predicted methyltransferase domain of scTrm9 shares a 46.7% identity with ABH8 and a 45.7% identity with hKIAA1456. About 71.7% of the scTrm9 methyltransferase domain amino acids are highly conserved in hABH8 and 69.6% of the scTrm9 methyltransferase domain amino acids are conserved in hKIAA1456.
FIG. 6.
Line diagrams of scTrm9 (A), scTrm11 (B), scTrm112 (C), and their human homolog candidates. ScTrm112 can dimerize with scTrm9 and scTrm11. hTRMT112 can dimerize with hTRMT11 and hABH8.
HABH8 was originally described as a homolog of the E. coli protein AlkB, a demethylator used in DNA damage repair. RNAi knockdown of hABH8 in urothelial carcinoma cells caused a significant increase in apoptotic cell death (Shimada et al., 2009). ABH8 knockout mice display no obvious phenotypes up to 20 months of age (Songe-Moller et al., 2010). LC-MS-MS analysis of the knockout mice's total tRNA revealed a loss of mcm5U, mcm5s2U, and mcm5Um (a modification unique to mammals) and an increase in cm5U in liver, brain, and testis samples. These mice also lack corresponding methyl modifications in tRNAGlu(UUC), tRNAArg(UCU), and tRNASeC. This study also analyzed the protein complexing potential of hABH8 by expressing a 6XHis-Tagged form of hABH8 in E. coli. HABH8 and hTRMT112 (the human scTrm112 homolog) were co-expressed in E. coli and were found to copurify with each other. This hABH8/hTRMT112 complex displayed methyltransferase activity in vitro. Analysis of hABH8 truncations showed that the methyltransferase domain was necessary for hTRMT112 interaction and methyltransferase activity but that the AlkB domain was not. It is interesting to note that hABH8-catalyzed tRNA modifications have been implicated in the incorporation of selenocysteine at UGA stop codons. Thus, hABH8 activity appears to be important for stop codon recoding and optimizing the levels of the selenocysteine containing proteins, with some of these proteins important for proper response to reactive oxygen species (Songe-Moller et al., 2010).
hABH8 activity was also studied in human tissue. HABH8 localizes to the cytoplasm in cultured cells (Fu et al., 2010a). Moreover, hABH8 was able to precipitate tRNAArg(UCU), Glu(UUC) as well as several proteins, including hTRMT112 and several TRiC (TCP Ring Complex) subunits. Notably, tRNALys(UUU), SeC(UGA) did not copurify with hABH8. HABH8 was also found to increase methylation of depleted human tRNA 2.5-fold in vitro. The methyltransferase domain of hABH8 was found to be sufficient for this behavior. shRNA depletion of hABH8 caused an increase in cm5U and a decrease in mcm5U, as well as susceptibility to DNA damaging agents. These results strongly imply that hABH8 is a scTrm9 homolog. In 2010, Fu et al. suggested that the AlkB domain is used to hydroxylate mcm5U and mcm5s2 into chm5U and mchm5U in tRNAGly(U*CC) in vitro (Fu et al., 2010b). A 2011 study has uncovered a new function for hABH8. In the study, hABH8 was shown in vitro and in vivo to generate one stereoisomer of 5-methylcarbonylhydroxymethyluridine (mchm5U) in mammalian tRNAArg(UCG), Gly(UCC) (van den Born et al., 2011).
HKIAA1456 has been labeled a putative tumor suppressor for colorectal cancer and was found to be downregulated in 9 of 12 primary tumors analyzed as well as in several colon cancer cell lines (Flanagan et al., 2004). However, hKIAA1456 mutations were only found in one of the 88 tumors studied and epigenetic silencing is a likely route to inactivation in tumors. Although mouse ABH8 appears to be a primary ScTrm9 homolog in mammalian cells, it remains to be seen if hABH8 is the only Trm9-like methyltransferase in humans. Based on complementation studies in trm9Δ yeast cells (Patil and Begley, unpublished results), it appears that hKIAA1456 is also a scTrm9 homolog and involved in tRNA modification. However, tRNA modification activity has not been confirmed for hKIAA1456 and is required to verify a functional similarity to scTrm9. We speculate that there may be tRNA isotype-specific, cell-specific, or developmental-specific roles for hKIAA1456 in wobble base modification.
scTrm10
ScTrm10 is likely responsible for the m1G9 tRNA methylation found in 10 different tRNA species. The first report of scTrm10 resulted from an S. cerevisiae library screen looking for the m1G9 methyltransferase (Jackman et al., 2003). Deletion analysis confirmed that scTrm10 depletion resulted in a loss of m1G9 and not m1G37. ScTrm10 was found to be both necessary and sufficient for m1G9 methylation in E. coli tRNA both in vitro and in vivo for 9 of the 10 known yeast tRNA substrates. tRNAAla m1G9 methylation could not be confirmed. A study of scTrm10 by Gustavsson and Ronne (2008) revealed that trm10Δ cells are more sensitive to 5-fluorouracil exposure than was wild-type yeast. The sensitivity to 5-fluorouracil was also observed for trm1Δ and trm8Δtrm82Δ cells. The 5-fluorouracil sensitivity was enhanced at 38°C. Mutational analysis also revealed a potential interaction between scTrm1 and scTrm10 and between scTrm8/scTrm82 and scTrm10, as those double mutants exhibited an increase in 5-fluorouracil sensitivity.
Our search for human homologs generated three candidates also found by Jackman et al. in their BLAST search: hRG9MTD (RNA(guanine-9-)methyltransferase domain containing) 1, 2, and 3. Based on amino acid sequence, HRG9MTD2 is the closest match of the three to scTrm10. The only structural difference between the three proteins is a small coiled-coil domain located near the N-terminus of hRG9MTD2 that is missing from the other two (Fig. 5b). The methyltransferase domain of scTrm10 shares a 34.1% identity with the hRG9MTD2 methyltransferase domain, 25.6% identity with the hRG9MTD1 methyltransferase domain and 27.7% identity with the hRG9MTD3 methyltransferase domain. About 67.7% of the scTrm10 methyltransferase domain amino acids are highly conserved in hRG9MTD2, 56% of the scTrm10 methyltransferase domain is highly conserved in hRG9MTD1, and 53.2% of the scTrm10 methyltransferase domain is conserved in hRG9MTD3. To date, we have been unable to find any publications studying hRG9MTD1 and hRG9MTD3. The only available study of hRG9MTD2 is a comparative genomic hybridization study suggesting a genetic link between hRG9MTD2 and colorectal cancer (Berg et al., 2010). hRG9MTD2 was one of seven genes displaying a significant difference in mRNA levels between early-onset and late-onset patients. No biochemical analysis was performed on hRG9MTD2. At this point, it is difficult to predict which protein is the most likely homolog since little is known about the scTrm10 methyltransferase domain.
scTrm11/112
ScTrm11 and scTrm112 form a heterodimeric complex to catalyze the formation of m2G10 modifications in at least 18 different tRNA species. scTrm11 (originally named Mtc12p [Bujnicki et al., 2004]) has a THUMP (thiouridine synthases, RNA methyltransferases and pseudouridine synthases) domain at the N-terminus (Fig. 6b). This THUMP domain is believed to be used for RNA binding. ScTrm11 also contains a characteristic Rossmann fold methyltransferase domain at the C-terminus.
ScTrm11 was initially 1 of 20 candidate methyltransferases tested for m2G10 activity (Purushothaman et al., 2005) and, after being the only one capable of methylating G10 in tRNAIle(UAU) in vitro, it was renamed scTrm11. Yeast cells deficient in scTrm11 lack m2G10 in tRNA. However, scTrm11 was not able to repeat this methylation in E. coli cells, suggesting that another protein was necessary. A subsequent review of the literature revealed that protein scTrm112 was found to interact with scTrm11 as well as with scTrm9, scMtc6p, and scLys9p (Gavin et al., 2002). ScTrm112 is comprised of 135 amino acids, with the only commonly recognized feature being a Trm112 family domain, a unique variant of a zinc finger (Purushothaman et al., 2005) (Fig. 6c). Deletion of TRM112 caused a severe reduction in growth rate and, importantly, a loss of m2G10 from purified tRNA (Purushothaman et al., 2005). A physical interaction between scTrm11 and scTrm112 was shown by co-immunoprecipitation (Purushothaman et al., 2005).
The human homolog candidates for scTrm11 and scTrm112 are named hTRMT11 and hTRMT112, respectively. HTRMT11 is very similar to scTrm11 in both size (463 amino acids) and domain architecture (Fig. 6b). HTRMT11 also has a THUMP domain at the N-terminus with a methyltransferase domain at the C-terminus. The predicted methyltransferase domain of hTRMT11 shares a 41.7% identity with scTrm11 and 62.9% of the amino acids are highly conserved. Currently, there are no published references to hTRMT11.
HTRMT112 contains a similar C-terminal domain to scTrm112 (Fig. 6c). HTRMT112 has been reported to interact with the human eukaryotic release factor-1 (heRF1) methyltransferase HemK2α (Figaro et al., 2008). This complex has also been shown to methylate heRF1 in the presence of eRF3 and GTP. HeRF1 is responsible for translation termination for eukaryotic cells. HeRF1 has a highly conserved GGQ tripeptide domain and methylation of the glutamine residue is critical to maintaining heRF1 function. Expression of HemK2α in the mtq2Δ strain of S. cerevisiae complemented the growth phenotype, suggesting that HemK2α is capable of interacting with scTrm112.
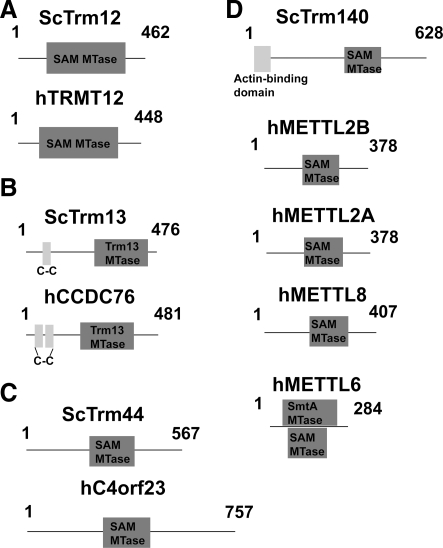
scTrm12/scTyw2
ScTrm12, also known as scTyw2, is responsible for the third chemical reaction necessary for the formation of wybutosine at position 37 of tRNAPhe (yW37). ScTyw2 was determined to be responsible for the conversion of the wybutosine precursor yW-187 to yW-86 via the transfer of an α-amino-α-carboxypropyl group from SAM to the C-7 position of yW-187 (Noma et al., 2006). Wybutosine is believed to be important for proper translation by stabilizing the codon–anticodon interaction (Grosjean et al., 1998; Konevega et al., 2004). Incomplete conversion of G37 to wybutosine may lead to increased frameshifting and increased susceptibility to retroviral protein expression (Carlson et al., 2001). ScTyw2 features the typical Rossmann fold methyltransferase domain. ScTrm12 mutant tRNA was shown to contain the yW precursor 4-demethylwyosine (ImG-14 aka yW-187), the second step to yW formation, in 2005 (Kalhor et al., 2005). The trm12 deletion mutant displayed no significant difference in growth rate when compared to wild-type strains. The full pathway to yW formation in yeast was clarified a year later by Noma et al.'s study utilized reverse genetic analysis and MS to identify four yeast deletion strains that lack wybutosine, one of which was the trm12 deletion strain described by Kalhor et al. The four proteins were renamed scTyw1-4 (tRNA-yW synthesizing protein 1–4) (Noma et al., 2006). Further in vitro biochemical analysis revealed that scTrm12 converted yW-187 to yW-86 in the presence of SAM, leading Noma et al. to rename the protein scTyw2.
Our search confirmed Kahlor et al.'s and Noma et al.'s identification of the human homolog candidate for scTrm12. The protein, called hTRMT12 in the PubMed database, is similar in size and domain structure to scTrm12 (Fig. 7a). The predicted methyltransferase domain of scTyw2 shares a 47.8% identity with hTRMT12 and 61.2% of the amino acids are highly conserved. A reference to hTRMT12 was found in a study by Rodriguez et al. (2007), suggesting a genetic link between hTRMT12 and breast cancer using a BAC array and FISH. HTRMT12 mRNA was amplified in 7 of 8 cancer cell lines examined and 26 of 30 tumors examined. No biochemical analysis of hTRMT12 has been reported.
FIG. 7.
Line diagrams of scTrm12 (A), scTrm13 (B), scTrm44 (C), scTrm140 (D), and their human homolog candidates. The protein C4ORF23 has recently been renamed METTL19 in its PubMed Gene entry.
scTrm13
ScTrm13 is responsible for the 2′-O ribomethylation at yeast tRNA position 4 for tRNAGly(GCC), His, Pro species. ScTrm13 generates Cm4 for tRNAPro and tRNAGly(GCC) and Am4 for tRNAHis. ScTrm13 contains one CHHC zinc finger commonly seen in two other eukaryotic protein families near the N-terminus (Fig. 7b). ScTrm13 has been confirmed to bind zinc 1:1, but the zinc finger domain is likely used for RNA binding (Andreeva and Tidow, 2008). ScTrm13 also has a typical methyltransferase domain taking up the remainder of the amino acid sequence, though it shares little similarity with the other known methyltransferases. Further research is required to identify the function of position 4 ribomethylation.
The human homolog candidate for scTrm13 is named hCCDC76 (coiled-coil domain-containing protein 76). HCCDC76 is similar in size and domain architecture to scTrm13, and the primary difference between the two proteins is that hCCDC76 contains two projected Zn finger domains instead of one (Fig. 7b). The predicted methyltransferase domain of scTrm13 shares a 26.0% identity with hCCDC76 and 51.0% of the amino acids are highly conserved. The first reference to hCCDC76 is in a 2002 study by Weinmann et al. where the CCDC76 gene (referenced by its accession number) was one of 68 targets pulled from a large-scale ChIP assay against the transcription factor αE2F (Weinmann et al., 2002). In an analysis of CpG island chromatin, the CCDC76 gene bound E2F4 strongly, E2F1 weakly, and RNA polymerase II weakly.
scTrm44
ScTrm44 is responsible for the Um44 2′-O ribomethylation on yeast tRNASer only. Position 44 ribomethylation occurs under specific conditions: only when nucleotide 44 is uridine and only if the tRNA has a long variable loop (Kotelawala et al., 2008). Despite its size, the only defined domain in scTrm44 is its C-terminal methyltransferase domain. The methyltransferase domain contains the classical GXGXG domain that alignment by BLAST indicated is highly conserved among eukaryotic homologs, including humans (Kotelawala et al., 2008). Based on a predicted interaction between m22G26 and Um44, researchers hypothesize that Um44 2′-O ribomethylation helps stabilize the tRNA tertiary structure (Sampson and Uhlenbeck, 1988).
In the same study that detailed the discovery of scTrm44, a human homolog candidate was mentioned (Kotelawala et al., 2008). This candidate has since been renamed hMETTL19 in the PubMed database. Three different versions of this protein appear in the NCBI database with amino acid lengths of 711, 411, and 365. All three contain the putative methyltransferase domain (Fig. 7c) and the middle variant was mentioned in a categorization of a potential new methyltransferase family (Knizewski and Ginalski, 2006). The methyltransferase domain of hMETTL19 is 45.5% identical to the methyltransferase domain of scTrm44. About 67.9% of the scTrm44 methyltransferase domain is highly conserved in hMETTL19.
scTrm140
The most recently discovered Trm is scTrm140, responsible for the m3C32 methylation on tRNASer, tRNAThr, and tRNAArg. ScTrm140 was originally identified as an actin-binding protein named Abp140 (Asakura et al., 1998). The protein is translated from two reading frames (YOR239W and YOR240W) when the mRNA sequence CTT-AGG-C initiates a programmed frameshift. ScTrm140 contains an N-terminal actin-binding domain in addition to a classical methyltransferase domain (Fig. 7d). Abp140 was identified as a methyltransferase by two groups simultaneously in 2011 (D'Silva et al., 2011; Noma et al., 2011). D'Silva et al. (2011) utilized a primer extension assay to identify the gene responsible for m3C32 methylation in tRNAThr(IGU). From 77 target deletion strains, only the abp140Δ strain lacked the m3C32 modification. D'Silva et al. also reported that scTrm140 was sufficient for methylation in vivo and in vitro and only the methyltransferase domain from the YOR240W ORF is necessary for methyltransferase activity. The abp140Δ trm1Δ strain displayed a slight sensitivity to cyclohexamide, indicating that scTrm140 may play a role in translation regulation.
Noma et al. (2011) identified scTrm140 by using the reverse genetic analysis method previously used by the group in their scTyw2 study. Similar to the D'Silva study, Noma et al. confirmed that scAbp140 contained the m3C32 methyltransferase and that the actin-binding domain was not needed for methyltransferase activity. However, Noma et al. reported that tRNASer1 could not be methylated by recombinant scTrm140 in vitro, suggesting that additional factors may be required for full activity in vivo. Interestingly, Noma et al. searched for human Trm140 homologs and found the protein hMETTL2B. When hMETTL2B was knocked down in HeLa cells with short-hairpin RNA, m3C levels were specifically decreased, suggesting that METTL2B is indeed the methyltransferase.
Our BLAST search also identified hMETTL2B as the closest human homolog candidate to scTrm140, with hMETTL2A, hMETTL6, and hMETTL8 also qualifying as homolog candidates according to our search parameters. All four candidates are smaller than scTrm140 (378, 378, 284, and 407 amino acids, respectively [Fig. 7d]), with all four seeming to contain only the YOR240W portion of scTrm140. All four proteins contain a methyltransferase domain with a GXGXG sequence highly conserved with scTrm140. All four candidates have nearly identical methyltransferase sequences, as shown in their alignments to scTrm140. The hMETTL2A and hMETTL2B methyltransferase domains are both 39.8% identical to scTrm140, and 67.6% of the scTrm140 methyltransferase domain is highly conserved in hMETTL2A and hMETTL2B. The scTrm140 methyltransferase domain is slightly more conserved in hMETTL6 (41.5% identical and 69.8% conserved) and slightly less conserved in hMETTL8 (39.4% identical and 62.4% conserved). There are no reported transcript variants for any of the four proteins. Besides the work done by Noma et al., the only published work referencing any of the four proteins comes from an evolutionary analysis of the scTrm140 CTT-AGG-C programmed frameshift (Farabaugh et al., 2006). In the study, hMETTL2B was identified as an scTrm140 homolog, with no further analysis performed.
Conclusions
Each yeast Trm has at least one potential human homolog and, in some cases, there are several candidates that may contribute to human tRNA methylation (Fig. 8). While virtually all known yeast tRNA methylations are accounted for, at least three unaccounted human tRNA methylations (m5C48, m5C49, and m5C40) are reported (Brzezicha et al., 2006). The Trms responsible for these unaccounted-for scTrm4 m5C methylation events may be among the unstudied NSUN proteins mentioned here. It is also likely, as the recent scTrm140 discovery shows, that there are human Trms yet to be discovered. At first glance, it seems obvious that yeast Trms would have a human homolog; however, there are examples in the literature where a tRNA modification is known to exist in a species, but there are no obvious homologs to known Trms in other species. For example, the Um44 modification is known to exist in plants but a BLAST search performed using scTrm44 found no candidate plant homologs (Kotelawala et al., 2008).
FIG. 8.
An overview of known (A) yeast and (B) predicted human homolog tRNA modifications and corresponding enzymes. Each known methylation is marked on the tRNA model with a filled circle and labeled with the enzyme potentially responsible for the methylation. For wybutosine, scTrm12 is one of five proteins responsible (scTyw1-5).
There are several explanations for finding more than one human homolog to some yeast Trms. One possibility is that some human methyltransferases have more stringent substrate specificity than their yeast counterparts, with scTrm4/hNSUN2 an example. While scTrm4 methylates four sites, hNSUN2 only methylates C34 for certain. It is possible that other members of the NSUN family methylate the other three targets or it may be that hNSUN2 methylates the other three under different (i.e., stress) conditions. Both possibilities are feasible and remain to be studied. Another reason for the increased number of Trm enzymes in humans could be due to more modified bases in humans, relative to yeast. For example, human tRNA contains mcm5Um, which is absent in yeast, necessitating further work to determine the enzyme(s) responsible for this modification. Another likely reason for the increased number of human methyltransferases is the added complexity of human cells, relative to budding yeast. While many basic cellular mechanisms are shared between these two species, the different needs of a multicellular organism may demand more specific or more varieties of tRNA methylation. We speculate that there may be developmental or organ-specific roles for specific human Trms, which necessitates a need for multiple homologs. Cumulatively, there are most likely multiple reasons for the increased number of Trms in humans, relative to S. cerevisiae, with future studies needed to discern each Trms role in human cells.
While six of the human methyltransferase candidates mentioned here have been characterized, the majority remain poorly studied after their initial discovery. Considering that some of the proteins—like scTrm44—were discovered very recently, it is not surprising that research on human methyltransferases still lags. One of the challenges is that while a consistent battery of experimental tests have been developed to characterize S. cerevisiae Trms, no such consistency exists in human studies thus far. The reports of scTrm13, scTrm44, and scTrm140 offer the current templates for characterizing an S. cerevisiae Trm. A screen is performed using the S. cerevisiae movable open reading frame proteome library to identify the protein capable of performing the target methylation. When a candidate displays evidence of Trm activity, its deletion strain is tested. High-performance liquid chromatography analysis of the deletion strain tRNA is performed to offer in vivo confirmation of the methyltransferase activity and crude lysate methyltransferase assays of wild-type lysate and deletion strain lysate offers in vitro confirmation. Finally, the candidate is expressed as an E. coli fusion protein. If the candidate expresses properly and shows at least 20% methylase activity, the candidate is likely necessary and sufficient for the tRNA methylation in question. In contrast, the studies confirming human Trm activity vary in protein expression platforms used and in the species of tRNA substrate tested. Methyltransferase activity can be tested in yeast deletion strains effectively but using the yeast model does not fully characterize the methyltransferase in human cells. RNAi knockdown and recovery of the human methyltransferase candidates may offer a human in vivo test of methyltransferase activity, as seen with hMETTL2B.
Another factor for the lag in human Trm study is that RNA analysis is notoriously difficult and the experimental methods used for Trm characterization are specialized techniques. Despite these hurdles, unlocking the mystery of tRNA methylation may reveal new insights into many human diseases from breast and colorectal cancer to mental retardation. Ten of the 34 homologs have some reference linking the homolog to a disease. These links go beyond cancer; diseases like Down's syndrome and AIDS have been connected to hTRMT2A and hTARBP1, respectively. The variety of disease links discussed in this review imply that improper tRNA modification may lead to more significant impaired cellular function in humans than in yeast. Further research is necessary to determine the extent of these links, but any opportunity to uncover the mechanics of a disease state is worth investigating. These studies may be a foreshadowing of new and interesting avenues of study related to post-transcriptional gene expression in human systems.
Acknowledgments
Financial support was provided by the National Institute of Environmental Health Sciences (R21 ES017146, R01 ES015037, and R01 ES017010).
Disclosure Statement
No competing financial interests exist.
References
- Alexandrov A. Chernyakov I., et al. Rapid tRNA decay can result from lack of nonessential modifications. Mol Cell. 2006;21:87–96. doi: 10.1016/j.molcel.2005.10.036. [DOI] [PubMed] [Google Scholar]
- Alexandrov A. Grayhack E.J., et al. tRNA m7G methyltransferase Trm8p/Trm82p: evidence linking activity to a growth phenotype and implicating Trm82p in maintaining levels of active Trm8p. RNA. 2005;11:821–830. doi: 10.1261/rna.2030705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov A. Martzen M.R., et al. Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. RNA. 2002;8:1253–1266. doi: 10.1017/s1355838202024019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J. Phan L., et al. The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev. 1998;12:3650–3662. doi: 10.1101/gad.12.23.3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J. Phan L., et al. The Gcd10p/Gcd14p complex is the essential two-subunit tRNA(1-methyladenosine) methyltransferase of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2000;97:5173–5178. doi: 10.1073/pnas.090102597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva A. Tidow H. A novel CHHC Zn-finger domain found in spliceosomal proteins and tRNA modifying enzymes. Bioinformatics. 2008;24:2277–2280. doi: 10.1093/bioinformatics/btn431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura T. Sasaki T., et al. Isolation and characterization of a novel actin filament-binding protein from Saccharomyces cerevisiae. Oncogene. 1998;16:121–130. doi: 10.1038/sj.onc.1201487. [DOI] [PubMed] [Google Scholar]
- Asefa B. Kauler P., et al. Genetic analysis of the yeast NUD1 endo-exonuclease: a role in the repair of DNA double-strand breaks. Curr Genet. 1998;34:360–367. doi: 10.1007/s002940050407. [DOI] [PubMed] [Google Scholar]
- Bahr A. Hankeln T., et al. Molecular analysis of METTL1, a novel human methyltransferase-like gene with a high degree of phylogenetic conservation. Genomics. 1999;57:424–428. doi: 10.1006/geno.1999.5780. [DOI] [PubMed] [Google Scholar]
- Bartlett J.M. Thomas J., et al. Mammostrat(R) as a tool to stratify breast cancer patients at risk of recurrence during endocrine therapy. Breast Cancer Res. 2010;12:R47. doi: 10.1186/bcr2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley T.J. Rosenbach A.S., et al. Hot spots for modulating toxicity identified by genomic phenotyping and localization mapping. Mol Cell. 2004;16:117–125. doi: 10.1016/j.molcel.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Begley U. Dyavaiah M., et al. Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol Cell. 2007;28:860–870. doi: 10.1016/j.molcel.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg M. Agesen T.H., et al. Distinct high resolution genome profiles of early onset and late onset colorectal cancer integrated with gene expression data identify candidate susceptibility loci. Mol Cancer. 2010;9:100. doi: 10.1186/1476-4598-9-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhasin M. Garg A., et al. PSLpred: prediction of subcellular localization of bacterial proteins. Bioinformatics. 2005;21:2522–2524. doi: 10.1093/bioinformatics/bti309. [DOI] [PubMed] [Google Scholar]
- Bhasin M. Raghava G.P. ESLpred: SVM-based method for subcellular localization of eukaryotic proteins using dipeptide composition and PSI-BLAST. Nucleic Acids Res. 2004;32(Web Server issue):W414–419. doi: 10.1093/nar/gkh350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjork G.R. Jacobsson K., et al. A primordial tRNA modification required for the evolution of life? EMBO J. 2001;20:231–239. doi: 10.1093/emboj/20.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brule H. Elliott M., et al. Isolation and characterization of the human tRNA-(N1G37) methyltransferase (TRM5) and comparison to the Escherichia coli TrmD protein. Biochemistry. 2004;43:9243–9255. doi: 10.1021/bi049671q. [DOI] [PubMed] [Google Scholar]
- Brzezicha B. Schmidt M., et al. Identification of human tRNA:m5C methyltransferase catalysing intron-dependent m5C formation in the first position of the anticodon of the pre-tRNA Leu (CAA) Nucleic Acids Res. 2006;34:6034–6043. doi: 10.1093/nar/gkl765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugl H. Fauman E.B., et al. RNA methylation under heat shock control. Mol Cell. 2000;6:349–360. doi: 10.1016/s1097-2765(00)00035-6. [DOI] [PubMed] [Google Scholar]
- Bujnicki J.M. Feder M., et al. Sequence-structure-function studies of tRNA:m5C methyltransferase Trm4p and its relationship to DNA:m5C and RNA:m5U methyltransferases. Nucleic Acids Res. 2004;32:2453–2463. doi: 10.1093/nar/gkh564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldas T. Binet E., et al. Translational defects of Escherichia coli mutants deficient in the Um(2552) 23S ribosomal RNA methyltransferase RrmJ/FTSJ. Biochem Biophys Res Commun. 2000;271:714–718. doi: 10.1006/bbrc.2000.2702. [DOI] [PubMed] [Google Scholar]
- Calvo O. Cuesta R., et al. GCD14p, a repressor of GCN4 translation, cooperates with Gcd10p and Lhp1p in the maturation of initiator methionyl-tRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:4167–4181. doi: 10.1128/mcb.19.6.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson B.A. Mushinski J.F., et al. 1-Methylguanosine in place of Y base at position 37 in phenylalanine tRNA is responsible for its shiftiness in retroviral ribosomal frameshifting. Virology. 2001;279:130–135. doi: 10.1006/viro.2000.0692. [DOI] [PubMed] [Google Scholar]
- Cartlidge R.A. Knebel A., et al. The tRNA methylase METTL1 is phosphorylated and inactivated by PKB and RSK in vitro and in cells. EMBO J. 2005;24:1696–1705. doi: 10.1038/sj.emboj.7600648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaille J. Chetouani F., et al. The yeast Saccharomyces cerevisiae YDL112w ORF encodes the putative 2′-O-ribose methyltransferase catalyzing the formation of Gm18 in tRNAs. RNA. 1999;5:66–81. doi: 10.1017/s1355838299981475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.T. Dyavaiah M., et al. A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet. 2010;6:e1001247. doi: 10.1371/journal.pgen.1001247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernyakov I. Whipple J.M., et al. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5′-3′ exonucleases Rat1 and Xrn1. Genes Dev. 2008;22:1369–1380. doi: 10.1101/gad.1654308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching Y.P. Zhou H.J., et al. Identification and characterization of FTSJ2, a novel human nucleolar protein homologous to bacterial ribosomal RNA methyltransferase. Genomics. 2002;79:2–6. doi: 10.1006/geno.2001.6670. [DOI] [PubMed] [Google Scholar]
- Chow T.Y. Perkins E.L., et al. Yeast RNC1 encodes a chimeric protein, RhoNUC, with a human rho motif and deoxyribonuclease activity. Nucleic Acids Res. 1992;20:5215–5221. doi: 10.1093/nar/20.19.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuesta R. Hinnebusch A.G., et al. Identification of GCD14 and GCD15, novel genes required for translational repression of GCN4 mRNA in Saccharomyces cerevisiae. Genetics. 1998;148:1007–1020. doi: 10.1093/genetics/148.3.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Silva S. Haider S.J., et al. A domain of the actin binding protein Abp140 is the yeast methyltransferase responsible for 3-methylcytidine modification in the tRNA anti-codon loop. RNA. 2011;17:1100–1110. doi: 10.1261/rna.2652611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunin-Horkawicz S. Czerwoniec A., et al. MODOMICS: a database of RNA modification pathways. Nucleic Acids Res. 2006;34(Database issue):D145–D149. doi: 10.1093/nar/gkj084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis S.R. Hopper A.K., et al. Amino-terminal extension generated from an upstream AUG codon increases the efficiency of mitochondrial import of yeast N2,N2-dimethylguanosine-specific tRNA methyltransferases. Mol Cell Biol. 1989;9:1611–1620. doi: 10.1128/mcb.9.4.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis S.R. Morales M.J., et al. Isolation and characterization of the TRM1 locus, a gene essential for the N2,N2-dimethylguanosine modification of both mitochondrial and cytoplasmic tRNA in Saccharomyces cerevisiae. J Biol Chem. 1986;261:9703–9709. [PubMed] [Google Scholar]
- Farabaugh P.J. Kramer E., et al. Evolution of +1 programmed frameshifting signals and frameshift-regulating tRNAs in the order Saccharomycetales. J Mol Evol. 2006;63:545–561. doi: 10.1007/s00239-005-0311-0. [DOI] [PubMed] [Google Scholar]
- Feder M. Pas J., et al. Molecular phylogenetics of the RrmJ/fibrillarin superfamily of ribose 2′-O-methyltransferases. Gene. 2003;302:129–138. doi: 10.1016/s0378-1119(02)01097-1. [DOI] [PubMed] [Google Scholar]
- Figaro S. Scrima N., et al. HemK2 protein, encoded on human chromosome 21, methylates translation termination factor eRF1. FEBS Lett. 2008;582:2352–2356. doi: 10.1016/j.febslet.2008.05.045. [DOI] [PubMed] [Google Scholar]
- Flanagan J.M. Healey S., et al. Mapping of a candidate colorectal cancer tumor-suppressor gene to a 900-kilobase region on the short arm of chromosome 8. Genes Chromosomes Cancer. 2004;40:247–260. doi: 10.1002/gcc.20039. [DOI] [PubMed] [Google Scholar]
- Freude K. Hoffmann K., et al. Mutations in the FTSJ1 gene coding for a novel S-adenosylmethionine-binding protein cause nonsyndromic X-linked mental retardation. Am J Hum Genet. 2004;75:305–309. doi: 10.1086/422507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye M. Dragoni I., et al. Genomic gain of 5p15 leads to over-expression of Misu (NSUN2) in breast cancer. Cancer Lett. 2010;289:71–80. doi: 10.1016/j.canlet.2009.08.004. [DOI] [PubMed] [Google Scholar]
- Frye M. Watt F.M. The RNA methyltransferase Misu (NSun2) mediates Myc-induced proliferation and is upregulated in tumors. Curr Biol. 2006;16:971–981. doi: 10.1016/j.cub.2006.04.027. [DOI] [PubMed] [Google Scholar]
- Fu D. Brophy J.A., et al. Human AlkB homolog ABH8 Is a tRNA methyltransferase required for wobble uridine modification and DNA damage survival. Mol Cell Biol. 2010a;30:2449–2459. doi: 10.1128/MCB.01604-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. Dai Q., et al. The AlkB domain of mammalian ABH8 catalyzes hydroxylation of 5-methoxycarbonylmethyluridine at the wobble position of tRNA. Angew Chem Int Ed Engl. 2010b;49:8885–8888. doi: 10.1002/anie.201001242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E. Hoogland C. Gattiker A. Duvaud S. Wilkins M.R. Appel R.D. Bairoch A. Humana Press; NewYork, NY: 2005. Protein Identification and Analysis Tools on the ExPASy Server. [DOI] [PubMed] [Google Scholar]
- Gavin A.C. Bosche M., et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Gong P. Li J., et al. Genetic variations in FTSJ1 influence cognitive ability in young males in the Chinese Han population. J Neurogenet. 2008;22:277–287. doi: 10.1080/01677060802337299. [DOI] [PubMed] [Google Scholar]
- Modulatory Role of Modified Nucleotides in RNA Loop-Loop Interactions Modification and Editing of RNA. In: Grosjean H., editor; Houssier C., editor; Romby P., editor; Marquet R., editor. ASM Press; Washington, DC: 1998. [Google Scholar]
- Gustavsson M. Ronne H. Evidence that tRNA modifying enzymes are important in vivo targets for 5-fluorouracil in yeast. RNA. 2008;14:666–674. doi: 10.1261/rna.966208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harashima S. Hinnebusch A.G. Multiple GCD genes required for repression of GCN4, a transcriptional activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:3990–3998. doi: 10.1128/mcb.6.11.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks D.G. Janarthanan B.R., et al. The expression of TRMT2A, a novel cell cycle regulated protein, identifies a subset of breast cancer patients with HER2 over-expression that are at an increased risk of recurrence. BMC Cancer. 2010;10:108. doi: 10.1186/1471-2407-10-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong B. Brockenbrough J.S., et al. Nop2p is required for pre-rRNA processing and 60S ribosome subunit synthesis in yeast. Mol Cell Biol. 1997;17:378–388. doi: 10.1128/mcb.17.1.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonowski D. Zink S., et al. tRNAGlu wobble uridine methylation by Trm9 identifies Elongator's key role for zymocin-induced cell death in yeast. Mol Microbiol. 2006;59:677–688. doi: 10.1111/j.1365-2958.2005.04972.x. [DOI] [PubMed] [Google Scholar]
- Jackman J.E. Montange R.K., et al. Identification of the yeast gene encoding the tRNA m1G methyltransferase responsible for modification at position 9. RNA. 2003;9:574–585. doi: 10.1261/rna.5070303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamsai D. Rijal S., et al. A novel protein, sperm head and tail associated protein (SHTAP), interacts with cysteine-rich secretory protein 2 (CRISP2) during spermatogenesis in the mouse. Biol Cell. 2010;102:93–106. doi: 10.1042/BC20090099. [DOI] [PubMed] [Google Scholar]
- Johansson M.J. Bystrom A.S. Dual function of the tRNA(m(5)U54)methyltransferase in tRNA maturation. RNA. 2002;8:324–335. doi: 10.1017/s1355838202027851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan R.M. Clarke S. Widespread occurrence of three sequence motifs in diverse S-adenosylmethionine-dependent methyltransferases suggests a common structure for these enzymes. Arch Biochem Biophys. 1994;310:417–427. doi: 10.1006/abbi.1994.1187. [DOI] [PubMed] [Google Scholar]
- Kalhor H.R. Clarke S. Novel methyltransferase for modified uridine residues at the wobble position of tRNA. Mol Cell Biol. 2003;23:9283–9292. doi: 10.1128/MCB.23.24.9283-9292.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalhor H.R. Penjwini M., et al. A novel methyltransferase required for the formation of the hypermodified nucleoside wybutosine in eucaryotic tRNA. Biochem Biophys Res Commun. 2005;334:433–440. doi: 10.1016/j.bbrc.2005.06.111. [DOI] [PubMed] [Google Scholar]
- King M. Ton D., et al. A conserved motif in the yeast nucleolar protein Nop2p contains an essential cysteine residue. Biochem J. 1999;337(Pt 1):29–35. [PMC free article] [PubMed] [Google Scholar]
- Knizewski L. Ginalski K. DUF1613 is a novel family of eucaryotic AdoMet-dependent methyltransferases. Cell Cycle. 2006;5:1580–1582. doi: 10.4161/cc.5.14.2978. [DOI] [PubMed] [Google Scholar]
- Konevega A.L. Soboleva N.G., et al. Purine bases at position 37 of tRNA stabilize codon-anticodon interaction in the ribosomal A site by stacking and Mg2+-dependent interactions. RNA. 2004;10:90–101. doi: 10.1261/rna.5142404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotelawala L. Grayhack E.J., et al. Identification of yeast tRNA Um(44) 2′-O-methyltransferase (Trm44) and demonstration of a Trm44 role in sustaining levels of specific tRNA(Ser) species. RNA. 2008;14:158–169. doi: 10.1261/rna.811008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurowski M.A. Sasin J.M., et al. Characterization of the cofactor-binding site in the SPOUT-fold methyltransferases by computational docking of S-adenosylmethionine to three crystal structures. BMC Bioinform. 2003;4:9. doi: 10.1186/1471-2105-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. Straby K.B. The human tRNA(m(2)(2)G(26))dimethyltransferase: functional expression and characterization of a cloned hTRM1 gene. Nucleic Acids Res. 2000;28:3445–3451. doi: 10.1093/nar/28.18.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J. Huang B., et al. The Kluyveromyces lactis gamma-toxin targets tRNA anticodons. RNA. 2005;11:1648–1654. doi: 10.1261/rna.2172105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazauric M.H. Dirick L., et al. Trm112p is a 15-kDa zinc finger protein essential for the activity of two tRNA and one protein methyltransferases in yeast. J Biol Chem. 2010;285:18505–18515. doi: 10.1074/jbc.M110.113100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud J. Kudoh J., et al. Isolation and characterization of a human chromosome 21q22.3 gene (WDR4) and its mouse homologue that code for a WD-repeat protein. Genomics. 2000;68:71–79. doi: 10.1006/geno.2000.6258. [DOI] [PubMed] [Google Scholar]
- Motorin Y. Grosjean H. Multisite-specific tRNA:m5C-methyltransferase (Trm4) in yeast Saccharomyces cerevisiae: identification of the gene and substrate specificity of the enzyme. RNA. 1999;5:1105–1118. doi: 10.1017/s1355838299982201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motorin Y. Helm M. tRNA stabilization by modified nucleotides. Biochemistry. 2010;49:4934–4944. doi: 10.1021/bi100408z. [DOI] [PubMed] [Google Scholar]
- Noma A. Kirino Y., et al. Biosynthesis of wybutosine, a hyper-modified nucleoside in eukaryotic phenylalanine tRNA. EMBO J. 2006;25:2142–2154. doi: 10.1038/sj.emboj.7601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma A. Yi S., et al. Actin-binding protein ABP140 is a methyltransferase for 3-methylcytidine at position 32 of tRNAs in Saccharomyces cerevisiae. RNA. 2011;17:1111–1119. doi: 10.1261/rna.2653411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordlund M.E. Johansson J.O., et al. Identification of the TRM2 gene encoding the tRNA(m5U54)methyltransferase of Saccharomyces cerevisiae. RNA. 2000;6:844–860. doi: 10.1017/s1355838200992422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura T. Tomoyasu T., et al. Structure and function of the ftsH gene in Escherichia coli. Res Microbiol. 1991;142:279–282. doi: 10.1016/0923-2508(91)90041-8. [DOI] [PubMed] [Google Scholar]
- Ozanick S. Krecic A., et al. The bipartite structure of the tRNA m1A58 methyltransferase from S. cerevisiae is conserved in humans. RNA. 2005;11:1281–1290. doi: 10.1261/rna.5040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozanick S.G. Wang X., et al. Rex1p deficiency leads to accumulation of precursor initiator tRNAMet and polyadenylation of substrate RNAs in Saccharomyces cerevisiae. Nucleic Acids Res. 2009;37:298–308. doi: 10.1093/nar/gkn925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintard L. Lecointe F., et al. Trm7p catalyses the formation of two 2′-O-methylriboses in yeast tRNA anticodon loop. EMBO J. 2002;21:1811–1820. doi: 10.1093/emboj/21.7.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purushothaman S.K. Bujnicki J.M., et al. Trm11p and Trm112p are both required for the formation of 2-methylguanosine at position 10 in yeast tRNA. Mol Cell Biol. 2005;25:4359–4370. doi: 10.1128/MCB.25.11.4359-4370.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaud A. Facca C., et al. Disruption and functional analysis of six ORFs of chromosome IV: YDL103c (QRI1), YDL105w (QRI2), YDL112w (TRM3), YDL113c, YDL116w (NUP84) and YDL167c (NRP1) Yeast. 2001;18:273–282. doi: 10.1002/1097-0061(200102)18:3<273::AID-YEA665>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Rodriguez V. Chen Y., et al. Chromosome 8 BAC array comparative genomic hybridization and expression analysis identify amplification and overexpression of TRMT12 in breast cancer. Genes Chromosomes Cancer. 2007;46:694–707. doi: 10.1002/gcc.20454. [DOI] [PubMed] [Google Scholar]
- Sadekova S. Chow T.Y. Over-expression of the NUD1-coded endo-exonuclease in Saccharomyces cerevisiae enhances DNA recombination and repair. Curr Genet. 1996;30:50–55. doi: 10.1007/s002940050099. [DOI] [PubMed] [Google Scholar]
- Saikia M. Fu Y., et al. Genome-wide analysis of N1-methyl-adenosine modification in human tRNAs. RNA. 2010;16:1317–1327. doi: 10.1261/rna.2057810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakita-Suto S. Kanda A., et al. Aurora-B regulates RNA methyltransferase NSUN2. Mol Biol Cell. 2007;18:1107–1117. doi: 10.1091/mbc.E06-11-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson J.R. Uhlenbeck O.C. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc Natl Acad Sci U S A. 1988;85:1033–1037. doi: 10.1073/pnas.85.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M. Hagemann S., et al. Azacytidine inhibits RNA methylation at DNMT2 target sites in human cancer cell lines. Cancer Res. 2009;69:8127–8132. doi: 10.1158/0008-5472.CAN-09-0458. [DOI] [PubMed] [Google Scholar]
- Schubert H.L. Blumenthal R.M., et al. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem Sci. 2003;28:329–335. doi: 10.1016/S0968-0004(03)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semionov A. Cournoyer D., et al. The effect of the Saccharomyces cerevisiae endo-exonuclease NUD1 gene expression on the resistance of HeLa cells to DNA-damaging agents. Mutat Res. 1999;433:169–181. doi: 10.1016/s0921-8777(99)00002-6. [DOI] [PubMed] [Google Scholar]
- Shimada K. Nakamura M., et al. A novel human AlkB homologue, ALKBH8, contributes to human bladder cancer progression. Cancer Res. 2009;69:3157–3164. doi: 10.1158/0008-5472.CAN-08-3530. [DOI] [PubMed] [Google Scholar]
- Songe-Moller L. van den Born E., et al. Mammalian ALKBH8 possesses tRNA methyltransferase activity required for the biogenesis of multiple wobble uridine modifications implicated in translational decoding. Mol Cell Biol. 2010;30:1814–1827. doi: 10.1128/MCB.01602-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood R. Bonner T.I., et al. Cloning and characterization of 13 novel transcripts and the human RGS8 gene from the 1q25 region encompassing the hereditary prostate cancer (HPC1) locus. Genomics. 2001;73:211–222. doi: 10.1006/geno.2001.6500. [DOI] [PubMed] [Google Scholar]
- Sprinzl M. Horn C., et al. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1998;26:148–153. doi: 10.1093/nar/26.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel M.C. Abelson J. Effect of intron mutations on processing and function of Saccharomyces cerevisiae SUP53 tRNA in vitro and in vivo. Mol Cell Biol. 1986;6:2663–2673. doi: 10.1128/mcb.6.7.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studte P. Zink S., et al. tRNA and protein methylase complexes mediate zymocin toxicity in yeast. Mol Microbiol. 2008;69:1266–1277. doi: 10.1111/j.1365-2958.2008.06358.x. [DOI] [PubMed] [Google Scholar]
- Takano K. Nakagawa E., et al. A loss-of-function mutation in the FTSJ1 gene causes nonsyndromic X-linked mental retardation in a Japanese family. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:479–484. doi: 10.1002/ajmg.b.30638. [DOI] [PubMed] [Google Scholar]
- Tkaczuk K.L. Dunin-Horkawicz S., et al. Structural and evolutionary bioinformatics of the SPOUT superfamily of methyltransferases. BMC Bioinform. 2007;8:73. doi: 10.1186/1471-2105-8-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trynka G. Zhernakova A., et al. Coeliac disease-associated risk variants in TNFAIP3 and REL implicate altered NF-kappaB signalling. Gut. 2009;58:1078–1083. doi: 10.1136/gut.2008.169052. [DOI] [PubMed] [Google Scholar]
- Tscherne J.S. Nurse K., et al. Purification, cloning, and characterization of the 16S RNA m5C967 methyltransferase from Escherichia coli. Biochemistry. 1999;38:1884–1892. doi: 10.1021/bi981880l. [DOI] [PubMed] [Google Scholar]
- Urbonavicius J. Qian Q., et al. Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J. 2001;20:4863–4873. doi: 10.1093/emboj/20.17.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbonavicius J. Stahl G., et al. Transfer RNA modifications that alter +1 frameshifting in general fail to affect −1 frameshifting. RNA. 2003;9:760–768. doi: 10.1261/rna.5210803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Born E. Vagbo C.B., et al. ALKBH8-mediated formation of a novel diastereomeric pair of wobble nucleosides in mammalian tRNA. Nat Commun. 2011;2:172. doi: 10.1038/ncomms1173. [DOI] [PubMed] [Google Scholar]
- Van Vliet-Reedijk J.C. Planta R.J. The RHO4a and NUD1 genes on Saccharomyces cerevisiae chromosome XI. Yeast. 1993;9:1139–1147. doi: 10.1002/yea.320091015. [DOI] [PubMed] [Google Scholar]
- Vauti F. Goller T., et al. The mouse Trm1-like gene is expressed in neural tissues and plays a role in motor coordination and exploratory behaviour. Gene. 2007;389:174–185. doi: 10.1016/j.gene.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Weinmann A.S. Yan P.S., et al. Isolating human transcription factor targets by coupling chromatin immunoprecipitation and CpG island microarray analysis. Genes Dev. 2002;16:235–244. doi: 10.1101/gad.943102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan J.T. Hollis S.E., et al. Post-transcriptional regulation of the Ras-ERK/MAPK signaling pathway. J Cell Physiol. 2011 doi: 10.1002/jcp.22899. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Wu F. Garcia J., et al. tat regulates binding of the human immunodeficiency virus trans-activating region RNA loop-binding protein TRP-185. Genes Dev. 1991;5:2128–2140. doi: 10.1101/gad.5.11.2128. [DOI] [PubMed] [Google Scholar]
- Wu H. Min J., et al. Crystal structure of the methyltransferase domain of human TARBP1. Proteins. 2008;72:519–525. doi: 10.1002/prot.22065. [DOI] [PubMed] [Google Scholar]
- Wu P. Brockenbrough J.S., et al. NCL1, a novel gene for a non-essential nuclear protein in Saccharomyces cerevisiae. Gene. 1998;220:109–117. doi: 10.1016/s0378-1119(98)00330-8. [DOI] [PubMed] [Google Scholar]
- Wu-Baer F. Lane W.S., et al. Identification of a group of cellular cofactors that stimulate the binding of RNA polymerase II and TRP-185 to human immunodeficiency virus 1 TAR RNA. J Biol Chem. 1996;271:4201–4208. doi: 10.1074/jbc.271.8.4201. [DOI] [PubMed] [Google Scholar]