Abstract
Eukaryotic cilia and flagella are motile organelles built on a scaffold of doublet microtubules and powered by dynein ATPase motors. Some thirty years ago, two competing views were presented to explain how the complex machinery of these motile organelles had evolved. Overwhelming evidence now refutes the hypothesis that they are the modified remnants of symbiotic spirochaete-like prokaryotes, and supports the hypothesis that they arose from a simpler cytoplasmic microtubule-based intracellular transport system. However, because intermediate stages in flagellar evolution have not been found in living eukaryotes, a clear understanding of their early evolution has been elusive. Recent progress in understanding phylogenetic relationships among present day eukaryotes and in sequence analysis of flagellar proteins have begun to provide a clearer picture of the origins of doublet and triplet microtubules, flagellar dynein motors, and the 9+2 microtubule architecture common to these organelles. We summarize evidence that the last common ancestor of all eukaryotic organisms possessed a 9+2 flagellum that was used for gliding motility along surfaces, beating motility to generate fluid flow, and localized distribution of sensory receptors, and trace possible earlier stages in the evolution of these characteristics.
Keywords: cilia, flagella, motility, axoneme, microtubule, dynein, intraflagellar transport, central pair, radial spoke, tubulin
Evidence for the presence of a 9+2, motile, sensory organelle in the last common eukaryotic ancestor
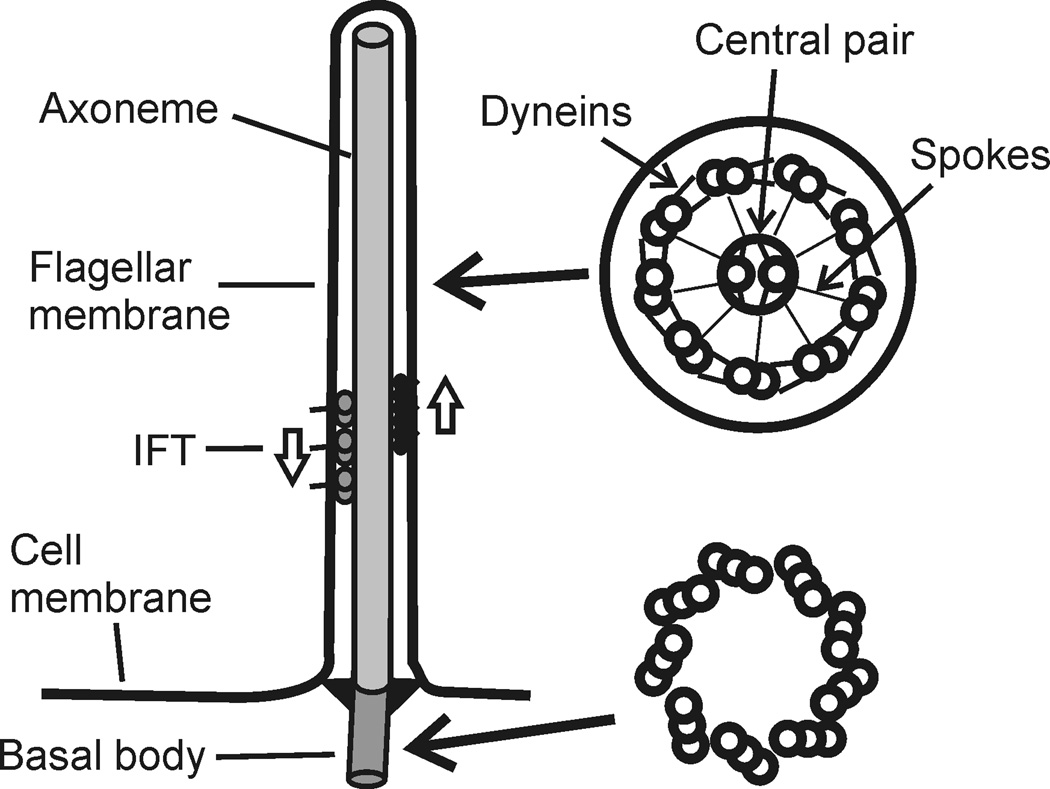
As summarized in Figure 1, typical cilia and flagella (hereafter called flagella, there being no consistent structural or functional difference between organelles with these two designations) are motile projections oriented perpendicular to the cell surface, but they vary in length, in number per cell, and in the patterns of motility that they produce. They are composed of a cylinder (the axoneme) of nine doublet microtubules surrounding two single microtubules and are covered by the cell membrane. Between each pair of flagellar doublets are rows of axonemal dynein ATPases, which power the bending of these organelles, and extending toward the center of the cylinder are radial spokes, which touch upon a central apparatus and regulate axonemal dyneins. This central element consists of two single microtubules, assembled from a unique nucleating site1, plus many interconnecting microtubule-associated proteins (reviewed in ref. 2). Together they form a structure that provides a cylindrical surface apposing the ends of the radial spokes3.
Figure 1.
Diagram of structures common to all motile cilia and flagella. Longitudinal view to the left shows the relationship between the axoneme and basal body, and the location of intraflagellar transport (IFT) motors between axonemal doublet microtubules and the flagellar membrane. Transition fibers attached to the basal body separate the flagellar membrane domain from the rest of the cell membrane. Cross sectional views to the right show structures in flagella, including the nine outer doublet and two single central pair microtubules (top) and the nine triplet microtubules of basal bodies (bottom).
The nine doublets assemble from a much shorter cylinder of nine triplet microtubules, the basal body or centriole, which is anchored to the cell surface and stabilized in the cytoplasm by other cytoskeletal elements. Basal bodies that anchor flagella are often interchangeable during the cell cycle with centrioles4, and these two names should be considered as two functional descriptions for the same structure. Between the doublets and the membrane are particles associated with intraflagellar transport (IFT), a process important for flagellar assembly and protein trafficking in this cellular compartment5,6. In the outward (anterograde) direction, IFT is powered by kinesins of the kinesin2 family; in the inward (retrograde) direction, power is provided by dyneins of the cytoplasmic dynein 2 (DHC1b) family. Many flagella act as sensory antennae through localization of receptors to the flagellar membrane. In extreme cases, termed primary cilia or sensory cilia, the motile function has been discarded and with it the dyneins, radial spokes, the central pair complex, and other proteins needed for bend formation. IFT is still required for the assembly and maintenance of these primary cilia, which play important sensory roles in metazoan organisms6.
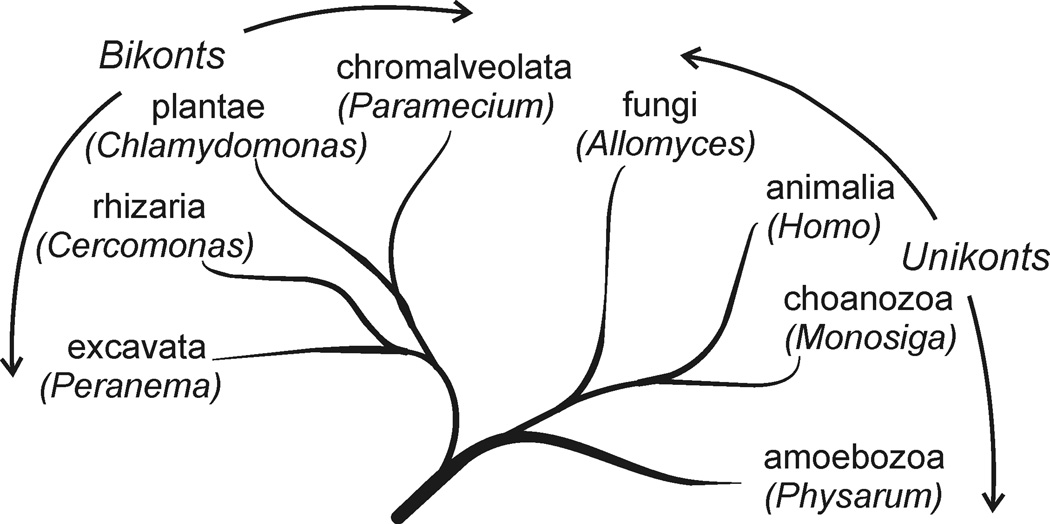
Some attempts have been made in the past to identify intermediates in the evolution of flagella by looking within existing branches of eukaryotes for organisms that may have diverged before the complete 9+2 flagellum had evolved. However, improved methods of analysis and the recent burst of sequence data are rapidly transforming long-held views of eukaryotic phylogeny to new schemes in which there are many branches that diverged within a relatively short period of time7,8 (Fig. 2). Many of these branches are represented today by single-celled protists, so that the true diversity of eukaryotes cannot be appreciated without some understanding of the relationships among these often less-studied organisms. Thus the nearest relatives of animals (metazoans) are single-celled choanoflagellates9,10. Fungi (some of which were once considered primitive because of their simplicity) turn out to be another twig of this same branch, the opisthokonts or unikonts. Allomyces, a chytridiomycete fungus with flagellated gametes and zoospores, is just one example of a fungal cell that swims, like a sperm, with a typical 9+2 flagellum. Amoebozoa, containing such amoeboflagellates as Physarum, are probably (based on a shared gene fusion and on mitochondrial sequences) the only other unikont twig, and branch somewhat earlier than fungi10–13. The other major superclade of eukaryotes, the bikonts, encompasses a great variety of flagellated and amoeboid organisms, including green plants and green and red algae (plantae), ciliates, dinoflagellates and their kin (chromalveolata), euglenids, trypanosomatids, diplomonads and their sister taxa (excavata), and the radiolaria, cercozoa, etc. (rhizaria)14,15. In the resulting tree (Fig. 2), one should note that most of the model organisms under intense study during the past twenty years reside on one branch (unikont), but fortunately for studies of flagellar evolution, additional attention has been focused on a few flagellated bikont organisms, most especially the green alga Chlamydomonas reinhardtii.
Figure 2.
Diagram of probable evolutionary divergence that generated all existing branches of eukaryotic organisms. Under the name of each branch or clade is a the name of a representative genus in that clade that contains species with typical motile 9+2 flagella. Based on recent studies of rare gene fusion events, as well as more traditional sequence comparisons, the entire tree is divided into two superclades, unikonts and bikonts.
One striking conclusion of these recent phylogenetic studies is that every extant branch of eukaryotes includes organisms with motile, 9+2 flagella. Even proteins of the central pair apparatus, such as products of the Chlamydomonas PF6, PF16, PF20, KLP1, and CPC1 genes, have been conserved between algae and humans16. From this we can only conclude that these organelles had evolved prior to the divergence of all extant eukaryotic clades from a common ancestor. In addition, IFT proteins, which are central to flagellar assembly and to the display of sensory receptors and flagellar surface motility, are also present in flagella from distant branches of eukaryotic phylogeny (e.g. trypanosomes17, insects18, and green algae19), and therefore must have evolved prior to the beginnings of eukaryotic radiation18. The microtubule rootlet structures that stabilize basal bodies in the cytoplasm do vary phylogenetically and therefore the nature of those that might have been present in the last common eukaryotic ancestor are difficult to determine20,21, but all of the elements of triplet microtubules, and the accessory proteins needed for basal body formation, must also have been present at the base of this tree.
Eukaryotes likely developed the nucleus, endomembrane system, and cytoskeleton, and then used the phagocytic ability that was provided by the combined cytoskeletal and endomembrane systems to obtain the precursors to mitochondria, long before the evolution of flagella. The framework of doublet microtubules upon which flagella depend must have evolved from simpler single microtubules which, as essential elements of the mitotic machinery, would have been needed to segregate a genome enclosed in a nucleus. Likewise, dyneins as microtubule-based motors undoubtedly functioned as transport motors on cytoplasmic and mitotic microtubules long before their use was adapted to flagella. If all of the essential elements of eukaryotic cells were in place for so long before the advent of flagella, one must ask why there are no branches of existing eukaryotes that lack flagella. The simplest explanation is that the branch of early eukaryotes that first developed a functional 9+2 flagellum possessed a tremendous selective advantage over its competitors, and was the only eukaryote whose descendants survive today.
Evolution of tubulin, dynein and kinesin
The closest prokaryotic relative of tubulins, FtsZ, functions during bacterial septation, and FtsZ homologs continue to perform a similar role in chloroplasts and perhaps some mitochondria22–24. In early eukaryotes, FtsZ gene duplication and modification led to alpha and beta tubulin, which form a stable dimer that retains FtsZ properties such as polymerization, GTP-binding, and GTP hydrolysis-dependent conformational change, but which gained the added ability to interact laterally to form tubes. Gamma tubulin is also ubiquitous and likely emerged early, to function as a nucleating site that helps determine microtubule polarity and distribution. Additional tubulin isoforms delta and epsilon are ubiquitous among organisms with triplet microtubules, and the formation of triplet microtubules, essential for the function of basal bodies, has been shown to require both delta and epsilon tubulin in Chlamydomonas25,26 and both epsilon27 and the less ubiquitous eta tubulin28 in Paramecium. The presence of these tubulin isoforms in members of both the unikont and bikont clades17,29 argues for their evolution prior to the divergence of eukaryotes.
The dynein motors that power both flagellar beating and retrograde IFT movements are members of the superfamily of AAA ATPases. In dyneins, six individual AAA domains have become fused into a single large polypeptide30, but only four of these six domains in dyneins retain the signature sequences of nucleotide binding pockets31. DNA pumping ATPases of archaea (HerA) and bacteria (FtsK), also AAA ATPases, are needed during prokaryotic cell division for correct daughter chromosome segregation32, and it would be tempting to assume that an interaction between FtsK and FtsZ could have evolved directly into an interaction between dynein and tubulin. However, dyneins apparently evolved from an entirely different branch of the AAA superfamily from the DNA pumping ATPases. The closest eukaryotic relatives of dyneins are midasins, similarly giant eukaryotic AAA ATPases, which function at the nuclear pore in 60S ribosome export. The closest prokaryotic homologs of dyneins and midasins are members of the MoxR family, single AAA domain ATPases that function as chaperones in the assembly of large protein complexes, such as methanol dehydrogenase and nitric oxide reductase30. It would thus appear that dyneins evolved as microtubule motors in the early eukaryotic lineage, and that their prokaryotic ancestors were proteins that performed conformational work linked to ATP binding and hydrolysis.
Sequence comparisons among extant dynein heavy chains divide dyneins into two broad families, cytoplasmic and axonemal33. Most organisms have only two cytoplasmic dyneins, one devoted to general cytoplasmic microtubule-based movements, present in all eukaryotes, and one for retrograde IFT movement on axonemal microtubules, absent from organisms such as Saccharomyces cerevisiae that lack axonemes. Each is thought to function as a homodimer of catalytic heavy chains. Organisms with motile flagella also have a large family of axonemal dyneins that can be further divided into three subfamilies, outer row dyneins, I1 inner row dyneins, and additional diverse inner row dyneins (reviewed in ref. 34). Outer row dyneins diverged before the common ancestor into two isoforms (alpha and beta) that form heavy chain heterodimers. A third isoform that diverged more recently is found in Chlamydomonas (plantae) and Tetrahymena (chromalveolata) but not in sea urchins or fruit flies (animalia). Outer row dyneins bind in a continuous row with 24 nm spacing (every third 8 nm tubulin dimer), whereas inner row dynein isoforms occur once every 96 nm along each doublet microtubule35. The I1 inner row dynein is a typical heterodimer of two heavy chain subunits, and both isoform subfamilies are represented in the genomes of all organisms that retain motile flagella. I1 dyneins appear to have become established early as major targets of signal-dependent regulation of flagellar bending parameters36. The many additional inner row dyneins present in, for example, sea urchins and ciliates, appear to have diverged more recently; several of the ciliate inner row dyneins are more closely related to each other than to any of the urchin inner row dyneins. Structural, genetic and biochemical analyses in Chlamydomonas indicate that these additional inner row dyneins function as monomers, rather than the dimers typical of all other dyneins, and some isoforms may be differentially distributed along the length of the organelle37. Their sequence relationships suggest that the last common eukaryote may have had a single isoform of this monomeric inner row dynein.
Among the members of the very large and diverse superfamily of kinesin ATPases are at least two families with members that function as flagellar proteins. The small kinesin9 family is represented by a sequence expressed in ciliated cells of mammals38 and by the Chlamydomonas Klp1 protein39. Klp1 has been localized to the central pair complex39 and shown to be important for normal flagellar motility in that organism40. Although evidence for the role of kinesin9 members in mammalian cells is not available, their expression patterns support an early evolving flagellar function for this protein. The kinesin2 family is larger and functionally more diverse. While some members of the kinesin2 family are anterograde motors for IFT, others are anterograde motors in other cytoplasmic compartments such as neurons of metazoans. The presence of kinesin2 homologs in ciliates and flagellates, but not in non-flagellated fungi, suggests that kinesin2 co-evolved with axonemes and was only co-opted for other transport functions in recent metazoan evolution6.
The origins of 9+2 flagella
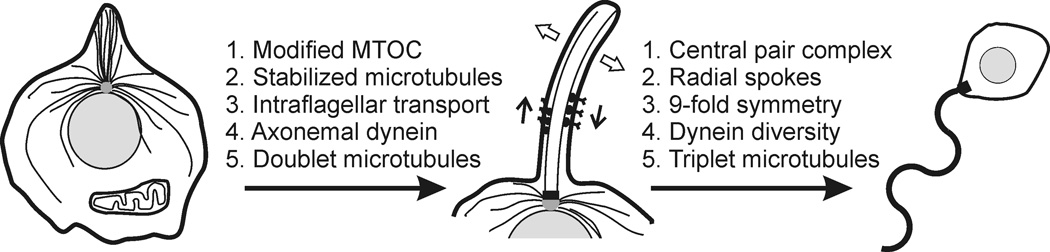
Given the evidence summarized above, the last common eukaryotic ancestor had a motile 9+2 flagellum, was anchored on a basal body of triplet microtubules, and required IFT for its assembly. How did such a complex system evolve? Clearly it must have been preceded by a microtubule cytoskeleton with dynein and kinesin motors. Strong arguments have been made that the driving force for the evolution of a microtubule cytoskeleton and its associated motor proteins was the ability to accurately segregate a large, nuclear membrane-enclosed genome by mitosis. Although the complicated checks and balances used to assure mitotic fidelity vary widely in extant eukaryotes, some aspects of mitosis have been sufficiently conserved and are so central to the process that they must have been present at an early stage. Other aspects of mitosis, assumed to be ancient because of their simplicity, reveal a minimal apparatus but may not reflect the ancestral condition. Although model organisms such as fungi have provided many details of such minimal systems, the tremendous variety of extant mitotic mechanisms (as reviewed in ref. 41) should not be forgotten. Mitosis requires a microtubule organizing center (MTOC) that duplicates once per cell cycle, a connection between each chromatid and one of the duplicated MTOCs, and separation of the MTOCs with their associated chromatids. These MTOCs vary structurally from the simple nuclear membrane-embedded spindle pole bodies of some unicellular organisms to the complex centriole-containing centrosomes of many metazoan cells. Most organisms form two microtubule arrays during mitosis, one oriented toward the chromosomes to link each chromatid to its MTOC, and a second that assembles between the two MTOCs to form a scaffold for MTOC separation. In G1 phase of the cell cycle, prior to DNA and MTOC duplication, the primitive cell would have had a single MTOC, with a single array of microtubules directed away from the nucleus that defined the polarity of the cell. It is this cytoplasmic microtubule array that most likely provided the raw material for evolution of the flagellar axoneme42–44.
A polarized array of microtubules projecting from one side of the nucleus, as seen in most cells today, need only become linked into a bundle to provide an organelle that could distort the cell membrane and form a protoflagellum (Fig. 3). Microtubules radiating from the MTOC that were not incorporated into this protoflagellar bundle would continue to provide a cytoskeleton for general cytoplasmic transport and organization of the endomembrane system, and the interaction of motors such as kinesin and dynein with this microtubule cytoskeleton would provide directed motility of associated vesicles. Such movement along the protoflagellar bundle could direct exocytosis and endocytosis to a specific region of the cell membrane, creating a new membrane domain. The similarity of IFT proteins to proteins involved in vesicle trafficking18 and the similarity of IFT kinesin and dynein to cytoplasmic versions of these motors, argues that axonemes evolved from proteins that were already in use in microtubule-based vesicular transport systems.
Figure 3.
Proposed steps in the transition from an early eukaryote, with a polarized morphology based on asymmetric placement of a microtubule organizing center, but lacking flagella (left), through an intermediate with a protoflagellum that supported gliding and limited bending (center), to the last common eukaryotic ancestor, with a fully developed, motile 9+2 flagellum (right).
Early eukaryotes, lacking any other means of locomotion, were presumably benthic amoeboid cells and could not yet swim. Microtubule-based motors moving along a parallel bundle of microtubules in the protoflagellum would have provided at least two specific advantages to these organisms. Simple coupling of retrograde movement to transmembrane proteins would convert the protoflagellum to a feeding organelle, bringing particles that adhered to its surface back toward the cell body by retrograde IFT for subsequent phagocytosis. Alternatively, if substrate adhesion through IFT-associated protoflagellar transmembrane proteins was strong, and cell body adhesion proportionately weak, retrograde IFT could support gliding motility. Flagellar gliding as a means of locomotion is common in many pelagic, benthic and soil flagellates today, and such surface motility has also been described for metazoan cilia, suggesting that it was either an early adaptation of the IFT system, or one that has happened repeatedly during subsequent evolution44. As the coupling mechanisms between flagellar adhesion molecules and gliding motors have not been widely studied, their evolutionary history remains unknown.
Movement of secretory vesicles along a polarized microtubule bundle would also provide a polarized distribution of cell surface molecules, including receptors. While the localization of receptors to ciliary and flagellar surfaces has been documented in both protists and in metazoan sensory cilia, including chemosensory cilia in C. elegans, kidney cilia in vertebrates, and the highly modified cilia in sensory neurons of the vertebrate retina, evidence that specific receptors have been localized to this membrane domain since before eukaryotic divergence is only fragmentary (reviewed in ref. 44). Receptor localization does not require the complex structure of a flagellum, but the ability to define a polarized cell surface domain for sensory signaling may have been one of the driving forces in early flagellar evolution.
Flagella are first and foremost organelles that beat, and their complex structure would not have evolved without strong selection for motility, as witnessed by the rapid loss of much of this complexity in non-motile sensory cilia, and the complete loss of flagella in non-motile organisms such as yeasts. However strong this advantage of a beating flagellum might have been to early eukaryotes, intermediate stages must have existed that provided intermediate levels of motion; a sudden jump from a benthic, amoeboid or gliding organism to one that can swim by flagellar beating is not plausible. To understand the advantages of less vigorous bending motility, one need only look at flagellar function among existing flagellates. Many organisms use flagella to generate feeding currents that increase the frequency with which food particles (bacteria, other eukaryotes, or detritus) can be ingested, while others use flagellar movements to aid in trapping food particles45,46. Feeding currents are common in organisms such as choanoflagellates, which attach to the substrate with stalks and use flagellar beating to create currents past the stationary cell, in mastigamoebae, which create currents while continuing to move by amoeboid activity, and in many biflagellates such as bodonids, which create currents with an anterior flagellum and glide on a posterior flagellum. Even a modest ability to vibrate or wave a protoflagellum could have provided the initial selective advantage that drove further development of single microtubules into doublet microtubules, and favored diversification of dyneins that could take advantage of this new doublet microtubule track (Fig. 3).
Along with increased motility came the increased need to anchor the axoneme, which may have driven both centriole/basal body evolution and the development of links between the flagellar base and the cell membrane. These links would segregate the flagellar compartment from the rest of the cytoplasm, requiring further refinements in the IFT sorting/trafficking mechanism so that standard vesicle fusion occurred at the flagellar base, and IFT movement transported both membrane and non-membrane components to and from the flagellar compartment. In addition, such links create a boundary between the flagellar membrane and the rest of the plasma membrane, sequestering receptors and adhesion molecules into a unique membrane domain.
The ability to bend does not require an axoneme with 9-fold symmetry, and many axonemes have been discovered that depart from this pattern, yet most of these departures appear to be more recent modifications of an ancestral 9+2 pattern. As argued in more detail elsewhere, I propose that motility regulation by a central apparatus provided a strong selective advantage to the organism in which it evolved, and that the most successful regulatory mechanism was based on an apparatus built on a scaffold of two central microtubules, with regulatory signals transmitted through radial spokes of a defined length44,47. The geometry of this regulatory mechanism presumably favors an outer cylinder of precisely nine doublet microtubules, with the distance between doublets determined by the reach of dyneins that must span each interdoublet gap. Whether central pair regulation was based on a fixed central pair orientation, as found today in metazoans such as bivalves48, sea urchins49, and ctenophores50 and possibly in excavates such as euglenids51, or a rotating central pair52, as commonly found in green algae such as Chlamydomonas53 and Micromonas54, and chromalveolates such as Paramecium55 and Synura56, remains to be determined. Central pair rotation may allow regulation of bends in different beat planes, and therefore be a more flexible regulatory system for organisms whose survival is most highly dependent on rapid changes in flagellar beat parameters47,53. The origin of radial spokes remains, at this time, one of the greater mysteries of flagellar evolution, as related proteins have not been identified in other microtubule-associated regulatory complexes.
Diversification of flagellar structure and function during eukaryotic radiation
Many changes to the basic (if complex) 9+2 flagellum that was present in the last common eukaryotic ancestor are seen in some present day organisms, whereas others appear to have retained the original model with few alterations. Changes include additions, such as mastigonemes that project from the membrane surface and increase effective hydrodynamic resistance, and accessory structures that increase axoneme stiffness (paraflagellar rods in euglenoids, outer dense fibers or extra microtubules in spermatozoa; for a more extensive survey of structural changes in spermatozoa, see Baccetti, ref. 57). Simplifications or deletions of structures no longer used by an organism or cell type include loss of the outer row dyneins, loss of the central apparatus and radial spokes (motile 9+0), loss of the central apparatus, radial spokes, and some doublet microtubules (6+0, 3+0). In the case of strictly non-motile sensory cilia, all components necessary for motility (central pair, spokes, dyneins) may be absent, leaving only the membrane and the nine doublet microtubules to support IFT and receptor localization. Motile flagella that lack the central pair and radial spokes appear to have a simplified bending pattern that only accommodates helical bending waves, and are found on parasitic flagellates that do not depend on flagellar motility for locomotion in a complex environment58, on nodal cilia in early vertebrate embryos, where they generate a unidirectional fluid current essential for establishing left-right asymmetry59, and on certain vertebrate spermatozoa60. Some insect sperm that lack central pair microtubules have cylinders of 12 or 14 doublets, or spirals of hundreds of doublets57, suggesting further that the original standard of a cylinder of nine doublet microtubules was selected to accommodate the radial spoke-central pair regulatory complex, and that if the central pair is not needed, successful axonemes can evolve with alternative doublet patterns.
More difficult to catalog are modifications of existing parts to meet new demands, often observable not at the structural level but as differences in average length, beat frequency, or waveform, or as the ability to change beat frequency, beat direction, or waveform in response to signaling cascades. These signaling pathways may in turn begin with stimulation of flagellar surface receptors, or with cascades that are transmitted from elsewhere in the cell. The evolution of some of these changes, especially those affecting signaling pathways, will be difficult to trace until many more genomes have been sequenced, and proteomic analysis confirms the location and function of putative flagellar gene products. Some changes probably occurred only once, and surveying the distribution of organisms that retain such features may clarify phylogenetic relationships. Other changes have occurred independently more than once and can be considered convergent evolution. For example, the ability to generate ATP within the flagellar compartment has evolved in several independent ways. Glycolysis is an important source of energy for sperm motility in mammals61 where fermentable sugars come directly from seminal fluid61,62, but has not been reported in vertebrate cilia and flagella other than sperm tails and some types of non-motile cilia (e.g. the outer segments of mammalian photoreceptor cells63). Mammalian sperm-specific isoforms of glycolytic enzymes such as glyceraldehyde 3-phosphate dehydrogenase64 and enolase65 have likely evolved recently, as the glycolytic enzymes identified in Chlamydomonas flagella66 are not closely related and appear to have been targeted for flagellar use specifically in algae. Completely different methods of flagellar ATP generation occur in other organisms, including phosphocreatine/creatine phosphokinase shuttles in sea urchin67 and mammalian68 spermatozoa, and in chicken photoreceptor outer segments69, and a phosphoarginine/arginine phosphokinase shuttle in Paramecium cilia70. Overall, the basic motile machinery of 9+2 organelles appears to be ancient and highly conserved, whereas the signaling cascades that regulate motility and accessory structures that modify its output have changed to suit specific organismal needs.
Summary
Typical 9+2 flagella likely evolved from bundling and extension of cytoplasmic microtubules that assembled on a microtubule organizing center and that generated a polarized cellular morphology. Close apposition of the plasma membrane created a separate membrane domain that could be used to localize receptors for sensory signal transduction, and required simultaneous evolution of intraflagellar transport to maintain this polarized structure. Membrane-associated IFT-based movement provided a mechanism for gliding motility, and addition of axonemal dynein motors allowed this extension to bend and generate currents past the cell. Formation of doublet microtubules allowed elaboration of dyneins to improve motility, and the addition of the radial spoke-central pair regulatory system provided responsive dynein control. The strong selective advantage of a motile 9+2 flagellum may have resulted in rapid diversification of the last common eukaryotic ancestor into all existing branches of eukaryotic organisms.
Acknowledgements
Robert Bloodgood contributed through thoughtful discussions on flagellar gliding, David Asai shared recent results on dynein evolution, and Gaspar Jekely provided insight into the evolutionary connections between IFT and vesicle trafficking.
Reference List
- 1.McKean PG, Baines A, Vaughan S, Gull K. Gamma-tubulin functions in the nucleation of a discrete subset of microtubules in the eukaryotic flagellum. Curr Biol. 2003;13(7):598–602. doi: 10.1016/s0960-9822(03)00174-x. [DOI] [PubMed] [Google Scholar]
- 2.Smith EF, Yang P. The radial spokes and central apparatus: mechano-chemical transducers that regulate flagellar motility. Cell Motil Cytoskeleton. 2004;57:8–17. doi: 10.1002/cm.10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mitchell DR. Reconstruction of the projection periodicity and surface architecture of the flagellar central pair complex. Cell Motil Cytoskeleton. 2003;55:188–199. doi: 10.1002/cm.10121. [DOI] [PubMed] [Google Scholar]
- 4.Beisson J, Wright M. Basal body/centriole assembly and continuity. Curr Opin Cell Biol. 2003;15(1):96–104. doi: 10.1016/s0955-0674(02)00017-0. [DOI] [PubMed] [Google Scholar]
- 5.Rosenbaum J. Intraflagellar transport. Curr Biol. 2002;12(4):R125. doi: 10.1016/s0960-9822(02)00703-0. [DOI] [PubMed] [Google Scholar]
- 6.Scholey JM. Intraflagellar transport. Annu Rev Cell Dev Biol. 2003;19:423–443. doi: 10.1146/annurev.cellbio.19.111401.091318. [DOI] [PubMed] [Google Scholar]
- 7.Simpson AG, Roger AJ. The real 'kingdoms' of eukaryotes. Curr Biol. 2004;14(17):R693–R696. doi: 10.1016/j.cub.2004.08.038. [DOI] [PubMed] [Google Scholar]
- 8.Baldauf SL. The deep roots of eukaryotes. Science. 2003;300(5626):1703–1706. doi: 10.1126/science.1085544. [DOI] [PubMed] [Google Scholar]
- 9.Cavalier-Smith T, Chao EE. Phylogeny of choanozoa, apusozoa, and other protozoa and early eukaryote megaevolution. J Mol Evol. 2003;56(5):540–563. doi: 10.1007/s00239-002-2424-z. [DOI] [PubMed] [Google Scholar]
- 10.Lang BF, O'Kelly C, Nerad T, Gray MW, Burger G. The closest unicellular relatives of animals. Curr Biol. 2002;12(20):1773–1778. doi: 10.1016/s0960-9822(02)01187-9. [DOI] [PubMed] [Google Scholar]
- 11.Bapteste E, Gribaldo S. The genome reduction hypothesis and the phylogeny of eukaryotes. Trends Genet. 2003;19(12):696–700. doi: 10.1016/j.tig.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 12.Stechmann A, Cavalier-Smith T. Phylogenetic analysis of eukaryotes using heat-shock protein Hsp90. J Mol Evol. 2003;57(4):408–419. doi: 10.1007/s00239-003-2490-x. [DOI] [PubMed] [Google Scholar]
- 13.Steenkamp ET, Baldauf SL. Origin and evolution of animals, fungi and their unicellular allies (Opisthokonta) In: Hirt RP, Horner DS, editors. Organelles, genomes and eukaryote phylogeny. Boca Raton: CRC Press; 2004. pp. 109–129. [Google Scholar]
- 14.Nikolaev SI, Berney C, Fahrni JF, Bolivar I, Polet S, Mylnikov AP, et al. The twilight of Heliozoa and rise of Rhizaria, an emerging supergroup of amoeboid eukaryotes. Proc Natl Acad Sci U S A. 2004;101(21):8066–8071. doi: 10.1073/pnas.0308602101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cavalier-Smith T, Chao EEY. Molecular phylogeny of centrohelid heliozoa, a novel lineage of bikont eukaryotes that arose by ciliary loss. J Mol Evol. 2003;56(4):387–396. doi: 10.1007/s00239-002-2409-y. [DOI] [PubMed] [Google Scholar]
- 16.Pazour GJ, Agrin N, Leszyk J, Witman GB. Proteomic analysis of a eukaryotic cilium. J Cell Biol. 2005;170:103–113. doi: 10.1083/jcb.200504008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309(5733):416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- 18.Avidor-Reiss T, Maer AM, Koundakjian E, Polyanovsky A, Keil T, Subramaniam S, et al. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell. 2004;117(4):527–539. doi: 10.1016/s0092-8674(04)00412-x. [DOI] [PubMed] [Google Scholar]
- 19.Li JB, Gerdes JM, Haycraft CJ, Fan Y, Teslovich TM, May-Simera H, et al. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 2004;117(4):541–552. doi: 10.1016/s0092-8674(04)00450-7. [DOI] [PubMed] [Google Scholar]
- 20.Cavalier-Smith T. The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int J Syst Evol Microbiol. 2002;52(Pt 2):297–354. doi: 10.1099/00207713-52-2-297. [DOI] [PubMed] [Google Scholar]
- 21.Moestrup O. The flagellate cytoskeleton. In: Leadbeater BCS, Green J, editors. The flagellates. London: Taylor and Francis; 2000. pp. 69–94. [Google Scholar]
- 22.McKean PG, Vaughan S, Gull K. The extended tubulin superfamily. J Cell Sci. 2001;114(15):2723–2733. doi: 10.1242/jcs.114.15.2723. [DOI] [PubMed] [Google Scholar]
- 23.Amos LA, van den EF, Lowe J. Structural/functional homology between the bacterial and eukaryotic cytoskeletons. Curr Opin Cell Biol. 2004;16(1):24–31. doi: 10.1016/j.ceb.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 24.Gitai Z. The new bacterial cell biology: Moving parts and subcellular architecture. Cell. 2005;120(5):577–586. doi: 10.1016/j.cell.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 25.Dutcher SK. The tubulin fraternity: alpha to eta. Curr Opin Cell Biol. 2001;13:49–54. doi: 10.1016/s0955-0674(00)00173-3. [DOI] [PubMed] [Google Scholar]
- 26.Dutcher SK. Elucidation of Basal Body and Centriole Functions in Chlamydomonas reinhardtii. Traffic. 2003;4(7):443–451. doi: 10.1034/j.1600-0854.2003.00104.x. [DOI] [PubMed] [Google Scholar]
- 27.Ruiz F, Krzywicka A, Klotz C, Keller A, Cohen J, Koll F, et al. The SM19 gene, required for duplication of basal bodies in Paramecium, encodes a novel tubulin, eta-tubulin. Curr Biol. 2000;10(22):1451–1454. doi: 10.1016/s0960-9822(00)00804-6. [DOI] [PubMed] [Google Scholar]
- 28.Dupuis-Williams P, Fleury-Aubusson A, de Loubresse NG, Geoffroy H, Vayssie L, Galvani A, et al. Functional role of epsilon-tubulin in the assembly of the centriolar microtubule scaffold. J Cell Biol. 2002;158(7):1183–1193. doi: 10.1083/jcb.200205028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dutcher SK. Long-lost relatives reappear: identification of new members of the tubulin superfamily. Curr Opin Microbiol. 2003;6(6):634–640. doi: 10.1016/j.mib.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 30.Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J Struct Biol. 2004;146(1–2):11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 31.Asai DJ, Koonce MF. The dynein heavy chain: structure, mechanics and evolution. TICB. 2001;11(5):196–202. doi: 10.1016/s0962-8924(01)01970-5. [DOI] [PubMed] [Google Scholar]
- 32.Iyer LM, Makarova KS, Koonin EV, Aravind L. Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res. 2004;32(17):5260–5279. doi: 10.1093/nar/gkh828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gibbons BH, Asai DJ, Tang W-JY, Hays TS, Gibbons IR. Phylogeny and expression of axonemal and cytoplasmic dynein genes in sea urchins. Mol Biol Cell. 1994;5:57–70. doi: 10.1091/mbc.5.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Asai DJ, Wilkes DE. The dynein heavy chain family. J Euk Microbiol. 2004;51(1):23–29. doi: 10.1111/j.1550-7408.2004.tb00157.x. [DOI] [PubMed] [Google Scholar]
- 35.Porter ME. Axonemal dyneins: Assembly, organization, and regulation. Curr Opin Cell Biol. 1996;8:10–17. doi: 10.1016/s0955-0674(96)80042-1. [DOI] [PubMed] [Google Scholar]
- 36.Porter ME, Sale WS. The 9+2 axoneme anchors multiple inner arm dyneins and a network of kinases and phosphatases that control motility. J Cell Biol. 2000;151(5):F37–F42. doi: 10.1083/jcb.151.5.f37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mitchell DR. Chlamydomonas flagella. J Phycol. 2000;36:261–273. [Google Scholar]
- 38.Miki H, Setou M, Hirokawa N. Kinesin superfamily proteins (KIFs) in the mouse transcriptome. Genome Res. 2003;13(6B):1455–1465. doi: 10.1101/gr.984503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bernstein M, Beech PL, Katz SG, Rosenbaum JL. A new kinesin-like protein (Klp1) localized to a single microtubule of the Chlamydomonas flagellum. J Cell Biol. 1994;125:1313–1326. doi: 10.1083/jcb.125.6.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yokoyama R, O'Toole E, Ghosh S, Mitchell DR. Regulation of flagellar dynein by a central pair kinesin. Proc Natl Acad Sci U S A. 2004;101:17398–17403. doi: 10.1073/pnas.0406817101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kubai DF. The evolution of the mitotic spindle. Int Rev Cytol. 1975;43:167–227. doi: 10.1016/s0074-7696(08)60069-8. [DOI] [PubMed] [Google Scholar]
- 42.Cavalier-Smith T. The evolutionary origin and phylogeny of eukaryote flagella. Symp Soc Exp Biol. 1982;35:465–493. [PubMed] [Google Scholar]
- 43.Cavalier-Smith T. The evolutionary origin and phylogeny of microtubules, mitotic spindles and eukaryote flagella. Biosystems. 1978;10(1–2):93–114. doi: 10.1016/0303-2647(78)90033-3. [DOI] [PubMed] [Google Scholar]
- 44.Mitchell DR. Speculations on the evolution of 9+2 organelles and the role of central pair microtubules. Biol Cell. 2004;96(9):691–696. doi: 10.1016/j.biolcel.2004.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arndt H, Dietrich D, Auer B, Cleven E-J, Grafenhan T, Weitere M, et al. Functional diversity of heterotrophic flagellates in aquatic ecosystems. In: Leadbeater BCS, Green J, editors. The flagellates. London: Taylor and Francis; 2000. pp. 240–268. [Google Scholar]
- 46.Sleigh MA. Trophic strategies. In: Leadbeater BCS, Green J, editors. The flagellates. London: Taylor and Francis; 2000. pp. 147–165. [Google Scholar]
- 47.Mitchell DR. Regulation of eukaryotic flagellar motility. Am Inst Phys Conf Proc. 2005;555:130–136. [Google Scholar]
- 48.Gibbons IR. The relationship between the fine structure and direction of beat in gill cilia of a lamellibranch mollusc. J Biophys Bioch Cyt. 1961;11:179–205. doi: 10.1083/jcb.11.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sale WS. The axonemal axis and calcium-induced asymmetry of active microtubule sliding in sea urchin sperm tails. J Cell Biol. 1986;102:2042–2052. doi: 10.1083/jcb.102.6.2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tamm SL, Tamm S. Ciliary reversal without rotation of axonemal structures in ctenophore comb plates. J Cell Biol. 1981;89:495–509. doi: 10.1083/jcb.89.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Melkonian M, Preisig HR. Twist of central pair microtubules in the flagellum of the green flagellate Scourfieldia caeca. Cell Biol Int Rep JID - 7708050. 1982;6(3):269–277. doi: 10.1016/0309-1651(82)90079-0. [DOI] [PubMed] [Google Scholar]
- 52.Omoto CK, Gibbons IR, Kamiya R, Shingyoji C, Takahashi K, Witman GB. Rotation of the central pair microtubules in eukaryotic flagella. Mol Biol Cell. 1999;10(1):1–4. doi: 10.1091/mbc.10.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mitchell DR, Nakatsugawa M. Bend propagation drives central pair rotation in Chlamydomonas reinhardtii flagella. J Cell Biol. 2004;166(5):709–715. doi: 10.1083/jcb.200406148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Omoto CK, Witman GB. Functionally significant central-pair rotation in a primitive eukaryotic flagellum. Nature. 1981;290:708–710. doi: 10.1038/290708a0. [DOI] [PubMed] [Google Scholar]
- 55.Omoto CK, Kung C. Rotation and twist of the central-pair microtubules in the cilia of Paramecium. J Cell Biol. 1980;87(1):33–46. doi: 10.1083/jcb.87.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jarosch R, Fuchs B. On the rotation of fibrils in the synura-flagellum (author's transl) Protoplasma. 1975;85(2–4):285–290. doi: 10.1007/BF01567953. [DOI] [PubMed] [Google Scholar]
- 57.Baccetti B. Evolutionary trends in sperm structure. Comp Biochem Physiol A. 1986;85(1):29–36. doi: 10.1016/0300-9629(86)90457-3. [DOI] [PubMed] [Google Scholar]
- 58.Prensier G, Vivier E, Goldstein S, Schrevel J. Motile flagellum with a "3 + 0" ultrastructure. Science. 1980;207:1493–1494. doi: 10.1126/science.7189065. [DOI] [PubMed] [Google Scholar]
- 59.Nonaka S, Tanaka Y, Okada Y, Takeda S, Harada A, Kanai Y, et al. Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell. 1998;95(6):829–837. doi: 10.1016/s0092-8674(00)81705-5. [DOI] [PubMed] [Google Scholar]
- 60.Gibbons BH, Gibbons IR, Baccetti B. Structure and motility of the 9 + 0 flagellum of eel spermatozoa. J Submicrosc Cytol. 1983;15:15–20. [PubMed] [Google Scholar]
- 61.Mukai C, Okuno M. Glycolysis plays a major role for adenosine triphosphate supplementation in mouse sperm flagellar movement. Biol Reprod. 2004;71(2):540–547. doi: 10.1095/biolreprod.103.026054. [DOI] [PubMed] [Google Scholar]
- 62.Lardy HA, Phillips PH. The interrelation of oxidative and glycolytic processes as sources of energy for bull spermatozoa. American Journal of Physiology. 1941;133:602–609. [Google Scholar]
- 63.Hsu SC, Molday RS. Glycolytic enzymes and a GLUT-1 glucose transporter in the outer segments of rod and cone photoreceptor cells. J Biol Chem. 1991;266(32):21745–21752. [PubMed] [Google Scholar]
- 64.Westhoff D, Kamp G. Glyceraldehyde 3-phosphate dehydrogenase is bound to the fibrous sheath of mammalian spermatozoa. J Cell Sci. 1997;110(Pt 15):1821–1829. doi: 10.1242/jcs.110.15.1821. [DOI] [PubMed] [Google Scholar]
- 65.Gitlits VM, Toh BH, Loveland KL, Sentry JW. The glycolytic enzyme enolase is present in sperm tail and displays nucleotide-dependent association with microtubules. Eur J Cell Biol. 2000;79:104–111. doi: 10.1078/S0171-9335(04)70012-6. [DOI] [PubMed] [Google Scholar]
- 66.Mitchell BF, Pedersen LB, Feely M, Rosenbaum JL, Mitchell DR. ATP production in Chlamydomonas reinhardtii flagella by glycotytic enzymes. Mol Biol Cell. 2005;16:4509–4518. doi: 10.1091/mbc.E05-04-0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tombes RM, Brokaw CJ, Shapiro BM. Creatine kinase-dependent energy transport in sea urchin spermatozoa. Flagellar wave attenuation and theoretical analysis of high energy phosphate diffusion. Biophys J. 1987;52(1):75–86. doi: 10.1016/S0006-3495(87)83190-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huszar G, Sbracia M, Vigue L, Miller DJ, Shur BD. Sperm plasma membrane remodeling during spermiogenetic maturation in men: relationship among plasma membrane beta 1,4-galactosyltransferase, cytoplasmic creatine phosphokinase, and creatine phosphokinase isoform ratios. Biol Reprod. 1997;56(4):1020–1024. doi: 10.1095/biolreprod56.4.1020. [DOI] [PubMed] [Google Scholar]
- 69.Wallimann T, Wegmann G, Moser H, Huber R, Eppenberger HM. High content of creatine kinase in chicken retina: compartmentalized localization of creatine kinase isoenzymes in photoreceptor cells. Proc Natl Acad Sci U S A. 1986;83(11):3816–3819. doi: 10.1073/pnas.83.11.3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Noguchi M, Sawadas T, Akazawa T. ATP-regenerating system in the cilia of Paramecium caudatum. J Exp Biol. 2001;204(6):1063–1071. doi: 10.1242/jeb.204.6.1063. [DOI] [PubMed] [Google Scholar]