Abstract
Tau aggregation in neurofibrillary tangles is a pathological hallmark in tauopathies including Alzheimer’s disease (AD). The predominant aggregation of certain MAPT (tau gene) isoforms, either the 4-repeat (4R tau) or the 3-repeat (3R tau) isoform has been widely described in tauopathies. Alterations of the 4R tau to 3R tau ratio may be a key for tau-related neurodegeneration. To study the biological consequences in expression between tau splicing isoforms 4R and 3R, we analyzed the main neurobiological effects of inclusion of the repeat region coded by exon 10 in MAPT. We compared the transcriptional profiles of the 4R tau isoforms to 3R tau isoforms using whole-genome gene expression profiling microarrays using human neuroblastoma SH-SY5Y cell lines overexpressing either human 4R tau or 3R tau isoforms. We identified 68 transcripts that differed significantly (at p < 0.001) between 4R and 3R isoforms as conditioned on a second variant, the so-called 2N inclusion. We extended these findings in a 2 × 2 ANOVA to examine interaction effects of these variants. Transcripts involved in embryonic development were downregulated when exon 10 was present, while transcripts related to outgrowth of neurites were generally upregulated. An important pathway implicated in AD also differed between the 3R and 4R cell lines, Wnt signaling. These studies demonstrate expression differences between MAPT isoforms 4R tau or 3R tau due to the inclusion/exclusion of the repeat region coded for by exon 10. Our data add to complex findings on the role of 3R/4R in normal and abnormal neuronal function and highlight several molecular mechanisms that might drive neurodegeneration, or perhaps, set the stage for it.
Keywords: Alzheimer’s disease, gene expression profiling, microarrays, tau 3R, tau 4R
INTRODUCTION
In the adult human brain, alternative splicing of tau generates six isoforms that differ by the regulated inclusion of two inserts near the N-terminus (or their absence, hereafter 2N and 0N) and either three or four imperfect repeat regions, corresponding to the microtubule-binding domains in the C-terminal [1]. Tau isoforms containing either three (3R) or four (4R) microtubule-binding repeats are approximately equal in normal brain i.e., they are in a 50/50 ratio [2,3]. These ratios can be substantially altered in most of the neurodegenerative tauopathies: increases in the 4R to 3R ratio have been described in frontotemporal dementia, progressive supranuclear palsy, and sporadic corticobasal ganglionic degeneration [4–6]. In Alzheimer’s disease (AD) an altered ratio is present due to an increase in 4R tau isoforms or decrease in 3R tau levels, and resulting in an approximately 2:1 4R:3R ratio [7, 8].
Alternative RNA splicing and phosphorylation of tau are cellular mechanisms that regulate microtubule stability. During embryonic and early developmental stages the 3R tau isoforms are predominant, while in adult human brain all six tau isoforms are present [2,3]. Increased phosphorylation occurs at embryonic stages when there is more neuronal plasticity while it is relatively reduced in adult human brain compared to that in embryonic brain. Increased tau phosphorylation reduces the amount of tau that binds to microtubules, and 3R tau isoforms also bind less tightly than 4R tau to microtubules [9]. Hyperphosphorylation of tau is thought to be pathogenic in tau-related toxicity in AD. The precise relationship between 3R:4R ratio, tau phosphorylation, tau microtubule stabilization, tau aggregation, and ultimately, cell death is unknown (see [10] for an overview). Impact of the N terminal was examined by Conrad and colleagues [8] who observed that AD-control classification was well above chance using exon 2 presence or absence in a postmortem study.
In this study, we analyzed the primary biological effects of inclusion of the imperfect repeat region coded for by exon 10 in MAPT using a human neuroblastoma cell line. When exon 10 is present, tau is considered 4R, when absent 3R. In order to further refine our analysis we conditioned 4R and 3R contrasts on N terminal presence or absence of exons 2 and 3 in tau. By comparing 4R tau isoforms to 3R tau isoforms using whole-genome gene expression profiling microarrays, we were able to comprehensively and systematically determine the downstream consequences of 4R and 3R at the level of individual transcripts and signaling pathways.
MATERIALS AND METHODS
Cell culture and stable transfection of human 4R and 3R cDNAs
Human neuroblastoma SH-SY5Y cells were maintained at 37°C and 5% CO2, in Dulbecco’s modified minimal essential medium (DMEM) (Life Technologies, Burlington, ON) supplemented with 10% (v/v) fetal bovine serum (FBS), 1% (v/v) penicillin and streptomycin (Life Technologies, Burlington, ON). SH-SY5Y cells were stable transfected with a four repeat (plasmids: pRcCMV2-Tau A and pRcCMV2-Tau C) and three repeat (plasmids: pRcCMV2-Tau B and pRcCMV2-Tau D) human tau cDNAs using Lipofectamine™ 2000 (Invitrogen, CA) as follows: cells were plated at density of 2.5 × 105 cells/well in six-well plates and grown until they reached 90% confluence. They were then transfected in serum-free medium with 10 µl of Lipofectamine plus 4µg of plasmid and left at 37°C for 5 h. After replacement of serum-free with complete medium, they were maintained for an additional 48 h, trypsinized and plated in the presence of 400 µg/mlG-418 (Invitrogen, CA) until all non-transfected cells were dead. Individual resistant colonies were picked and maintain in the presence of G-418 (200 µg/ml).
Microarray experiments
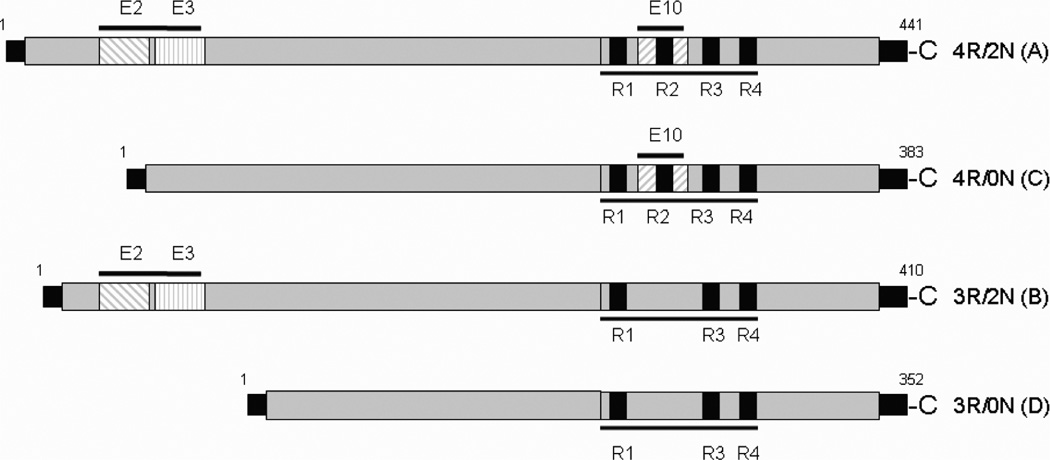
Six experiments per each condition were performed using the stably transfected SH-SY5Y cells overexpressing Tau A (4R/2N), Tau B (3R/2N), Tau C (4R/0N), and Tau D (3R/0N). As noted when exon 10 is present, tau is considered 4R, when absent 3R. Tau isoforms A and B have inclusions of two inserts near the N-terminus (exons 2 and 3), and will be hereafter called 2N. Tau C and Tau D lack those two inserts, and are hereafter called 0N. Thus, four isoforms were included in this study (Tau A, B, C, D).
Cells were plated at a density of 3.0 × 105 cells/well in six-well plates and grown for 24 h at 37°C and 5% CO2, in DMEM supplemented with 10% (v/v) FBS, 1% (v/v) penicillin and streptomycin. Total RNA from cell lines was extracted using standard procedures as described previously [11]. Briefly, total RNA (DNAse treated) was isolated using an RNeasy kit with a QI-Ashredder column (Quiagen) and measured for quality using Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). RIN values in this study were ≥ 9.0. Microarrays were utilized according to the manufacturer’s guidelines (Illumina). Total RNA (260 ng) was converted to cDNA by reverse transcription using ArrayScript™ reverse transcriptase and T7-Oligo (dT)24 primers, followed by second-strand synthesis to generate double-stranded cDNA. After purification, the cDNA was converted to biotin-labeled cRNA (Totalprep™ RNA Labeling Kit, Ambion), hybridized to the microarray platform: HumanWG-6_V2 Expression BeadChip (Illumina, San Diego, CA), and stained with streptavidin-Cy3 for visualization (1.5 µg/20 µl cRNA loaded on the chip). A total of 24 microarrays were run (six experiments of neuroblastoma cells overexpressing Tau A, six overexpressing Tau B, six overexpressing Tau C, and six overexpressing Tau D).
Data preprocessing
After the probe arrays were scanned, the resulting images were first pre-processed using the BeadStudio software (Illumina, San Diego, CA), which calculates the mean fluorescence signal across all 30-replicates of each gene/transcript (AVG_Signal) along with a detection score that represents the probability that the mean signal for each gene/transcript on the chip is greater than background (i.e., detection p-value). Genes/transcripts were defined as being significantly expressed above background (as detected by the array) when each gene’s detection p-value was ≤ 0.001. The expression data were then normalized within quantiles across samples of the distribution of gene expression values.
BeadStudio calculates background as the average signal intensity estimated from the negative control bead types (~700) and removes outliers using the median absolute deviation method (Illumina, BeadStudio). However, previous studies [12] and our own pilot studies indicated that background subtraction had a negative impact on data quality (e.g., it lowered correlation coefficients between technical replicates), so we therefore exported data that were normalized, but that did not undergo background subtraction. As a result of this processing 20107 of 48701 possible transcripts met quality control criteria and were used in all subsequent analyses in BRB Array Tools 3.7, a statistical software package designed for microarray analysis developed by NIH (http://linus.nci.nih.gov/BRB-ArrayTools.html).
Statistical analysis
Normalized data were imported to BRB-Array Tools and processed within the univariate module. First we examined the isoforms using a conservative Venn-type approach designed to yield robust and consistent differences between 3R and 4R isoforms. We thus conditioned 3R versus 4R contrasts on the presence or absence of 2N (i.e., 2N and 0N).We identified those transcripts which both differed significantly (p ≤ 0.001) and differed in the same direction (i.e., up or down regulated) in the two separate 3R-4R contrasts. Thus we contrasted Tau A versus Tau B (2N present) and Tau C versus Tau D (2N absent). Six biological replicates were averaged in each group. Likelihood ratio test statistics (F test) were used to investigate the significance of the difference score between the classes i.e., an F-test was computed separately for each gene using the normalized intensities. We report p values for the class variable only. Expression differences were considered statistically significant if their p values were less than 0.001. We limited the proportion of false discoveries using a multivariate permutation test in which the FDR was set at 0.10. We then selected the transcripts that differed significantly and in the same direction for further heat map analyses, pathway analyses, and discussion.
We also conducted a second set of parametric statistical analyses by examining the data in a 2 (3R/4R) × 2 (2N/0N) ANOVA in the BRB statistical environment using the A*B model. As a result we could detect main effects of 3R-4R isoforms, main effects of 2N-0N isoforms, and interactions.
Hierarchical clustering of expression changes
Hierarchical clustering of transcript difference scores was performed using Euclidean distance as a distance metric and complete linkage. The cluster analysis of transcripts (and cases) produced a heat map image in which the rows in the image plot represent the transcripts, and the columns in the image plot represent the neuroblastoma cell line samples.
Biological and signaling pathways analyses (KEGG)
Goeman’s Global Test is a logistic regression model that tests the null hypothesis that there are no genes within a given set (pathway) that are differentially expressed using an exact permutation method (n = 10000) to determine an exact p value [13,14].
Real-time quantitative polymerase chain reaction (RTq-PCR)
For selected transcripts (10 genes), showing differential expression between the 3R and 4R groups, cD-NA synthesis was generated for each sample with the Ambion reverse transcription Kit and oligo dT primer. For each sample, amplified product differences were measured with two replicates with locked nucleic acid (LNA) chemistry-based detection. For housekeeping genes, primers, and probes used, see Supplementary Table 1 (available online: http://www.j-alz.com/issues/22/vol22-4.html# supplementarydata05). The RT-qPCR reactions were carried out in an ABI Prism 7900HT thermal cycler (Applied Biosystems Inc.), determining the ΔΔCt and fold changes. Statistical analysis on 3R and 4R difference scores was performed using a Student’s t-test in Microsoft Excel.
IPA analysis: functional analysis of the 68 transcripts
We ascertained the biological significance of the transcripts found to be significant by F test and FDR (n = 68, see below) for both comparisons (Tau A vs. Tau B; Tau C vs. Tau D) using Ingenuity Pathway Analysis application (IPA/Ingenuity Systems, http://www.ingenuity.com) [15].
We first entered our transcripts of interest. Fold changes for each transcript were also included in order to refine the specificity of the analysis for both comparisons A vs. B and C vs. D. The Functional Analysis identified the biological functions and/or pathways that were most significant to the data set. Genes from the dataset that were associated with biological functions and/or pathways in the Ingenuity Pathways Knowledge Base were considered for the analysis. Fisher’s exact test was used to calculate a p-value determining the probability that each biological function assigned to that data set was found by chance.
RESULTS
Transcripts differentially expressed between 4R and 3R isoforms (3R-4R contrast conditioned on 2N-0N)
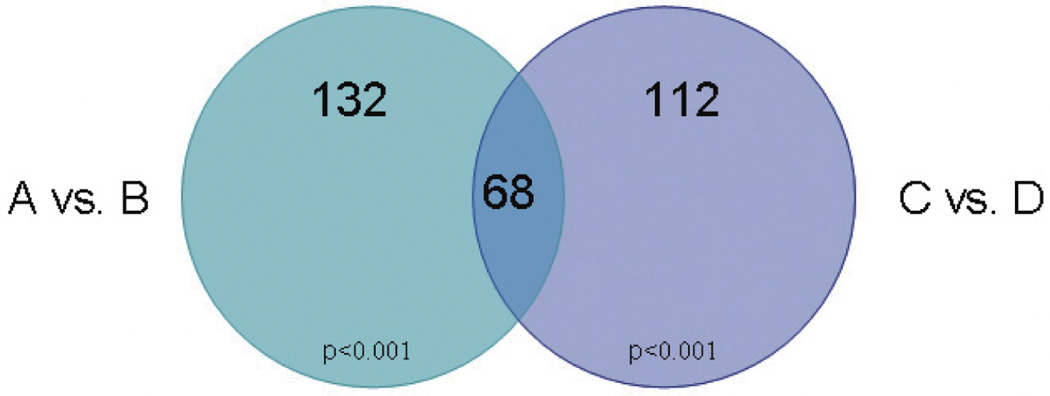
In order to identify the main biological effects of 3R-4R we compared the effects of overexpressing 4R isoforms vs. 3R isoforms. We first compared transcripts differentially expressed between a stable SH-SY5Y cell line overexpressing Tau A isoform (4R/2N) and a stable SH-SY5Y cell line overexpressing Tau B isoform (3R/2N), we called A vs. B comparison. We identified 132 differentially expressed transcripts that differed significantly (at p < 0.001) between A and B groups on the univariate test. The global p value for the analysis was significant (p = 0.002). We next compared transcripts differentially expressed between a stable SH-SY5Y cell line overexpressing Tau C isoform (4R/0N) and a stable SH-SY5Y cell line overexpressing Tau D isoform (3R/0N). We identified 112 differentially expressed transcripts that differed significantly (at p < 0.001) between C and D groups on the univariate test. The global p value for the analysis was significant (p = 0.002).
Last and critically in this conservative approach, we determined transcripts in common between both the A vs. B and the C vs. D comparisons that were also regulated in the same direction using these criteria (Figs 1 and 2).
Fig. 1.
Tau isoforms overexpressed on SH-SY5Y cell lines. Tau 4R (A and C) and Tau 3R (B and D).
Fig. 2.
Transcripts Differentially Expressed Due to Exon 10: Venn diagram of comparisons A vs. B and C vs. D. Left circle: A vs. B comparison identifying 132 transcripts differentially expressed that differed significantly (at p < 0.001) between A and B groups on the univariate test. Right circle: C vs. D comparison identifying 112 transcripts differentially expressed that differed significantly (at p < 0.001) between C and D groups on the univariate test. Overlapping area: 68 transcripts in common between both comparisons: A vs. B and C vs. D that were also regulated in the same direction of up or down-regulated.
We identified 68 transcripts corresponding to genes from RefSeq and UniGene. The 68 transcripts, their exact p values, and fold change score for both comparisons are listed in Table 1 (Differentially expressed transcripts between both comparisons A vs. B and C vs. D are listed in Tables 2 and 3 in Supplementary Material).
Table 1.
Transcripts differentially expressed in common between both A vs. B and C vs. D comparisons
| RefSeq transcript ID |
Gene symbol |
Entrez Gene name | Fold change (A vs. B) |
Parametric p-value |
Fold change (C vs. D) |
Parametric p-value |
|---|---|---|---|---|---|---|
| NM_001613 | ACTA2 | actin, alpha 2, smooth muscle, aorta | 2.59 | < 1e-07 | 1.59 | < 1e-07 |
| NM_017637 | BNC2 | basonuclin 2 | −2.86 | < 1e-07 | −2.03 | < 1e-07 |
| NM_174941 | CD163L1 | CD163 molecule-like 1 | 3.10 | < 1e-07 | 4.07 | < 1e-07 |
| NM_001769 | CD9 | CD9 molecule | −2.94 | < 1e-07 | −4.09 | < 1e-07 |
| NM_014141 | CNTNAP2 | contactin associated protein like 2 | 13.44 | < 1e-07 | 2.38 | < 1e-07 |
| NM_001915 | CYB561 | cytochrome b-561 | 1.52 | < 1e-07 | 2.83 | < 1e-07 |
| NM_080760 | DACH1 | dachshund homolog 1 (Drosophila) | −6.67 | < 1e-07 | −2.38 | < 1e-07 |
| NM_012242 | DKK1 | dickkopf homolog 1 (Xenopus laevis) | −14.29 | < 1e-07 | −2.01 | < 1e-07 |
| NM_139072 | DNER | delta/notch-like EGF repeat | 3.87 | < 1e-07 | 2.40 | < 1e-07 |
| NM_018962 | DSCR6 | Down syndrome critical region gene 6 | 3.51 | < 1e-07 | 1.56 | < 1e-07 |
| NM_001946 | DUSP6 | dual specificity phosphatase 6 | −2.50 | < 1e-07 | −1.43 | 2.00E-07 |
| NM_182801 | EGFLAM | EGF-like, fibronectin III and laminin | 2.39 | < 1e-07 | 1.55 | < 1e-07 |
| NM_032048 | EMILIN2 | elastin microfibril interfacer 2 | 3.65 | < 1e-07 | 4.52 | < 1e-07 |
| NM_004100 | EYA4 | eyes absent homolog 4 (Drosophila) | −3.23 | < 1e-07 | −1.81 | < 1e-07 |
| NM_207334 | FAM43B | family with sequence similarity 43,B | 4.99 | < 1e-07 | 3.52 | < 1e-07 |
| NM_001453 | FOXC1 | forkhead box C1 | −4.76 | < 1e-07 | −2.18 | < 1e-07 |
| NM_018027 | FRMD4A | FERM domain containing 4A | 2.33 | < 1e-07 | 3.28 | < 1e-07 |
| NM_152330 | FRMD6 | FERM domain containing 6 | −3.33 | < 1e-07 | −1.73 | < 1e-07 |
| NM_001475 | GAGE5 | G antigen 5 | −1.14 | 0.0002161 | −2.34 | < 1e-07 |
| NM_001475 | GAGE5 | G antigen 5 | −1.15 | 0.0007225 | −2.34 | < 1e-07 |
| NM_033258 | GNG8 | guanine nucleotide binding protein, 8 | −7.69 | < 1e-07 | −2.10 | < 1e-07 |
| NM_001001995 | GPM6B | glycoprotein M6B | −1.70 | < 1e-07 | −3.73 | < 1e-07 |
| NM_001001995 | GPM6B | glycoprotein M6B | −1.32 | 1.00E-07 | −3.49 | < 1e-07 |
| NM_144594 | GTSF1 | gametocyte specific factor 1 | 15.14 | < 1e-07 | 15.35 | < 1e-07 |
| NM_005519 | HMX2 | H6 family homeobox 2 | −5.56 | < 1e-07 | −4.28 | < 1e-07 |
| NM_002148 | HOXD10 | homeobox D10 | −1.89 | < 1e-07 | −5.71 | < 1e-07 |
| AB073882 | Hs.1832 | human neuroblastoma cDNA | 3.89 | < 1e-07 | 3.08 | < 1e-07 |
| BX100246 | Hs.38132 | Soares fetal liver spleen 1NFLS | −2.70 | < 1e-07 | −1.28 | 3.15E-05 |
| BX537518 | Hs.7023 | H.S. mRNA; cDNA DKFZp686N1989 | 3.05 | < 1e-07 | 1.85 | < 1e-07 |
| NM_014817 | KIAA0644 | KIAA0644 gene product | −3.33 | < 1e-07 | −1.45 | 1.00E-07 |
| NM_020775 | KIAA1324 | KIAA1324 | 4.04 | < 1e-07 | 1.30 | 1.81E-05 |
| NM_001290 | LDB2 | LIM domain binding 2 | 4.39 | < 1e-07 | 2.13 | < 1e-07 |
| NM_057159 | LPAR1 | lysophosphatidic acid receptor 1 | −4.76 | < 1e-07 | −1.76 | < 1e-07 |
| NM_002334 | LRP4 | LDL receptor-related protein 4 | −1.85 | < 1e-07 | −2.21 | < 1e-07 |
| NM_001013653 | LRRC26 | leucine rich repeat containing 26 | 5.94 | < 1e-07 | 2.46 | < 1e-07 |
| NM_201630 | LRRN2 | leucine rich repeat neuronal 2 | 4.13 | < 1e-07 | 1.95 | < 1e-07 |
| NM_002345 | LUM | lumican | −2.94 | < 1e-07 | −1.27 | 6.91E-05 |
| NM_021048 | MAGEA10 | melanoma antigen family A, 10 | 16.37 | < 1e-07 | 15.06 | < 1e-07 |
| NM_145764 | MGST1 | microsomal glutath. S-transferase 1 | 2.83 | < 1e-07 | 1.48 | 6.00E-07 |
| NM_006157 | NELL1 | NEL-like 1 (chicken) | 1.44 | < 1e-07 | 3.43 | < 1e-07 |
| NM_000905 | NPY | neuropeptide Y | 9.93 | < 1e-07 | 15.57 | < 1e-07 |
| XM 001133042 | PDZRN3 | PDZ domain containing RING finger 3 | −5.26 | < 1e-07 | −4.54 | < 1e-07 |
| XM 001133042 | PDZRN3 | PDZ domain containing RING finger 3 | −5.55 | < 1e-07 | −4.00 | < 1e-07 |
| NM_001039582 | PNCK | preg. up-reg non-ubiq. CaM kinase | 4.46 | < 1e-07 | 1.62 | < 1e-07 |
| NM_013364 | PNMA3 | paraneoplastic antigen MA3 | 5.68 | < 1e-07 | 2.55 | < 1e-07 |
| NM_181676 | PPP2R2B | protein phosphatase 2, subunit B, β | 3.65 | < 1e-07 | 1.29 | 1.93E-05 |
| NM_181876 | PPP2R2C | protein phosphatase 2, subunit B, γ | 5.33 | < 1e-07 | 1.62 | < 1e-07 |
| NM_002737 | PRKCA | protein kinase C, alpha | −1.89 | < 1e-07 | −2.89 | < 1e-07 |
| NM_006017 | PROM1 | prominin 1 | −2.22 | < 1e-07 | −1.24 | 0.0001516 |
| NM_003619 | PRSS12 | protease, serine, 12 | −2.63 | < 1e-07 | −1.41 | 7.00E-07 |
| NM_002872 | RAC2 | ras-related C3 botulinum toxin sub. 2 | 2.83 | < 1e-07 | 1.43 | < 1e-07 |
| NM_020975 | RET | ret proto-oncogene, variant 2 | −2.38 | < 1e-07 | −1.81 | < 1e-07 |
| NM_020630 | RET | ret proto-oncogene, variant 4 | −2.70 | < 1e-07 | −1.43 | 2.40E-06 |
| NM_020975 | RET | ret proto-oncogene, variant 2 | −2.86 | < 1e-07 | −1.20 | 0.000377 |
| NM_173662 | RNF175 | ring finger protein 175 | 4.03 | < 1e-07 | 1.92 | < 1e-07 |
| NM_002942 | ROBO2 | roundabout, axon guidance recept, 2 | −4.76 | < 1e-07 | −5.85 | < 1e-07 |
| NM_003020 | SCG5 | secretogranin V (7B2 protein) | 3.09 | < 1e-07 | 1.99 | < 1e-07 |
| NM_000624 | SERPINA5 | serpin peptidase inhibitor, clade A, 5 | 9.89 | < 1e-07 | 3.90 | < 1e-07 |
| NM_005627 | SGK1 | serum/glucocorticoid regulat-kinase 1 | 1.27 | 1.00E-06 | 3.41 | < 1e-07 |
| NM_007374 | SIX6 | SIX homeobox 6 | 5.08 | < 1e-07 | 1.39 | 5.40E-06 |
| NR_002784 | SMEK3P | SMEK homolog 3 | 3.89 | < 1e-07 | 4.33 | < 1e-07 |
| NM_013322 | SNX10 | sorting nexin 10 | 4.50 | < 1e-07 | 1.95 | < 1e-07 |
| NM_020826 | SYT13 | synaptotagmin XIII | 5.05 | < 1e-07 | 1.73 | < 1e-07 |
| NM_080390 | TCEAL2 | transcription elongation factor A | 7.28 | < 1e-07 | 1.38 | 5.00E-07 |
| NM_018286 | TMEM100 | transmembrane protein 100 | −1.79 | < 1e-07 | −2.89 | < 1e-07 |
| NM_007115 | TNFAIP6 | tumor necrosis factor, α-ind. protein 6 | −3.70 | < 1e-07 | −3.93 | < 1e-07 |
| NM_003283 | TNNT1 | troponin T type 1 (skeletal, slow) | 3.62 | < 1e-07 | 1.72 | 1.00E-07 |
| NM_194435 | VIP | vasoactive intestinal peptide | −20.00 | < 1e-07 | −5.24 | < 1e-07 |
Table 2.
RT-qPCR p-values and fold changes for twelve transcripts
| RefSeq transcript ID |
Gene symbol |
Fold change Microarray (A vs. B) |
Parametric p-value |
Fold change Microarray (C vs. D) |
Parametric p-value |
Fold change RT-qPCR (A vs. B) |
Parametric p-value |
Fold change RT-qPCR (C vs. D) |
Parametric p-value |
|---|---|---|---|---|---|---|---|---|---|
| NM_014141 | CNTNAP2 | 13.44 | < 1e-07 | 2.38 | < 1e-07 | 30.50 | 5.98E-06 | 4.50 | 0.00041961 |
| NM_012242 | DKK1 | −14.29 | < 1e-07 | −2.01 | < 1e-07 | −25.00 | 0.002791324 | −1.63 | 0.008044056 |
| NM_139072 | DNER | 3.87 | < 1e-07 | 2.40 | < 1e-07 | 43.60 | 1.42567E-06 | 6.55 | < 1e-07 |
| NM_032048 | EMILIN2 | 3.65 | < 1e-07 | 4.52 | < 1e-07 | 6.18 | 0.000289114 | 11.08 | < 1e-07 |
| NM_001453 | FOXC1 | −4.76 | < 1e-07 | −2.18 | < 1e-07 | −5.20 | < 1e-07 | −1.42 | 0.000207761 |
| NM_000905 | NPY | 9.93 | < 1e-07 | 15.57 | < 1e-07 | 16.79 | < 1e-07 | 28.54 | < 1e-07 |
| NM_181876 | PPP2R2C | 5.33 | < 1e-07 | 1.62 | < 1e-07 | 11.58 | 0.001891617 | 2.55 | 0.000132606 |
| NM_002737 | PRKCA | −1.89 | < 1e-07 | −2.89 | < 1e-07 | −1.58 | 0.000433749 | −2.70 | < 1e-07 |
| NM_002942 | ROBO2 | −4.76 | < 1e-07 | −5.85 | < 1e-07 | −33.30 | < 1e-07 | −12.50 | < 1e-07 |
| NM_194435 | VIP | −20.00 | < 1e-07 | −5.24 | < 1e-07 | −50.00 | < 1e-07 | −16.66 | 1.56897E-07 |
| NM_004655 | AXIN2 | 1.09 | 0.02 | −1.00 | 0.69 | 1.16 | 0.10 | 1.01 | 0.92 |
| NM_001904 | CTNNB1 | 1.06 | 0.45 | −1.06 | 0.71 | −1.75 | 0.01 | 1.26 | 0.11 |
Table 3.
Biological functions as determined by 3R/4R contrast conditioned by 2N/0N
| Embryonic development |
Neuronal cell morphology |
Cellular growth proliferation |
Cell death |
|
|---|---|---|---|---|
| p-value | 1.52E-04-4.45E-02 | 9.84E-06-4.96E-02 | 4.78E-04-4.35E-02 | 9.07E-04-4.98E-02 |
| Molecules | RAC2 | CNTNAP2 | NPY | NELL1 |
| LRP4 | SGK1 | RAC2 | MGST1 | |
| LPAR1 | SYT13 | EMILIN2 | RAC2 | |
| CD9 | TNNT1 | VIP | SGK1 | |
| ROBO2 | DNER | HMX2 | EMILIN2 | |
| DKK1 | ACTA2 | FOXC1 | DUSP6 | |
| VIP | VIP | CD9 | VIP | |
| HMX2 | CD9 | LPAR1 | LPAR1 | |
| RET | LPAR1 | LUM | CD9 | |
| FOXC1 | SERPINA5 | SIX6 | SCG5 | |
| HOXD10 | SCG5 | DACH1 | PPP2R2B | |
| LUM | PPP2R2C | DACH1 | ||
| PPP2R2C | DKK1 | DKK1 | ||
| ROBO2 | TNFAIP6 | RET | ||
| DKK1 | RET | PRKCA | ||
| RET | PRKCA |
Hierarchical clustering of expression changes
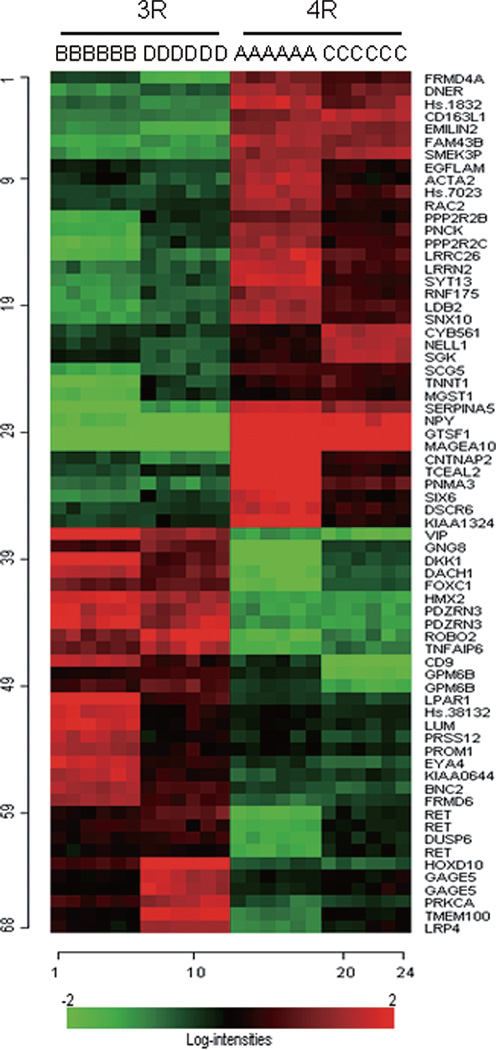
As shown in the heat map image (Fig. 3) the hierarchical clustering of expression changes demonstrated that in the two 4R groups (A and C), 36 of the significant transcripts identified by univariate statistics demonstrated differences in expression that were associated with strong upregulation (i.e., “red” cells). The 3R groups (B and D) demonstrated downregulation for the same 36 transcripts. The reverse pattern was shown for the 32 remaining transcripts. The A and C groups and the B and D groups both demonstrated similar magnitude of expression differences. Similarly, as shown in the Fig. 1 in Supplementary Material, the clustering dendrogram indicated that 3R cells, irrespective of 2N/0N, formed one large cluster (i.e., labeled Bs and Ds), and 4R cells (labeled As and Cs) formed a second large cluster.
Fig. 3.
Heat map showing up-and down-regulation for the 68 transcripts identified as significant between the two 4R groups (A and C) and the 3R groups (B and D). The 68 transcripts identified as significant (all p-values < 0.001) in the univariate analysis are on the y axis and cell lines overexpressing tau isoforms are one the x axis. Log intensities of 68 transcripts are represented in colored cells. Red cells indicate relatively strong upregulation for the transcripts in that particular cell line overexpressing the tau 4R or tau 3R, whereas green cells indicate the converse.
RT-qPCR
We validated our microarray findings in 10 transcripts that were selected from the list of 68 transcripts: CNTNAP2, DKK1, DNER, EMILIN2, FOXC1, NPY, PPP2R2C, PRKCA, ROBO2, and VIP. These transcripts were chosen on the basis of their statistical significance, role in key biological or signaling pathways, and possible relevance to AD. They are listed in Table 2, along with p value significance levels as determined by t-test and fold changes. Critically, for each and every positive finding, we found that the pattern of RT-qPCR results was consistent with that of the microarray results in terms of regional up-or down-regulation for the 4R and 3R groups. We also examined two more transcripts, AXIN2 and CTNNB1, the end products of the Wnt signaling canonical pathway for RT-qPCR (see below).
Functional analysis of the 68 transcripts: biological functions
In order to examine the biological significance of our findings we conducted a functional analysis of the interrelationships among the 68 transcripts identified as differing significantly between the 4R and 3R cell line groups on the univariate test using IPA. Results indicate that these 68 transcripts are implicated in several biological functions as follows below. These transcripts and their biological functions are listed in Table 3.
Transcripts involved in embryonic development
Of the 68 transcripts, eleven were present in the Embryonic Development biological function with Fisher’s Exact Test p values ranging from 1.52E-04 to 4.45E-02 for subclassifications. From the eleven transcripts, one transcript (RAC2) was upregulated in the 4R isoform group with respect to the 3R isoform group, while ten transcripts were consistently downregulated in the 4R isoform group with respect to the 3R isoform group: CD9, DKK1, FOXC1, HMX2, HOXD10, LPAR1, LRP4, RET, ROBO2, and VIP. Of these ten transcripts, we selected four of them for validation by RTqPCR: DKK1, FOXC1, ROBO and VIP (see section in RT-qPCR below).
Transcripts involved in neuronal cell morphology: outgrowth of neurites
Sixteen transcripts were present in the Cell Morphology biological function with Fisher’s Exact Test p values ranging from 9.84E-06 to 4.96E-02 for subclassifications. From these sixteen transcripts, seven were directly involved in outgrowth of neurites. One of the common markers for neuronal cell differentiation is the increase in neurite outgrowth. Five of these transcripts were upregulated by the 4R isoformin our study: CNT-NAP2, DNER, PPP2R2C, SGK1, and SYT13. Two of them, CD9 and RET, were downregulated.
Transcripts involved in cellular growth and proliferation
Sixteen transcripts were present in the Cellular Growth and Proliferation biological function with Fisher’s Exact Test p values ranging from 4.78E-04 to 4.35E-02 for subclassifications. Of the sixteen transcripts, five were upregulated: EMILIN2, NPY, PPP2R2C, RAC2, and SIX6. Eleven transcripts were consistently downregulated in the 4R isoform group with respect to the 3R isoform group, for both comparisons: CD9, DACH1, DKK1, FOXC1, HMX2, LPAR1, LUM, PRKCA, RET, TNFAIP6, and VIP. Most of these downregulated transcripts are involved in neuronal proliferation and survival, and suggest that those functions are decreased in Tau 4R.
Transcripts involved in cell death
For the Cell Death biological function, fifteen transcripts were present with Fisher’s Exact Test p values from 9.07E-04 to 4.98E-02 for subclassifications. Of the fifteen transcripts, seven transcripts were consistently upregulated in the 4R isoform group with respect to the 3R isoform group, for both comparisons: EMILIN2, MGST1, NELL1, PPP2R2B, RAC2, SCG5, and SGK1. Eight transcripts were downregulated: CD9, DACH1, DKK1, DUSP6, LPAR1, PRKCA, RET, and VIP. Four (VIP, LPAR1, RET and PRKCA) have been implicated in neuronal survival.
KEGG signaling pathways
One significant gene set was identified with Goeman’s global test p values of p < 0.002 within the 68 differentially expressed transcripts: the Wnt signaling pathway. Significant transcripts (p < 0.005) that differed between 4R and 3R included: RAC2, PPP2R2B, PPP2R2C, DKK1, and PRKCA. This Kyoto pathway is of interest because has been implicated in prior studies of AD.
2 × 2 ANOVA Analysis
The ANOVA identified 102 transcripts as a main effect of the 3R-4R contrast. These are listed in Table 4. Critically, but not unexpectedly, all 68 of the genes that we identified above using our conservative Venn-type approach were included in this set of transcripts.
Table 4.
Table of significant transcripts for testing “Class 3R vs. 4R” main effect (102 transcripts were found significant at level 0.001)
| RefSeq transcript ID |
Gene symbol | Entrez Gene name | p-value |
|---|---|---|---|
| NM_174941 | CD163L1 | Homo sapiens CD163 molecule-like 1 (CD163L1), mRNA. | 0.00E+00 |
| NM_032048 | EMILIN2 | Homo sapiens elastin microfibril interfacer 2 (EMILIN2), mRNA. | 0.00E+00 |
| NM_207334 | FAM43B | Homo sapiens family with sequence similarity 43, member B (FAM43B), mRNA. | 0.00E+00 |
| NM_144594 | GTSF1 | Homo sapiens gametocyte specific factor 1 (GTSF1), mRNA. | 0.00E+00 |
| NM_005519 | HMX2 | Homo sapiens H6 family homeobox 2 (HMX2), mRNA. | 0.00E+00 |
| AB073882 | Hs.1832 | Homo sapiens primary neuroblastoma cDNA, clone:Nbla00830, full insert sequence | 0.00E+00 |
| NM_021048 | MAGEA10 | Homo sapiens melanoma antigen family A, 10 (MAGEA10), transcript variant 2, mRNA. | 0.00E+00 |
| NM_000905 | NPY | Homo sapiens neuropeptide Y (NPY), mRNA. | 0.00E+00 |
| XM_001133042 | PDZRN3 | PREDICTED: Homo sapiens PDZ domain containing RING finger 3 (PDZRN3), mRNA. | 0.00E+00 |
| XM_001133042 | PDZRN3 | PREDICTED: Homo sapiens PDZ domain containing RING finger 3 (PDZRN3), mRNA. | 0.00E+00 |
| NM_002942 | ROBO2 | Homo sapiens roundabout, axon guidance receptor, homolog 2 (Drosophila) (ROBO2), mRNA. | 0.00E+00 |
| NR_002784 | SMEK3P | Homo sapiens SMEK homolog 3, suppressor of mek1 (Dictyostelium) pseudogene (SMEK3P) on chromosome X. | 0.00E+00 |
| NM_007115 | TNFAIP6 | Homo sapiens tumor necrosis factor, alpha-induced protein 6 (TNFAIP6), mRNA. | 0.00E+00 |
| NM_018027 | FRMD4A | Homo sapiens FERM domain containing 4A (FRMD4A), mRNA. | 0.00E+00 |
| NM_001769 | CD9 | Homo sapiens CD9 molecule (CD9), mRNA. | 0.00E+00 |
| NM_002334 | LRP4 | Homo sapiens low density lipoprotein receptor-related protein 4 (LRP4), mRNA. | 0.00E+00 |
| NM_017637 | BNC2 | Homo sapiens basonuclin 2 (BNC2), mRNA. | 0.00E+00 |
| NM_139072 | DNER | Homo sapiens delta/notch-like EGF repeat containing (DNER), mRNA. | 0.00E+00 |
| NM_000624 | SERPINA5 | Homo sapiens serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 (SERPINA5), mRNA. | 0.00E+00 |
| NM_003020 | SCG5 | Homo sapiens secretogranin V (7B2 protein) (SCG5), mRNA. | 0.00E+00 |
| NM_194435 | VIP | Homo sapiens vasoactive intestinal peptide (VIP), transcript variant 2, mRNA. | 0.00E+00 |
| NM_018286 | TMEM100 | Homo sapiens transmembrane protein 100 (TMEM100), transcript variant 2, mRNA. | 0.00E+00 |
| NM_020975 | RET | Homo sapiens ret proto-oncogene (RET), transcript variant 2, mRNA. | 0.00E+00 |
| NM_013364 | PNMA3 | Homo sapiens paraneoplastic antigen MA3 (PNMA3), mRNA. | 0.00E+00 |
| NM_002737 | PRKCA | Homo sapiens protein kinase C, alpha (PRKCA), mRNA. | 0.00E+00 |
| NM_001290 | LDB2 | Homo sapiens LIM domain binding 2 (LDB2), mRNA. | 0.00E+00 |
| BX537518 | Hs.7023 | Homo sapiens mRNA; cDNA DKFZp686N1989 (from clone DKFZp686N1989) | 0.00E+00 |
| NM_001013653 | LRRC26 | Homo sapiens leucine rich repeat containing 26 (LRRC26), mRNA. | 0.00E+00 |
| NM_001453 | FOXC1 | Homo sapiens forkhead box C1 (FOXC1), mRNA. | 0.00E+00 |
| NM_182801 | EGFLAM | Homo sapiens EGF-like, fibronectin type III and laminin G domains (EGFLAM), transcript variant 4, mRNA. | 0.00E+00 |
| NM_004100 | EYA4 | Homo sapiens eyes absent homolog 4 (Drosophila) (EYA4), transcript variant 1, mRNA. | 0.00E+00 |
| NM_201630 | LRRN2 | Homo sapiens leucine rich repeat neuronal 2 (LRRN2), transcript variant 2, mRNA. | 0.00E+00 |
| NM_080760 | DACH1 | Homo sapiens dachshund homolog 1 (Drosophila) (DACH1), transcript variant 2, mRNA. | 0.00E+00 |
| NM_152330 | FRMD6 | Homo sapiens FERM domain containing 6 (FRMD6), transcript variant 2, mRNA. | 0.00E+00 |
| NM_173662 | RNF175 | Homo sapiens ring finger protein 175 (RNF175), mRNA. | 0.00E+00 |
| NM_013322 | SNX10 | Homo sapiens sorting nexin 10 (SNX10), mRNA. | 0.00E+00 |
| NM_001613 | ACTA2 | Homo sapiens actin, alpha 2, smooth muscle, aorta (ACTA2), mRNA. | 0.00E+00 |
| NM_001001995 | GPM6B | Homo sapiens glycoprotein M6B (GPM6B), transcript variant 1, mRNA. | 0.00E+00 |
| NM_003283 | TNNT1 | Homo sapiens troponin T type 1 (skeletal, slow) (TNNT1), mRNA. | 0.00E+00 |
| NM_001915 | CYB561 | Homo sapiens cytochrome b-561 (CYB561), transcript variant 1, mRNA. | 0.00E+00 |
| NM_033258 | GNG8 | Homo sapiens guanine nucleotide binding protein (G protein), gamma 8 (GNG8), mRNA. | 0.00E+00 |
| NM_002148 | HOXD10 | Homo sapiens homeobox D10 (HOXD10), mRNA. | 0.00E+00 |
| NM_057159 | LPAR1 | Homo sapiens lysophosphatidic acid receptor 1 (LPAR1), transcript variant 2, mRNA. | 0.00E+00 |
| NM_001946 | DUSP6 | Homo sapiens dual specificity phosphatase 6 (DUSP6), transcript variant 1, mRNA. | 0.00E+00 |
| NM_145764 | MGST1 | Homo sapiens microsomal glutathione S-transferase 1 (MGST1), transcript variant 1d, mRNA. | 0.00E+00 |
| NM_018962 | DSCR6 | Homo sapiens Down syndrome critical region gene 6 (DSCR6), mRNA. | 0.00E+00 |
| NM_014141 | CNTNAP2 | Homo sapiens contactin associated protein-like 2 (CNTNAP2), mRNA. | 0.00E+00 |
| NM_002872 | RAC2 | Homo sapiens ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) (RAC2), mRNA. | 0.00E+00 |
| NM_020826 | SYT13 | Homo sapiens synaptotagmin XIII (SYT13), mRNA. | 0.00E+00 |
| NM_001039582 | PNCK | Homo sapiens pregnancy upregulated non-ubiquitously expressed CaM kinase (PNCK), mRNA. | 0.00E+00 |
| NM_020630 | RET | Homo sapiens ret proto-oncogene (RET), transcript variant 4, mRNA. | 0.00E+00 |
| NM_003619 | PRSS12 | Homo sapiens protease, serine, 12 (neurotrypsin, motopsin) (PRSS12), mRNA. | 0.00E+00 |
| NM_006157 | NELL1 | Homo sapiens NEL-like 1 (chicken) (NELL1), mRNA. | 0.00E+00 |
| NM_014817 | KIAA0644 | Homo sapiens KIAA0644 gene product (KIAA0644), mRNA. | 0.00E+00 |
| XM_939093 | FAM89A | PREDICTED: Homo sapiens family with sequence similarity 89, member A (FAM89A), mRNA. | 0.00E+00 |
| NM_181876 | PPP2R2C | protein phosphatase 2, subunit B, gamma isoform (PPP2R2C), transcript variant 2, mRNA. | 0.00E+00 |
| NM_012242 | DKK1 | Homo sapiens dickkopf homolog 1 (Xenopus laevis) (DKK1), mRNA. | 0.00E+00 |
| NM_006017 | PROM1 | Homo sapiens prominin 1 (PROM1), mRNA. | 0.00E+00 |
| BX100246 | Hs.38132 | BX100246 Soares fetal liver spleen 1NFLS Homo sapiens cDNA clone IMAGp998N04398, mRNA sequence | 0.00E+00 |
| NM_001001995 | GPM6B | Homo sapiens glycoprotein M6B (GPM6B), transcript variant 1, mRNA. | 0.00E+00 |
| NM_002345 | LUM | Homo sapiens lumican (LUM), mRNA. | 0.00E+00 |
| NM_007374 | SIX6 | Homo sapiens SIX homeobox 6 (SIX6), mRNA. | 0.00E+00 |
| NM_181676 | PPP2R2B | Homo sapiens protein phosphatase 2 (formerly 2A), regulatory subunit B, beta isoform (PPP2R2B), transcript variant 4, mRNA. | 0.00E+00 |
| NM_005627 | SGK | Homo sapiens serum/glucocorticoid regulated kinase (SGK), mRNA. | 0.00E+00 |
| NM_020975 | RET | Homo sapiens ret proto-oncogene (RET), transcript variant 2, mRNA. | 0.00E+00 |
| NM_020775 | KIAA1324 | Homo sapiens KIAA1324 (KIAA1324), mRNA. | 0.00E+00 |
| NM_002930 | RIT2 | Homo sapiens Ras-like without CAAX 2 (RIT2), mRNA. | 0.00E+00 |
| NM_080390 | TCEAL2 | Homo sapiens transcription elongation factor A (SII)-like 2 (TCEAL2), mRNA. | 0.00E+00 |
| NM_021170 | HES4 | Homo sapiens hairy and enhancer of split 4 (Drosophila) (HES4), mRNA. | 0.00E+00 |
| NM_001475 | GAGE5 | Homo sapiens G antigen 5 (GAGE5), mRNA. | 0.00E+00 |
| NM_130467 | PAGE5 | Homo sapiens P antigen family, member 5 (prostate associated) (PAGE5), transcript variant 1, mRNA. | 0.00E+00 |
| NM_004431 | EPHA2 | Homo sapiens EPH receptor A2 (EPHA2), mRNA. | 0.00E+00 |
| NM_182898 | CREB5 | Homo sapiens cAMP responsive element binding protein 5 (CREB5), transcript variant 1, mRNA. | 0.00E+00 |
| NM_001476 | GAGE6 | Homo sapiens G antigen 6 (GAGE6), mRNA. | 0.00E+00 |
| NM_001475 | GAGE5 | Homo sapiens G antigen 5 (GAGE5), mRNA. | 0.00E+00 |
| NM_001099660 | LRRN3 | Homo sapiens leucine rich repeat neuronal 3 (LRRN3), transcript variant 1, mRNA. | 0.00E+00 |
| NM_004192 | ASMTL | Homo sapiens acetylserotonin O-methyltransferase-like (ASMTL), mRNA. | 0.00E+00 |
| NM_004192 | ASMTL | Homo sapiens acetylserotonin O-methyltransferase-like (ASMTL), mRNA. | 0.00E+00 |
| NM_001474 | GAGE4 | Homo sapiens G antigen 4 (GAGE4), mRNA. | 0.00E+00 |
| NM_020822 | KCNT1 | Homo sapiens potassium channel, subfamily T, member 1 (KCNT1), mRNA. | 0.00E+00 |
| NM_001098409 | GAGE12G | Homo sapiens G antigen 12G (GAGE12G), mRNA. | 0.00E+00 |
| XM_938742 | SGPP2 | PREDICTED: Homo sapiens sphingosine-1-phosphate phosphotase 2 (SGPP2), mRNA. | 0.00E+00 |
| NM_014550 | CARD10 | Homo sapiens caspase recruitment domain family, member 10 (CARD10), mRNA. | 0.00E+00 |
| NM_001098411 | LOC645037 | Homo sapiens similar to GAGE-2 protein (G antigen 2) (LOC645037), mRNA. | 0.00E+00 |
| NM_001477 | GAGE12I | Homo sapiens G antigen 12I (GAGE12I), mRNA. | 0.00E+00 |
| NM_004617 | TM4SF4 | Homo sapiens transmembrane 4 L six family member 4 (TM4SF4), mRNA. | 1.00E−04 |
| NM_012202 | GNG3 | Homo sapiens guanine nucleotide binding protein (G protein), gamma 3 (GNG3), mRNA. | 1.00E−04 |
| NM_014220 | TM4SF1 | Homo sapiens transmembrane 4 L six family member 1 (TM4SF1), mRNA. | 1.00E−04 |
| NM_001031733 | CALML4 | Homo sapiens calmodulin-like 4 (CALML4), transcript variant 2, mRNA. | 1.00E−04 |
| NM_020116 | FSTL5 | Homo sapiens follistatin-like 5 (FSTL5), mRNA. | 2.00E−04 |
| NM_006528 | TFPI2 | Homo sapiens tissue factor pathway inhibitor 2 (TFPI2), mRNA. | 2.00E−04 |
| NM_207336 | ZNF467 | Homo sapiens zinc finger protein 467 (ZNF467), mRNA. | 2.00E−04 |
| NM_004864 | GDF15 | Homo sapiens growth differentiation factor 15 (GDF15), mRNA. | 2.00E−04 |
| NM_152780 | MAP7D2 | Homo sapiens MAP7 domain containing 2 (MAP7D2), mRNA. | 3.00E−04 |
| NM_133505 | DCN | Homo sapiens decorin (DCN), transcript variant C, mRNA. | 3.00E−04 |
| NM_022450 | RHBDF1 | Homo sapiens rhomboid 5 homolog 1 (Drosophila) (RHBDF1), mRNA. | 3.00E−04 |
| NM_172081 | CAMK2B | Homo sapiens calcium/calmodulin-dependent protein kinase (CaM kinase) II beta (CAMK2B), transcript variant 5, mRNA. | 4.00E−04 |
| NM_177526 | PPAP2C | Homo sapiens phosphatidic acid phosphatase type 2C (PPAP2C), transcript variant 2, mRNA. | 5.00E−04 |
| NM_000582 | SPP1 | Homo sapiens secreted phosphoprotein 1 (SPP1), transcript variant 2, mRNA. | 6.00E−04 |
| NM_183337 | RGS11 | Homo sapiens regulator of G-protein signaling 11 (RGS11), transcript variant 1, mRNA. | 7.00E−04 |
| NM_194439 | RNF212 | Homo sapiens ring finger protein 212 (RNF212), mRNA. | 8.00E−04 |
| NM_002414 | CD99 | Homo sapiens CD99 molecule (CD99), transcript variant 1, mRNA. | 9.00E−04 |
For the main effect of 2N-0N, we identified 56 genes that differed significantly. These are listed in Table 5. Of these genes, 52 also demonstrated 3R-4R main effect differences.
Table 5.
Table of significant transcripts for testing “Class 2N vs. 0N” main effect (56 transcripts were found significant at level 0.001)
| RefSeq transcript ID |
Gene symbol | Entrez Gene name | p-value |
|---|---|---|---|
| NM_002334 | LRP4 | Homo sapiens low density lipoprotein receptor-related protein 4 (LRP4), mRNA. | 0.00E+00 |
| XM_939093 | FAM89A | PREDICTED: Homo sapiens family with sequence similarity 89, member A (FAM89A), mRNA. | 0.00E+00 |
| NM_001769 | CD9 | Homo sapiens CD9 molecule (CD9), mRNA. | 0.00E+00 |
| NM_018286 | TMEM100 | Homo sapiens transmembrane protein 100 (TMEM100), transcript variant 2, mRNA. | 0.00E+00 |
| NM_182898 | CREB5 | Homo sapiens cAMP responsive element binding protein 5 (CREB5), transcript variant 1, mRNA. | 0.00E+00 |
| NM_020975 | RET | Homo sapiens ret proto-oncogene (RET), transcript variant 2, mRNA. | 0.00E+00 |
| NM_018027 | FRMD4A | Homo sapiens FERM domain containing 4A (FRMD4A), mRNA. | 0.00E+00 |
| NM_007115 | TNFAIP6 | Homo sapiens tumor necrosis factor, alpha-induced protein 6 (TNFAIP6), mRNA. | 0.00E+00 |
| NM_001613 | ACTA2 | Homo sapiens actin, alpha 2, smooth muscle, aorta (ACTA2), mRNA. | 0.00E+00 |
| NM_182801 | EGFLAM | Homo sapiens EGF-like, fibronectin type III and laminin G domains (EGFLAM), transcript variant 4, mRNA. | 0.00E+00 |
| NM_002942 | ROBO2 | Homo sapiens roundabout, axon guidance receptor, homolog 2 (Drosophila) (ROBO2), mRNA. | 0.00E+00 |
| NM_000905 | NPY | Homo sapiens neuropeptide Y (NPY), mRNA. | 0.00E+00 |
| NM_004100 | EYA4 | Homo sapiens eyes absent homolog 4 (Drosophila) (EYA4), transcript variant 1, mRNA. | 0.00E+00 |
| NM_000582 | SPP1 | Homo sapiens secreted phosphoprotein 1 (SPP1), transcript variant 2, mRNA. | 0.00E+00 |
| NM_194435 | VIP | Homo sapiens vasoactive intestinal peptide (VIP), transcript variant 2, mRNA. | 0.00E+00 |
| NM_017637 | BNC2 | Homo sapiens basonuclin 2 (BNC2), mRNA. | 0.00E+00 |
| NM_014141 | CNTNAP2 | Homo sapiens contactin associated protein-like 2 (CNTNAP2), mRNA. | 0.00E+00 |
| NM_003619 | PRSS12 | Homo sapiens protease, serine, 12 (neurotrypsin, motopsin) (PRSS12), mRNA. | 0.00E+00 |
| NM_022450 | RHBDF1 | Homo sapiens rhomboid 5 homolog 1 (Drosophila) (RHBDF1), mRNA. | 0.00E+00 |
| NM_001475 | GAGE5 | Homo sapiens G antigen 5 (GAGE5), mRNA. | 0.00E+00 |
| NM_033258 | GNG8 | Homo sapiens guanine nucleotide binding protein (G protein), gamma 8 (GNG8), mRNA. | 0.00E+00 |
| NM_001477 | GAGE12I | Homo sapiens G antigen 12I (GAGE12I), mRNA. | 0.00E+00 |
| NM_001098411 | LOC645037 | Homo sapiens similar to GAGE-2 protein (G antigen 2) (LOC645037), mRNA. | 0.00E+00 |
| NM_001476 | GAGE6 | Homo sapiens G antigen 6 (GAGE6), mRNA. | 0.00E+00 |
| NM_145764 | MGST1 | Homo sapiens microsomal glutathione S-transferase 1 (MGST1), transcript variant 1d, mRNA. | 0.00E+00 |
| NM_001474 | GAGE4 | Homo sapiens G antigen 4 (GAGE4), mRNA. | 0.00E+00 |
| NM_001946 | DUSP6 | Homo sapiens dual specificity phosphatase 6 (DUSP6), transcript variant 1, mRNA. | 0.00E+00 |
| NM_002930 | RIT2 | Homo sapiens Raslike without CAAX 2 (RIT2), mRNA. | 0.00E+00 |
| NM_002737 | PRKCA | Homo sapiens protein kinase C, alpha (PRKCA), mRNA. | 0.00E+00 |
| NM_002213 | ITGB5 | Homo sapiens integrin, beta 5 (ITGB5), mRNA. XM_944688 XM_944693 | 0.00E+00 |
| NM_001475 | GAGE5 | Homo sapiens G antigen 5 (GAGE5), mRNA. | 0.00E+00 |
| NM_004864 | GDF15 | Homo sapiens growth differentiation factor 15 (GDF15), mRNA. | 0.00E+00 |
| BX100246 | Hs.38132 | BX100246 Soares fetal liver spleen 1NFLS Homo sapiens cDNA clone IMAGp998N0 4398, mRNA sequence | 0.00E+00 |
| AB073882 | Hs.1832 | Homo sapiens primary neuroblastoma cDNA, clone:Nbla00830, full insert sequence | 0.00E+00 |
| NM_006017 | PROM1 | Homo sapiens prominin 1 (PROM1), mRNA. | 0.00E+00 |
| NM_002345 | LUM | Homo sapiens lumican (LUM), mRNA. | 0.00E+00 |
| NM_001098409 | GAGE12G | Homo sapiens G antigen 12G (GAGE12G), mRNA. | 0.00E+00 |
| NM_020630 | RET | Homo sapiens ret proto-oncogene (RET), transcript variant 4, mRNA. | 0.00E+00 |
| NM_002414 | CD99 | Homo sapiens CD99 molecule (CD99), transcript variant 1, mRNA. | 1.00E−04 |
| NM_001915 | CYB561 | Homo sapiens cytochrome b-561 (CYB561), transcript variant 1, mRNA. | 1.00E−04 |
| NM_057159 | LPAR1 | Homo sapiens lysophosphatidic acid receptor 1 (LPAR1), transcript variant 2, mRNA. | 1.00E−04 |
| NM_003283 | TNNT1 | Homo sapiens troponin T type 1 (skeletal, slow) (TNNT1), mRNA. | 1.00E−04 |
| NM_020975 | RET | Homo sapiens ret proto-oncogene (RET), transcript variant 2, mRNA. | 1.00E−04 |
| NM_001031733 | CALML4 | Homo sapiens calmodulin-like 4 (CALML4), transcript variant 2, mRNA. | 1.00E−04 |
| NM_013364 | PNMA3 | Homo sapiens paraneoplastic antigen MA3 (PNMA3), mRNA. | 1.00E−04 |
| NM_014220 | TM4SF1 | Homo sapiens transmembrane 4 L six family member 1 (TM4SF1), mRNA. | 1.00E−04 |
| NM_032048 | EMILIN2 | Homo sapiens elastin microfibril interfacer 2 (EMILIN2), mRNA. | 2.00E−04 |
| XM_001132569 | LOC730130 | PREDICTED: Homo sapiens hypothetical protein LOC730130 (LOC730130), mRNA. | 2.00E−04 |
| NM_004617 | TM4SF4 | Homo sapiens transmembrane 4 L six family member 4 (TM4SF4), mRNA. | 3.00E−04 |
| NM_001001391 | CD44 | Homo sapiens CD44 molecule (Indian blood group) (CD44), transcript variant 4, mRNA. | 3.00E−04 |
| NM_023940 | RASL11B | Homo sapiens RAS-like, family 11, member B (RASL11B), mRNA. | 4.00E−04 |
| NM_130467 | PAGE5 | Homo sapiens P antigen family, member 5 (prostate associated) (PAGE5), transcript variant 1, mRNA. | 4.00E−04 |
| NM_144594 | GTSF1 | Homo sapiens gametocyte specific factor 1 (GTSF1), mRNA. | 5.00E−04 |
| NM_194439 | RNF212 | Homo sapiens ring finger protein 212 (RNF212), mRNA. | 6.00E−04 |
| NM_006528 | TFPI2 | Homo sapiens tissue factor pathway inhibitor 2 (TFPI2), mRNA. | 8.00E−04 |
| BX537518 | Hs.7023 | Homo sapiens mRNA; cDNA DKFZp686N1989 (from clone DKFZp686N1989) | 9.00E−04 |
Last, 130 transcripts demonstrated an interaction effect listed in Table 6. (The majority of these also were subject to 3R-4R main effects). The interactions did not show a clear pattern.
Table 6.
Significant ANOVA interactions between 0N/2N and 3R/4R Classes
| RefSeq transcript ID |
Gene symbol | Mean1 4R/2N (A) |
Mean1 3R/2N (B) |
Mean1 4R/0N (C) |
Mean1 3R/0N (D) |
|---|---|---|---|---|---|
| NM_000685 | AGTR1 | 6.02 | 6.44 | 7.63 | 5.93 |
| NM_004192 | ASMTL | 8.92 | 6.50 | 7.85 | 7.61 |
| NM_018354 | C20orf46 | 9.39 | 6.71 | 8.04 | 8.42 |
| NM_002414 | CD99 | 5.26 | 5.54 | 9.10 | 5.72 |
| NM_014141 | CNTNAP2 | 10.81 | 7.06 | 7.79 | 6.54 |
| NM_133505 | DCN | 5.51 | 7.57 | 5.81 | 5.77 |
| NM_012242 | DKK1 | 8.30 | 12.18 | 9.92 | 10.92 |
| NM_001387 | DPYSL3 | 8.84 | 9.25 | 9.50 | 7.85 |
| NM_001430 | EPAS1 | 6.33 | 8.65 | 8.64 | 7.87 |
| NM_144967 | ARHGAP36 | 6.35 | 7.43 | 8.08 | 5.92 |
| NM_001458 | FLNC | 7.83 | 6.45 | 5.81 | 7.33 |
| NM_001001995 | GPM6B | 8.91 | 9.30 | 7.62 | 9.52 |
| NM_003918 | GYG2 | 5.37 | 5.69 | 7.11 | 5.50 |
| NR_002196 | H19 | 11.47 | 13.03 | 13.45 | 11.78 |
| AK095715 | Hs.155736 | 5.67 | 7.58 | 7.70 | 6.66 |
| CD678339 | TMEM229A | 8.28 | 9.64 | 9.80 | 9.34 |
| NM_000598 | IGFBP3 | 5.53 | 7.66 | 6.07 | 5.56 |
| NM_002213 | ITGB5 | 5.92 | 6.39 | 7.64 | 6.56 |
| NM_020822 | KCNT1 | 9.99 | 6.48 | 7.75 | 7.43 |
| NM_152780 | MAP7D2 | 8.43 | 6.37 | 8.17 | 8.21 |
| NM_024019 | NEUROG2 | 5.68 | 9.88 | 7.72 | 6.47 |
| NM_181689 | NNAT | 9.23 | 11.75 | 11.61 | 11.30 |
| NM_005386 | NNAT | 6.39 | 9.02 | 8.86 | 8.46 |
| NM_130467 | PAGE5 | 6.02 | 5.57 | 9.03 | 5.45 |
| NM_080390 | TCEAL2 | 8.92 | 6.05 | 6.76 | 6.30 |
| NM_003598 | TEAD2 | 5.45 | 8.72 | 8.60 | 5.85 |
| NM_004617 | TM4SF4 | 5.34 | 8.41 | 5.40 | 5.52 |
| NM_002166 | ID2 | 8.80 | 10.29 | 10.19 | 8.43 |
| NM_080415 | 4-Sep | 8.36 | 6.50 | 6.83 | 7.38 |
| NM_000316 | PTH1R | 7.84 | 6.17 | 6.56 | 6.70 |
| NM_032594 | INSM2 | 9.63 | 11.50 | 11.23 | 10.89 |
| NM_001001391 | CD44 | 5.74 | 7.57 | 5.77 | 5.52 |
| NM_032808 | LINGO1 | 7.53 | 5.95 | 7.12 | 7.37 |
| NM_006157 | NELL1 | 6.89 | 6.37 | 7.85 | 6.07 |
| NM_194439 | RNF212 | 7.11 | 5.37 | 5.35 | 5.47 |
| NM_020826 | SYT13 | 8.65 | 6.31 | 7.53 | 6.74 |
| NM_000419 | ITGA2B | 8.09 | 6.63 | 6.73 | 7.35 |
| NM_033258 | GNG8 | 5.90 | 8.84 | 8.04 | 9.11 |
| XM_001132569 | LOC730130 | 6.55 | 7.79 | 8.25 | 7.78 |
| NM_012202 | GNG3 | 9.88 | 6.83 | 7.48 | 7.36 |
| NM_003161 | FAM20C | 7.38 | 5.80 | 5.90 | 6.09 |
| NM_004192 | ASMTL | 8.37 | 6.16 | 7.52 | 7.29 |
| NM_001099660 | LRRN3 | 8.29 | 10.03 | 9.29 | 9.50 |
| NM_194435 | VIP | 6.27 | 10.53 | 5.84 | 8.23 |
| NM_020775 | KIAA1324 | 7.49 | 5.47 | 6.04 | 5.66 |
| NM_020851 | ISLR2 | 7.61 | 8.51 | 9.52 | 8.24 |
| NM_014220 | TM4SF1 | 5.27 | 6.37 | 5.23 | 5.28 |
| NM_001031733 | CALML4 | 8.88 | 8.90 | 7.76 | 8.86 |
| NM_005627 | SGK1 | 7.45 | 7.11 | 8.45 | 6.68 |
| NM_032229 | SLITRK6 | 5.52 | 7.28 | 7.01 | 6.58 |
| NM_057159 | LPAR1 | 7.23 | 9.50 | 7.15 | 7.96 |
| NM_181676 | PPP2R2B | 9.48 | 7.61 | 8.93 | 8.56 |
| NM_001039582 | PNCK | 9.85 | 7.70 | 9.21 | 8.51 |
| NM_000728 | CALCB | 9.55 | 8.24 | 8.15 | 8.72 |
| NM_002148 | HOXD10 | 6.06 | 6.97 | 5.94 | 8.45 |
| NM_174947 | C19ORF30 | 7.65 | 6.17 | 6.25 | 6.66 |
| NM_023940 | RASL11B | 7.69 | 8.79 | 8.94 | 8.76 |
| NM_012155 | EML2 | 9.94 | 8.17 | 8.73 | 8.97 |
| NM_033504 | TMEM54 | 7.68 | 6.14 | 6.42 | 6.79 |
| NM_001476 | GAGE6 | 5.36 | 5.51 | 5.51 | 6.69 |
| NM_181876 | PPP2R2C | 8.52 | 6.10 | 7.81 | 7.12 |
| NM_003053 | SLC18A1 | 7.38 | 7.62 | 8.81 | 7.28 |
| NM_177526 | PPAP2C | 8.54 | 6.92 | 8.04 | 8.10 |
| NM_007374 | SIX6 | 8.55 | 6.21 | 7.20 | 6.72 |
| XM_938742 | SGPP2 | 7.45 | 7.59 | 6.17 | 7.82 |
| NR_003512 | UBE2J2 | 5.68 | 6.79 | 7.82 | 5.79 |
| NM_000582 | SPP1 | 5.72 | 6.88 | 5.44 | 5.39 |
| NM_002872 | RAC2 | 7.06 | 5.56 | 6.22 | 5.70 |
| NM_002224 | ITPR3 | 8.19 | 6.72 | 6.73 | 7.38 |
| NM_014550 | CARD10 | 7.51 | 5.94 | 6.46 | 6.31 |
| NM_207336 | ZNF467 | 6.66 | 5.34 | 5.54 | 5.52 |
| NM_004864 | GDF15 | 8.23 | 6.85 | 6.68 | 6.68 |
| NM_001290 | LDB2 | 8.07 | 5.94 | 7.52 | 6.43 |
| NM_000624 | SERPINA5 | 9.21 | 5.91 | 8.81 | 6.84 |
| NM_018286 | TMEM100 | 8.97 | 9.80 | 9.68 | 11.22 |
| NM_015557 | CHD5 | 7.90 | 6.17 | 6.11 | 6.36 |
| NM_002345 | LUM | 5.76 | 7.31 | 5.64 | 5.98 |
| CN304251 | Hs.436189 | 8.18 | 5.89 | 7.27 | 7.47 |
| NM_172081 | CAMK2B | 9.60 | 7.79 | 8.88 | 8.92 |
| NM_080760 | DACH1 | 6.85 | 9.57 | 7.74 | 9.00 |
| NM_013322 | SNX10 | 9.13 | 6.96 | 8.34 | 7.38 |
| NM_018962 | DSCR6 | 7.75 | 5.94 | 6.69 | 6.04 |
| BX100246 | Hs.38132 | 6.02 | 7.48 | 5.89 | 6.26 |
| NM_021170 | HES4 | 10.17 | 8.26 | 8.91 | 8.60 |
| NM_004431 | EPHA2 | 8.62 | 6.99 | 7.55 | 7.32 |
| NM_001453 | FOXC1 | 6.68 | 8.94 | 7.73 | 8.85 |
| NM_022450 | RHBDF1 | 7.04 | 5.99 | 5.76 | 5.77 |
| NM_020975 | RET | 5.90 | 7.14 | 6.95 | 7.21 |
| NM_152330 | FRMD6 | 6.71 | 8.43 | 6.96 | 7.75 |
| NM_006528 | TFPI2 | 7.06 | 7.01 | 8.67 | 6.93 |
| NM_201630 | LRRN2 | 7.80 | 5.76 | 6.92 | 5.96 |
| NM_001013653 | LRRC26 | 8.92 | 6.35 | 8.23 | 6.93 |
| NM_001474 | GAGE7 | 5.24 | 5.41 | 5.43 | 6.69 |
| NM_014817 | TRIL | 8.01 | 9.73 | 8.16 | 8.69 |
| NM_001475 | GAGE7 | 5.27 | 5.47 | 5.43 | 6.66 |
| NM_183337 | RGS11 | 9.75 | 7.64 | 8.15 | 8.21 |
| NM_182801 | EGFLAM | 7.59 | 6.34 | 6.71 | 6.08 |
| NM_001098409 | GAGE6 | 5.41 | 5.55 | 5.53 | 6.75 |
| NM_001001995 | GPM6B | 6.92 | 7.68 | 6.09 | 7.90 |
| NM_013364 | PNMA3 | 9.28 | 6.77 | 8.04 | 6.69 |
| NM_004100 | EYA4 | 6.59 | 8.30 | 6.30 | 7.16 |
| NM_006017 | PROM1 | 5.54 | 6.68 | 5.43 | 5.74 |
| NM_000787 | DBH | 11.04 | 12.34 | 12.60 | 12.10 |
| NM_001477 | GAGE12I | 5.19 | 5.31 | 5.39 | 6.55 |
| NM_001946 | DUSP6 | 6.13 | 7.46 | 7.09 | 7.60 |
| NM_002930 | RIT2 | 6.88 | 5.66 | 5.76 | 5.50 |
| NM_001475 | GAGE7 | 5.45 | 5.64 | 5.67 | 6.90 |
| NM_001915 | CYB561 | 9.21 | 8.61 | 10.19 | 8.69 |
| NM_145764 | MGST1 | 8.21 | 6.71 | 8.39 | 7.82 |
| NM_020116 | FSTL5 | 8.11 | 6.84 | 6.96 | 6.88 |
| NM_173662 | RNF175 | 8.68 | 6.66 | 7.99 | 7.05 |
| NM_001098411 | GAGE2B | 5.38 | 5.52 | 5.58 | 6.71 |
| NM_003020 | SCG5 | 11.59 | 9.96 | 11.47 | 10.48 |
| NM_020630 | RET | 6.77 | 8.19 | 7.80 | 8.31 |
| NM_003619 | PRSS12 | 7.92 | 9.31 | 7.66 | 8.15 |
| NM_003283 | TNNT1 | 9.89 | 8.03 | 10.01 | 9.23 |
| NM_020975 | RET | 6.60 | 8.12 | 7.71 | 8.57 |
| NR_003655 | POLR2J4 | 5.85 | 7.38 | 7.37 | 5.72 |
| NM_207334 | FAM43B | 8.03 | 5.71 | 7.71 | 5.89 |
| NM_139072 | DNER | 7.47 | 5.52 | 7.12 | 5.86 |
| NM_018027 | FRMD4A | 7.94 | 6.72 | 7.62 | 5.91 |
| BX537518 | LOC100130155 | 6.62 | 5.01 | 5.89 | 5.00 |
| NM_001613 | ACTA2 | 7.95 | 6.58 | 6.84 | 6.18 |
| NM_017637 | BNC2 | 5.48 | 7.00 | 5.29 | 6.31 |
| NM_005519 | HMX2 | 5.48 | 7.98 | 5.50 | 7.60 |
| NM_000905 | NPY | 12.53 | 9.22 | 12.19 | 8.23 |
| NM_174941 | CD163L1 | 7.56 | 5.93 | 7.93 | 5.91 |
| NM_182898 | CREB5 | 9.37 | 9.63 | 7.91 | 8.95 |
| NM_002737 | PRKCA | 8.89 | 9.81 | 9.09 | 10.62 |
| XM_939093 | FAM89A | 7.42 | 6.51 | 5.88 | 5.49 |
Signal intensity values log2 transformed.
DISCUSSION
To the best of our knowledge, this is the first study to examine the downstream consequences of 3R and 4R isoforms, controlling for the presence or absence of 2N. In this study, we focused on and demonstrated expression differences between MAPT isoforms 4R tau and 3R tau in multiple transcripts, independent of inclusion/exclusion of 2N. As based on our analytic plan sixty eight transcripts were found to be common to both 3R-4R comparisons and were also identical in direction of regulation.
Comments about specific transcripts and signaling pathways
We found consistent evidence that transcripts involved in embryonic development were downregulated in the presence of 4R isoforms, and that transcripts related to outgrowth of neurites were generally upregulated. These data suggested that inclusion of the exon 10 may play a key role in neuronal maturation. Additionally, transcripts implicated in cell proliferation and survival were usually downregulated in 4R isoforms. Our data are in agreement with previous work by Sennvik et al. [16] that established a role for 4R tau isoforms in promoting neuronal differentiation and suppressing proliferation in hippocampus using a tau knockin/knockout mouse.
Embryonic development
ROBO2 has a crucial role in axon guidance in the mammalian central nervous system. It is strongly expressed in the fetal human brain but weakly expressed in adult brain [17]. ROBO2 mediates the function of Slit proteins in guiding commissural axons of the major forebrain projections [18]. LPAR1 activation induces a range of cellular responses: cell proliferation and survival, cell migration, and cytoskeletal changes [19]. In cortical neurons, LPA signaling inhibits migration by inducing neurite retraction and growth cone collapse [20,21]. LPA signaling has also been reported to influence survival [22]. VIP protein plays a role as neuromodulator in cell growth and differentiation during neurodevelopment [23]. It has been widely shown to have a neuroprotective effect decreasing the death of neurons in a wide range of experimental models [24,25]. RET is a member of the cadherin family, and encodes one of the receptor tyrosine kinases, which are cell-surface molecules that transduce signals for cell growth and differentiation. This gene plays a crucial role in neural crest development. As the transcripts involved in embryonic development were generally downregulated in Tau 4R, this isoform may play a role in reducing neuronal growth or plasticity.
Cell morphology
SYT13, synaptotagmin XIII, is active throughout the entire synapse formation process, being involved in vesicle docking, exocytosis, and endocytosis of synaptic vesicles, and contributing to neurite extension [26]. In neuroblastoma and PC12 cells, overexpression of human synaptotagmin protein(s) increases outgrowth of neurites and plays a role in neuronal differentiation [27, 28]. PPP2R2C, protein phosphatase 2 (formerly 2A), regulatory subunit B, gamma isoform is one of the four major Ser/Thr phosphatases, and it is implicated in the negative control of cell growth and division. Overexpression of PPP2R2C promotes neurite outgrowth through the MAPK pathway, a key mediator of neuronal differentiation [29]. Contactin associated protein-like 2 (CNTNAP2) protein is a cell adhesion molecule member of the neurexin family that contains EGF repeats and laminin G domains. CNTNAP2 is a neuronal membrane protein expressed in the axonal membrane and in the somatodendritic compartment [30]. It may play a role in the local differentiation of the axon into distinct functional subdomains. Serum and glucocorticoid inducible kinase 1 (SGK1) has been shown to increase neurite outgrowth in cultured hippocampal neurons through microtubule depolarization [31]. SGK1 expression is transcriptionally regulated by the MAPK/ERK pathway [32]. Blocking of CD9, a cell surface glycoprotein that participates in growth and differentiation in the nervous system, has been shown to promote neurite outgrowth [33]. This complex pattern of results may be consistent with increased neuronal outgrowth when 4R isoform is present.
Cell proliferation
Of the 4R upregulated transcripts, the neuropeptide Y(NPY) is widely expressed in the central nervous system and has recently been shown to stimulate neurogenesis in the hippocampus and subventricular zone. Intracerebroventricular injection of NPY stimulates proliferation of neural precursors in the mice dentate gyrus and in the subventricular zone, and promotes differentiation of neuronal progenitors in adult mice in vivo [34, 35]. In neuroblastoma cell lines, NPY protein has been shown to decrease in a dose-dependent manner proliferation [36]. Another upregulated transcript was RAC2. This gene has been implicated in axon growth, guidance and dendrite spine formation and maintenance [37,38]. Rho GTPases are essential regulators of cytoskeletal reorganization during neuronal morphogenesis. Activation of the non-canonical Wnt pathway through disheveled, RAC and JNK affects dendritic development [39]. Our data suggest that the non-canonical wnt pathway is regulated by the presence/absence of tau exon 10 through the activation of RAC2 (see below KEGG signaling pathway).
Cell death
The data also suggest a complex role for 3R-4R differences in the cell death process. At the level of individual transcripts involved in cell death, EMILIN2, MGST1, NELL1, PPP2R2B, RAC2, SCG5, and SGK1 were upregulated in 4R, while some transcripts implicated in neuronal survival processes were downregulated (e.g., VIP, LPAR1, RET, and PRKCA).
To summarize, transcripts involved in cellular growth and proliferation were generally downregulated in 4R isoforms while transcripts involved in neurite outgrowth were generally upregulated. Exon 10 thus may diminish proliferation and survival while increasing neurite outgrowth. Additionally cell death transcripts involved in neuronal survival were downregulated in 4R tau, in concordance with our results.
A major pathway that was differentially activated by depending on 3R or 4R status was the Wnt/β-catenin signaling implicated in AD. Wnt proteins can signal through disheveled (Dvl): 1) to inhibit GSK-3β (canonical pathway); 2) to regulate RAC (non-canonical pathway); or 3) to activate a Ca2+ dependent pathway (non-canonical pathway). For the canonical pathway β-catenin mediated transcriptional activation is involved; for the non-canonical pathway RAC and Ca2+ signaling are involved. If the canonical pathway was activated we would be able to detect increased expression of the Wnt canonical pathway main products, β-catenin and the endogenous target gene Axin2 [40]. We conducted RT-qPCR for β-catenin andAxin2 in order to determine if differential expression between 4R and 3R isoforms was present (Table 2). Our results were consistent with the microarray study in which we showed no significant differences for both genes between isoforms 4R and 3R, suggesting no activation of the final products of the Wnt canonical pathway. While activation of the canonical Wnt pathway does not affect dendritic development, activation of the non-canonical Wnt pathway through disheveled, RAC and JNK does so [39]. Our data suggest involvement of the wnt non-canonical pathway regulated by the presence/absence of exon 10 through the activation of RAC2. The Rho family of GTPases including RAC2, play an important role in various aspects of neuronal development such as neurite outgrowth and differentiation, axon pathfinding, and dendritic spine formation and maintenance [38]. Thus, our data suggest that 4R isoforms engage the non-canonical wnt signaling pathway, perhaps through RAC2. This pathway may affect dendritic development through disheveled, RAC, and JNK.
Nevertheless, the relatively large number of transcripts that were involved in neurodevelopment and maturation was unexpected. We had predicted that the majority of our findings would involve transcripts or pathways ostensibly involved in neurodegeneration. When we compared our findings to a postmortem brain microarray study of tauopathies we found a single gene in common (FOXC1) [41]. When we contrasted our results to a postmortem microarray study that examined tangle bearing neurons in AD [42] we did not find any genes in common.
Our study is also subject to a number of technical limitations. We used a neuroblastoma cell culture, which may yield slightly different results than if primary cultured neurons were used. For the sake of incisive experimental design we used cells which produced either 3R or 4R isoforms, which may not fully reflect more subtle differences in 3R/4R ratios in vivo. Additionally, our FDR approach was conservative, based on presumptive independence among all probes.
In conclusion, this study demonstrated that expression differences between 4R tau and 3R tau isoforms are present independent of the inclusion/exclusion of 2N (exons 2–3). Our data add to complex findings on the role of 3R/4R in normal and abnormal neuronal function and highlight several molecular mechanisms that might drive neurodegeneration, or perhaps, set the stage for it.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by The Litwin-Zucker Research Center for Study of Alzheimer’s Disease. Analysis was performed using BRB ArrayTools developed by Dr. Richard Simon and Amy Peng, Molecular Statistics and Bioinformatics Section, National Cancer Institute. We also thank Dr. Franak Batliwalla and Ms. Aarti Damle, members of the Feinstein Institute’s microarray core facility, for their assistance.
Footnotes
Supplementary data available online: http://www.j-alz.com/issues/22/vol22-4. html#supplementarydata05
Authors’ disclosures available online (http://www.j-alz.com/disclosures/view.php?id=597).
REFERENCES
- 1.Goedert M, Wischik CM, Crowther RA, Walker JE, Klug A. Cloning and sequencing of the cDNA encoding a core protein of the paired helical filament of Alzheimer disease: identification as the microtubule-associated protein tau. Proc Natl Acad Sci U S A. 1988;85:4051–4055. doi: 10.1073/pnas.85.11.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goedert M, Spillantini MG, Jakes R, Rutherford D, Crowther RA. Multiple isoforms of human microtubule-associated protein tau: sequences and localization in neurofibrillary tangles of Alzheimer’s disease. Neuron. 1989;3:519–526. doi: 10.1016/0896-6273(89)90210-9. [DOI] [PubMed] [Google Scholar]
- 3.Kosik KS, Orecchio LD, Bakalis S, Neve RL. Developmentally regulated expression of specific tau sequences. Neuron. 1989;2:1389–1397. doi: 10.1016/0896-6273(89)90077-9. [DOI] [PubMed] [Google Scholar]
- 4.Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H, Pickering-Brown S, Chakraverty S, Isaacs A, Grover A, Hackett J, Adamson J, Lincoln S, Dickson D, Davies P, Petersen RC, Stevens M, de Graaff E, Wauters E, van Baren J, Hillebrand M, Joosse M, Kwon JM, Nowotny P, Che LK, Norton J, Morris JC, Reed LA, Trojanowski J, Basun H, Lannfelt L, Neystat M, Fahn S, Dark F, Tannenberg T, Dodd PR, Hayward N, Kwok JB, Schofield PR, Andreadis A, Snowden J, Craufurd D, Neary D, Owen F, Oostra BA, Hardy J, Goate A, van Swieten J, Mann D, Lynch T, Heutink P. Association of missense and 5’-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- 5.Spillantini MG, Murrell JR, Goedert M, Farlow MR, Klug A, Ghetti B. Mutation in the tau gene in familial multiple system tauopathy with presenile dementia. Proc Natl Acad Sci U S A. 1998;95:7737–7741. doi: 10.1073/pnas.95.13.7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ingelsson M, Ramasamy K, Russ C, Freeman SH, Orne J, Raju S, Matsui T, Growdon JH, Frosch MP, Ghetti B, Brown RH, Irizarry MC, Hyman BT. Increase in the relative expression of tau with four microtubule binding repeat regions in frontotemporal lobar degeneration and progressive supranuclear palsy brains. Acta Neuropathol. 2007;114:471–479. doi: 10.1007/s00401-007-0280-z. [DOI] [PubMed] [Google Scholar]
- 7.Ginsberg SD, Che S, Counts SE, Mufson EJ. Shift in the ratio of three-repeat tau and four-repeat tau mRNAs in individual cholinergic basal forebrain neurons in mild cognitive impairment and Alzheimer’s disease. J Neurochem. 2006;96:1401–1408. doi: 10.1111/j.1471-4159.2005.03641.x. [DOI] [PubMed] [Google Scholar]
- 8.Conrad C, Zhu J, Conrad C, Schoenfeld D, Fang Z, Ingelsson M, Stamm S, Church G, Hyman BT. Single molecule profiling of tau gene expression in Alzheimer’s disease. J Neurochem. 2007;103:1228–1236. doi: 10.1111/j.1471-4159.2007.04857.x. [DOI] [PubMed] [Google Scholar]
- 9.Butner KA, Kirschner MW. Tau protein binds to microtubules through a flexible array of distributed weak sites. J Cell Biol. 1991;115:717–730. doi: 10.1083/jcb.115.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Avila J. Intracellular and extracellular tau. Front Neurosci. 2010;4:49. doi: 10.3389/fnins.2010.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conejero-Goldberg C, Wang E, Yi C, Goldberg TE, Jones-Brando L, Marincola FM, Webster MJ, Torrey EF. Infectious pathogen detection arrays: viral detection in cell lines and postmortem brain tissue. Biotechniques. 2005;39:741–751. doi: 10.2144/000112016. [DOI] [PubMed] [Google Scholar]
- 12.Barnes M, Freudenberg J, Thompson S, Aronow B, Pavlidis P. Experimental comparison and cross-validation of the Affymetrix and Illumina gene expression analysis platforms. Nucleic Acids Res. 2005;33:5914–5923. doi: 10.1093/nar/gki890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goeman JJ, van de Geer SA, de Kort F, van Houwelingen HC. A global test for groups of genes: testing association with a clinical outcome. Bioinformatics. 2004;20:93–99. doi: 10.1093/bioinformatics/btg382. [DOI] [PubMed] [Google Scholar]
- 14.Goeman JJ, Oosting J, Cleton-Jansen AM, Anninga JK, van Houwelingen HC. Testing association of a pathway with survival using gene expression data. Bioinformatics. 2005;21:1950–1957. doi: 10.1093/bioinformatics/bti267. [DOI] [PubMed] [Google Scholar]
- 15.Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, Chen RO, Brownstein BH, Cobb JP, Tschoeke SK, MillerGraziano C, Moldawer LL, Mindrinos MN, Davis RW, Tompkins RG, Lowry SF Inflamm and Host Response to Injury Large Scale Collab. Res P. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–1037. doi: 10.1038/nature03985. [erratum appears in Nature. 2005 Dec 1;438(7068):696]. [DOI] [PubMed] [Google Scholar]
- 16.Sennvik K, Boekhoorn K, Lasrado R, Terwel D, Verhaeghe S, Korr H, Schmitz C, Tomiyama T, Mori H, Krugers H, Joels M, Ramakers GJ, Lucassen PJ, Van Leuven F. Tau4R suppresses proliferation and promotes neuronal differentiation in the hippocampus of tau knockin/knockout mice. FASEB J. 2007;21:2149–2161. doi: 10.1096/fj.06-7735com. [DOI] [PubMed] [Google Scholar]
- 17.Yue Y, Grossmann B, Galetzka D, Zechner U, Haaf T. Isolation and differential expression of two isoforms of the ROBO2/Robo2 axon guidance receptor gene in humans and mice. Genomics. 2006;88:772–778. doi: 10.1016/j.ygeno.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 18.Long H, Sabatier C, Ma L, Plump A, Yuan W, Ornitz DM, Tamada A, Murakami F, Goodman CS, Tessier-Lavigne M. Conserved roles for Slit and Robo proteins in midline commissural axon guidance. Neuron. 2004;42:213–223. doi: 10.1016/s0896-6273(04)00179-5. [DOI] [PubMed] [Google Scholar]
- 19.Choi JW, Herr DR, Noguchi K, Yung YC, Lee C, Mutoh T, Lin M, Teo ST, Park KE, Mosley AN, Chun J. LPA Receptors: Subtypes and Biological Actions. Annu. Rev. Pharmacol. Toxicol. 2010;50:157–186. doi: 10.1146/annurev.pharmtox.010909.105753. [DOI] [PubMed] [Google Scholar]
- 20.Fukushima N, Weiner JA, Kaushal D, Contos JJ, Rehen SK, Kingsbury MA, Kim KY, Chun J. Lysophosphatidic acid influences the morphology and motility of young, post-mitotic cortical neurons. Mol Cell Neurosci. 2002;20:271–282. doi: 10.1006/mcne.2002.1123. [DOI] [PubMed] [Google Scholar]
- 21.Tigyi G, Fischer DJ, Sebok A, Yang C, Dyer DL, Miledi R. Lysophosphatidic acid-induced neurite retraction in PC12 cells: control by phosphoinositide-Ca2+ signaling and Rho. J Neurochem. 1996;66:537–548. doi: 10.1046/j.1471-4159.1996.66020537.x. [DOI] [PubMed] [Google Scholar]
- 22.Zheng ZQ, Fang XJ, Qiao JT. Dual action of lysophosphatidic acid in cultured cortical neurons: survival and apoptogenic. Sheng Li Hsueh Bao. 2004;56:163–171. [PubMed] [Google Scholar]
- 23.Muller JM, Lelievre V, Becq-Giraudon L, Meunier AC. VIP as a cell-growth and differentiation neuromodulator role in neurodevelopment. Mol Neurobiol. 1995;10:115–134. doi: 10.1007/BF02740671. [DOI] [PubMed] [Google Scholar]
- 24.Wu D. Neuroprotection in experimental stroke with targeted neurotrophins. NeuroRx. 2005;2:120–128. doi: 10.1602/neurorx.2.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brenneman DE. Neuroprotection: a comparative view of vasoactive intestinal peptide and pituitary adenylate cyclase-activating polypeptide. Peptides. 2007;28:1720–1726. doi: 10.1016/j.peptides.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 26.Detrait ER, Yoo S, Eddleman CS, Fukuda M, Bittner GD, Fishman HM. Plasmalemmal repair of severed neurites of PC12 cells requires Ca(2+). and synaptotagmin. J Neurosci Res. 2000;62:566–573. doi: 10.1002/1097-4547(20001115)62:4<566::AID-JNR11>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 27.Di Giovanni S, De Biase A, Yakovlev A, Finn T, Beers J, Hoffman EP, Faden AI. In Vivo and in Vitro Characterization of Novel Neuronal Plasticity Factors Identified following Spinal Cord Injury. J Biol Chem. 2005;280:2084–2091. doi: 10.1074/jbc.M411975200. [DOI] [PubMed] [Google Scholar]
- 28.Fukuda M, Mikoshiba K. Expression of synaptotagmin I or II promotes neurite outgrowth in PC12 cells. Neurosci Lett. 2000;295:33–36. doi: 10.1016/s0304-3940(00)01585-8. [DOI] [PubMed] [Google Scholar]
- 29.Strack S. Overexpression of the protein phosphatase 2A regulatory subunit Bgamma promotes neuronal differentiation by activating the MAP kinase (MAPK) cascade. J Biol Chem. 2002;277:41525–41532. doi: 10.1074/jbc.M203767200. [DOI] [PubMed] [Google Scholar]
- 30.Bel C, Oguievetskaia K, Pitaval C, Goutebroze L, Faivre-Sarrailh C. Axonal targeting of Caspr2 in hippocampal neurons via selective somatodendritic endocytosis. J Cell Sci. 2009;122:3403–3413. doi: 10.1242/jcs.050526. [DOI] [PubMed] [Google Scholar]
- 31.Yang YC, Lin CH, Lee EH. Serum- and glucocorticoid-inducible kinase 1 (SGK1) increases neurite formation through microtubule depolymerization by SGK1 and by SGK1 phosphorylation of tau. Mol Cell Biol. 2006;26:8357–8370. doi: 10.1128/MCB.01017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mizuno H, Nishida E. The ERK MAP kinase pathway mediates induction of SGK (serum- and glucocorticoid-inducible kinase) by growth factors. Genes Cells. 2001;6:261–268. doi: 10.1046/j.1365-2443.2001.00418.x. [DOI] [PubMed] [Google Scholar]
- 33.Banerjee SA, Hadjiargyrou M, Patterson PH. An antibody to the tetraspan membrane protein CD9 promotes neurite formation in a partially alpha3beta1 integrin-dependent manner. J Neurosci. 1997;17:2756–2765. doi: 10.1523/JNEUROSCI.17-08-02756.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Decressac M, Wright B, David B, Tyers P, Jaber M, Barker RA, Gaillard A. Exogenous neuropeptide Y promotes in vivo hippocampal neurogenesis. Hippocampus. 2010 doi: 10.1002/hipo.20765. in press. [DOI] [PubMed] [Google Scholar]
- 35.Decressac M, Prestoz L, Veran J, Cantereau A, Jaber M, Gaillard A. Neuropeptide Y stimulates proliferation, migration and differentiation of neural precursors from the subventricular zone in adult mice. Neurobiol Dis. 2009;34:441–449. doi: 10.1016/j.nbd.2009.02.017. [DOI] [PubMed] [Google Scholar]
- 36.Reubi JC, Gugger M, Waser B, Schaer JC. Y(1)-mediated effect of neuropeptide Y in cancer: breast carcinomas as targets. Cancer Res. 2001;61:4636–4641. [PubMed] [Google Scholar]
- 37.Ng J, Nardine T, Harms M, Tzu J, Goldstein A, Sun Y, Dietzl G, Dickson BJ, Luo L. Rac GTPases control axon growth, guidance and branching. Nature. 2002;416:442–447. doi: 10.1038/416442a. [DOI] [PubMed] [Google Scholar]
- 38.Govek EE, Newey SE, Van Aelst L. The role of the Rho GTPases in neuronal development. Genes Dev. 2005;19:1–49. doi: 10.1101/gad.1256405. [DOI] [PubMed] [Google Scholar]
- 39.Rosso SB, Sussman D, Wynshaw-Boris A, Salinas PC. Wnt signaling through Dishevelled, Rac and JNK regulates dendritic development. Nat Neurosci. 2005;8:34–42. doi: 10.1038/nn1374. [DOI] [PubMed] [Google Scholar]
- 40.Jho EH, Zhang T, Domon C, Joo CK, Freund JN, Costantini F. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol. 2002;22:1172–1183. doi: 10.1128/MCB.22.4.1172-1183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bronner IF, Bochdanovits Z, Rizzu P, Kamphorst W, Ravid R, van Swieten JC, Heutink P. Comprehensive mRNA expression profiling distinguishes tauopathies and identifies shared molecular pathways. PLoS ONE. 2009;4:e6826. doi: 10.1371/journal.pone.0006826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dunckley T, Beach TG, Ramsey KE, Grover A, Mastroeni D, Walker DG, LaFleur BJ, Coon KD, Brown KM, Caselli R, Kukull W, Higdon R, McKeel D, Morris JC, Hulette C, Schmechel D, Reiman EM, Rogers J, Stephan DA. Gene expression correlates of neurofibrillary tangles in Alzheimer’s disease. Neurobiol Aging. 2006;27:1359–1371. doi: 10.1016/j.neurobiolaging.2005.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.