Abstract
With the aim of selecting LAB strains with antilisterial activity to be used as protective cultures to enhance the safety of dairy products, the antimicrobial properties of 117 Lactococcus lactis subsp. lactis isolated from artisanal Sardinian dairy products were evaluated, and six strains were found to produce bacteriocin-like substances. The capacity of these strains to antagonize Listeria monocytogenes during cocultivation in skimmed milk was evaluated, showing a reduction of L. monocytogenes counts of approximately 4 log units compared to the positive control after 24 h of incubation. In order for a strain to be used as bioprotective culture, it should be carefully evaluated for the presence of virulence factors, to determine what potential risks might be involved in its use. None of the strains tested was found to produce biogenic amines or to possess haemolytic activity. In addition, all strains were sensitive to clinically important antibiotics such as ampicillin, tetracycline, and vancomycin. Our results suggest that these bac+ strains could be potentially applied in cheese manufacturing to control the growth of L. monocytogenes.
1. Introduction
In modern societies, the increasing consumer demand for natural and additive-free products has led food industry to research novel and alternative technologies in food preservation, with the objective to improve quality and safety of their products.
The use of microorganisms and/or their natural metabolites to inhibit the growth of pathogenic and spoilage bacteria has appeared as a promising tool and is also perceived by the consumer as a lower-risk food preservation [1]. In particular, lactic acid bacteria (LAB) have acquired considerable relevance in the food industry and in public health, since they are widely used in fermented foods, have a long history of safe use, and are commonly given the Generally Recognized As Safe (GRAS) status [2]. The preservative ability of LAB in food is attributed mainly to the production of antimicrobial substances, including organic acid, hydrogen peroxide, and bacteriocins [3, 4]. The latter are ribosomally synthesized, biologically active peptides or proteins with antagonistic activity against specific microorganisms [5, 6]. Many bacteriocin-producing LAB strains have proven effective against spoilage and pathogenic microorganisms in food products [7–9].
In the last decades, Listeria monocytogenes has become one of the most significant foodborne pathogens due to its widespread occurrence and its ability to tolerate environmental stresses such as low pH, low temperatures, and salt concentration up to 10% [10, 11]. These characteristics enable its frequent contamination of food products, particularly those minimally processed and refrigerated. Foodborne listeriosis is known to pose a serious health hazard when it occurs in newborns, pregnant women, and immunocompromised subjects [12, 13].
Since dairy products have been frequently reported as contaminated and associated with listeriosis outbreaks [11, 14], new preservation strategies to control growth of L. monocytogenes have been developed, including the direct application of bacteriocins as purified compound or the inoculation with a bacteriocin-producer LAB strain under conditions that favour production of the bacteriocin in situ [15–17]. Nisin, produced by Lactococcus lactis, is currently the only bacteriocin approved as preservative for utilization as direct human food ingredient [18, 19]. The in-situ production of a bacteriocin by potential adjuncts or starter cultures in fermentation processes requires a bacteriocinogenic strain that is well adapted to the particular food environment in which it will be used, that is able to grow under the food processing and/or storage conditions, and that produces bacteriocin in sufficient amounts to inhibit the target bacteria. LAB originally isolated from certain food products would be the best choice as starter cultures for these same products, because they would be more competitive than LAB from other sources [20].
With the aim of selecting LAB strains with antilisterial activity to be used as protective cultures to enhance the safety of dairy products, the antimicrobial properties of 117 L. lactis subsp. lactis isolated from artisanal Sardinian dairy products were evaluated, and six strains were found to produce bacteriocin-like substances. The capacity of these strains to antagonize L. monocytogenes during cocultivation in skimmed milk was also assessed. In addition, basic safety aspects of the strains such as production of biogenic amines, haemolytic activity, and antibiotic susceptibility were addressed. To our knowledge, this is the first report on the isolation of bacteriocin-like inhibitory substances from LAB strains isolated from artisanal Sardinian dairy products.
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
Bacteriocin producer and indicator strains used in this study are listed in Table 1. Lactococcus lactis subsp. lactis strains were identified on the basis of their morphological and biochemical characteristics as previously reported [21]; the identification was confirmed by PCR analysis using species-specific primers derived from 16S rRNA sequences, according to Pu et al. [22]. They were maintained at −20°C in M17 broth (Microbiol, Cagliari, Italy) with 15% (v/v) glycerol (Microbiol) and subcultured twice as 1% inoculums in M17 broth at 30°C for 24 h prior to experimental use. Listeria monocytogenes ATCC 7644, Escherichia coli ATCC 35150, Enterococcus faecalis ATCC 29212, Staphylococcus aureus ATCC 25923, Lactobacillus plantarum DSMZ 20174, and Lactobacillus sakei subsp. sakei DSMZ 20017 were used as indicators. All indicator strains were stored on nutrient broth (Microbiol) plus 20% (v/v) glycerol at −20°C except LAB strains which were maintained in MRS broth (Microbiol) with 15% (v/v) glycerol. Before use, they were subcultured twice in appropriate medium.
Table 1.
Bacteriocinogenic strains of Lactococcus lactis subsp. lactis and indicator bacteria used in this study.
| Species | Strain | Origin |
|---|---|---|
| Bacteriocinogenic strains | ||
| Lactococcus lactis subsp. lactis | 9FS16 | Ewes cheese |
| Lactococcus lactis subsp. lactis | 16FS16 | Ewes cheese |
| Lactococcus lactis subsp. lactis | 9/20234 | Raw ewes milk |
| Lactococcus lactis subsp. lactis | 6LS5 | Raw ewes milk |
| Lactococcus lactis subsp. lactis | 3LC39 | Raw goat milk |
| Lactococcus lactis subsp. lactis | 1LC18 | Raw goat milk |
|
| ||
| Indicator strains | ||
| Listeria monocytogenes | 7644 | ATCC |
| Escherichia coli | 35150 | ATCC |
| Enterococcus faecalis | 29212 | ATCC |
| Staphylococcus aureus | 25923 | ATCC |
| Lactobacillus plantarum | 20174 | DSMZ |
| Lactobacillus sakei subsp. sakei | 20017 | DSMZ |
2.2. Screening of Lactococcus lactis Strains for Antimicrobial Compound Production
A total of 117 L. lactis subsp. lactis strains, previously isolated from Sardinian dairy products including raw ewes and goat milk, and artisanal ewes and goat cheeses, were preliminarily screened for antimicrobial compound production against the indicator strains using an agar spot method [23]. Overnight cultures of lactococci were spotted (10 μL) onto the surface of MRS agar (1.2% (w/v) agar—0.2% (w/v) glucose) plates, which were then incubated anaerobically for 24 h at 37°C. The indicator strains were inoculated into 7 mL of soft agar medium (MRS or nutrient broth containing 0.7% w/v agar) to a final concentration of approximately 107 CFU/mL. The soft media were poured on the plates which were incubated for 24 h at the optimal growth temperature and atmosphere for the indicator strains. Inhibition was scored positive in the presence of a detectable clear zone around the colony of the producer strain.
2.3. Antimicrobial Activity Assay
The lactococcal strains exhibiting an inhibitory activity against at least three indicator strains, among which are L. monocytogenes ATCC 7644 and the bacteriocin-sensitive strain L. sakei subsp. sakei DSMZ 20017, were further tested for their antimicrobial activity against L. monocytogenes ATCC 7644 using the well-diffusion method as described by Shillinger and Lücke [23] with some modifications. Briefly, 1% (v/v) aliquot of overnight culture of the indicator strain was inoculated into 20 mL of appropriate soft agar medium and poured into Petri dishes. After cooling, wells (6 mm diameter) were cut into the agar and filled with 100 μL aliquots of cell-free supernatant of the potential producer strain collected by centrifugation (10000 ×g, 15 min). In order to eliminate the inhibitory effect of lactic acid and/or H2O2, the supernatants were adjusted to pH 6.5 with 5 M NaOH, treated with catalase (1 mg/mL, Sigma, Milan, Italy), and then filtered through a 0.45 μm pore-size cellulose acetate filter (Millipore, Bedford MA, USA) prior to use. The plates were refrigerated for 4 h to allow the radial diffusion of the compounds contained in the supernatant prior to incubation for 24 h at 37°C. The antimicrobial activity was expressed as the diameter of the inhibition zones around the wells. The nisin-positive L. lactis subsp. lactis ATCC 11454 was used as positive control. Sterile M17 broth was used as negative control.
Sensitivity to proteolytic enzymes of the cell-free supernatants of bacteriocin producer strains was tested by treatment with pronase E, proteinase K, α-chymotrypsin, trypsin, and papain (Sigma). All enzymes (10 mg/mL in sterile distilled water) were filter sterilized and added to supernatants at a final concentration of 1 mg/mL in phosphate buffer (pH 6.5). Following incubation at 37°C for 2 h, enzymes were denatured by heating at 100°C for 5 min. Untreated samples were used as controls. The residual activity of enzyme-treated samples against L. monocytogenes ATCC 7644 was determined by the well-diffusion method.
2.4. Antilisterial Activity: Coculture Tests in Skimmed Milk
Bacteriocinogenic L. lactis subsp. lactis strains were separately cocultured with L. monocytogenes ATCC 7644 in 100 mL of 10% reconstituted skimmed milk (RSM; Oxoid, Basingstoke, UK) at 30°C for 24 h. Bacteriocinogenic strains were inoculated at about 5 × 106 CFU/mL, and L. monocytogenes had a final count of 106 CFU/mL. In each experiment, negative control without bacteria, control inoculated with L. monocytogenes alone, and control inoculated with bacteriocinogenic strain alone were included. After 0, 5, 10, and 24 h, samples were taken, serially diluted in sterile saline solution, and plated onto M17 agar plates for the enumeration of L. lactis and on PALCAM (Microbiol) agar plates for the enumeration of L. monocytogenes. Values of pH were also monitored by using a HI8520 pH meter (P.B.I., Milan, Italy).
2.5. Safety Assessment of Strains
The method of Bover-Cid and Holzapfel [24] was used to screen Lactococcus strains for the production of biogenic amines. Briefly, the test strains were subcultured twice at 24 h intervals in M17 broth containing 1% of each precursor amino acid: tyrosine disodium salt, L-histidine monohydrochloride, L-ornithine monohydrochloride and lysine monohydrochloride (Sigma), and 0.005% pyridoxal-5-phosphate (Sigma) as a codecarboxylase factor. All strains were then streaked in duplicate on decarboxylase medium plates each containing only one of the above-mentioned amino acids and bromocresol purple as pH indicator and incubated for 4 days in anaerobic conditions at 37°C. Decarboxylase medium without amino acids was used as control. A colour change from brown to purple in the medium indicated an increase in pH and was considered a positive result.
Haemolytic activity was determined by streaking the strains on Columbia Blood (Microbiol) agar plates supplemented with 5% defibrinated sheep blood after 48 h of incubation at 37°C. The haemolytic reaction was recorded by observation of a clear zone of hydrolysis around the colonies (β-haemolysis), a partial hydrolysis, and greenish zone (α-haemolysis) or no reaction.
Antibiotic susceptibility testing was carried out by disc diffusion method according to the Clinical and Laboratory Standards Institute (CLSI) guidelines [25], but Mueller-Hinton agar was replaced by M17 agar. The following antibiotics (Oxoid or BBL) were tested: ampicillin (AM; 10 μg), vancomycin (VA; 30 μg) (inhibitors of cell wall synthesis), streptomycin (S; 10 μg), tetracycline (TE; 30 μg), gentamicin (GM; 10 μg), kanamycin (K; 30 μg), erythromycin (E; 15 μg), chloramphenicol (C; 30 μg), clindamycin (CM; 2 μg) (inhibitors of protein synthesis), ciprofloxacin (CIP; 5 μg), and trimethoprim/sulphamethoxazole (SXT; 25 μg) (inhibitors of nucleic acids). A suspension from fresh overnight cultures with a density of McFarland 0.5 in buffered saline was plated on M17 agar plates; then, antibiotic discs were dispensed onto the plates. After incubation at 37°C for 24 h in anaerobiosis, the diameters of the bacterial-free zone were measured, and results were expressed in terms of resistance according to the interpretative criteria issued by the CLSI [26].
3. Results and Discussion
The antagonistic effect of LAB dairy strains on pathogenic microorganisms could be used for expanding the range of healthful dairy foods. LAB, originally isolated from raw milk or artisanal dairy products, are probably the best candidate for improving the microbiological safety of these foods, because they are well adapted to the conditions of the substrate.
In this study a total of 117 Lactococcus lactis subsp. lactis strains, isolated from artisanal Sardinian dairy products, were preliminarily screened for antimicrobial compound production against six indicator strains, including well-recognized foodborne pathogens, by means of an agar spot method (Figure 1). Twenty-eight strains were found to produce an inhibition zone against at least three indicators, among which are Listeria monocytogenes ATCC 7644 and the bacteriocin-sensitive strain Lactobacillus sakei subsp. sakei DSMZ 20017, and were selected for further investigation. Subsequently, the cell-free supernatants from these strains were treated with catalase, neutralized, sterilized by filtration, and tested by well-diffusion assay against L. monocytogenes ATCC 7644. Six L. lactis subsp. lactis, representing 5% of the strain tested, were found to retain antimicrobial activity, showing around the well a measurable clear zone with mean values ranging from 2.3 to 3.7 mm (Table 2, control). The substances produced by these strains were neither hydrogen peroxide nor organic acid since the inhibitory activity was not affected by catalase and was maintained in neutralized supernatants.
Figure 1.
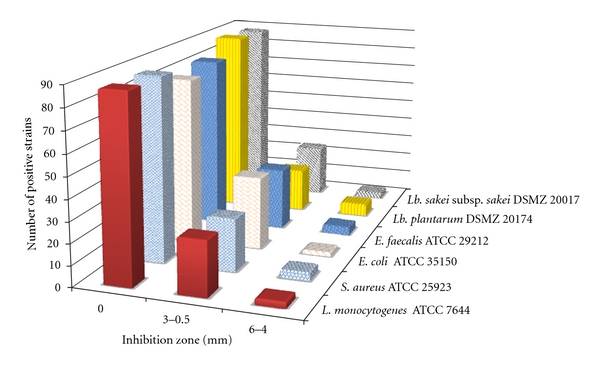
Preliminary screening for antibacterial activity of Lactococcus lactis subsp. lactis strains isolated from artisanal dairy products.
Table 2.
Enzyme sensitivity of the antibacterial compounds produced by the six Lactococcus lactis subsp. lactis determined by well-diffusion assay. Results are expressed as mean ± standard deviation of three independent experiments.
| Producer strains | Control | Residual activity against L. monocytogenes ATCC 7644 after enzymes treatment* | ||||
|---|---|---|---|---|---|---|
| Pronase E | Proteinase K | a-Chymotrypsin | Trypsin | Papain | ||
| 9FS16 | 3.7 | 0 | 0 | 2 | 3.7 | 3.7 |
| 16FS16 | 3.7 | 0 | 0 | 2 | 3.7 | 3.7 |
| 9/20234 | 2.3 | 0 | 0 | 0.8 | 2.3 | 2.3 |
| 6LS5 | 3.7 | 0 | 0 | 0.5 | 3.7 | 3.7 |
| 3LC39 | 3.0 | 0 | 0 | 0.5 | 3.0 | 3.0 |
| 1LC18 | 2.5 | 0 | 0 | 0.7 | 2.5 | 2.5 |
| Lc. lactis ATCC 11454§ | 2.7 | 0 | 0 | 1.7 | 2.7 | 2.7 |
*Inhibition zone in mm.
§Nisin A-producer.
Several studies have demonstrated the antagonistic activity of autochthonous cultures isolated from dairy products against L. monocytogenes [27–29]. The occurrence of bacteriocin-producing Lactococcus strains found in our study is lower than previously reported [27, 28]. On the other hand, the frequency of isolation of bacteriocin-producing strains is variable and could be attributed to differences such as the origin of the strains, the isolation media and technique used to detect antibacterial activity, and the diversity of indicator microorganisms used for initial screening.
The cell-free supernatants from the six strains producing antimicrobial compounds were assayed for sensitivity to proteolytic enzymes. The antimicrobial substances from all strains were completely inactivated by treatment with pronase E and proteinase K and partially eliminated with α-chymotrypsin. However, no loss of activity was observed when the supernatants were treated with trypsin and papain (Table 2). The sensitivity to proteolytic enzymes of the strains was similar to that observed in the nisin A-producer L. lactis subsp. lactis ATCC 11454 used as experimental control, thus suggesting that the inhibitory activity is related to heat-stable proteinaceous compounds and may be due to nisin-like molecules which many strains of L. lactis are known to produce [29, 30]. Further studies including purification, molecular characterization, and sequence determination of nisin genes are being currently carried out in our laboratory to confirm these findings.
The effects of the bacteriocin-producing strains on the growth of L. monocytogenes ATCC 7644 in skimmed milk are presented in Figure 2. Growth of L. monocytogenes increased from 106 CFU/mL to 108 CFU/mL within 10 h in control samples, reaching about 109 CFU/mL after 24 h of incubation. When L. monocytogenes was grown in co-culture with the bacteriocin-producing strains, different trends in the growth were obtained. As can be seen, the bacteriocin-producing strains reduced L. monocytogenes population within 10 h, although differences in the degree of inhibition were observed among the strains. The L. monocytogenes counts were reduced by approximately 4 log units compared to the positive control and by 2 log unit compared to the initial inoculum. The inhibition did not seem to be correlated with the reduction in pH during the first 10 hours of fermentation, confirming that the antimicrobial activity of the strains is not due to the production of organic acid. Our results suggest that these Bac+ strains could be potentially applied in cheese-manufacturing to control the growth of L. monocytogenes. In a previous study, a Bac+ L. lactis strain used in the manufacture of Jben cheese was able to reduce the growth of L. monocytogenes by 2.7 log units after 30 h of processing when an initial inoculum of 107 CFU/mL was used [31].
Figure 2.
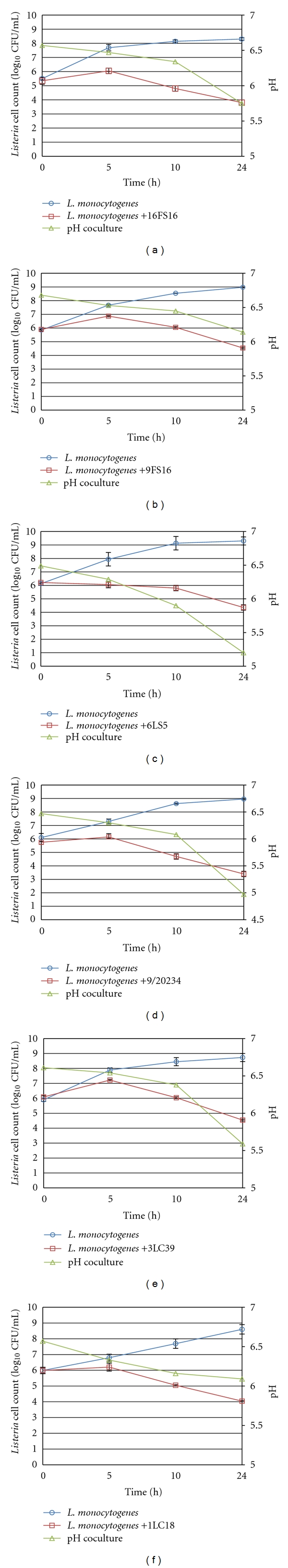
Growth of Listeria monocytogenes in co-culture with Lactococcus lactis subsp. lactis bacteriocin-producing strains. Microbial counts were calculated as the number of colony-forming units (CFU) per mL and reported as log10 CFU/mL. Data are expressed as mean ± standard errors from two independent experiments each with two replicates.
In order for a strain to be used as bioprotective culture, it should be carefully evaluated for the presence of virulence factors, to determine what potential risks might be involved in its use. The formation of biogenic amines is of concern in terms of food safety and quality. Biogenic amines are produced by LAB during the process of fermentation of foods and beverages by amino acid decarboxylation. Bover-Cid and Holzapfel [24] suggested that the capability to produce biogenic amines in a synthetic medium might be strain dependent rather than being related to specific species. In our screening, none of the strains tested was found to decarboxylate lysine, histidine, ornithine, or tyrosine (data not shown), in agreement with other findings [32, 33].
Haemolytic activity is a trait associated with virulence in some food-associated microorganisms, such as enterococci [34, 35], but it has not been frequently reported for lactococci of dairy origin [36]. In our study no strain showed haemolytic activity on sheep blood (data not shown).
The antimicrobial resistance of Lactococcus strains is reported in Table 3. Within the group of antimicrobial agents that inhibit the cell wall synthesis, all strains were susceptible to ampicillin and vancomycin. All strains were also susceptible to tetracycline, erythromycin, chloramphenicol, and clindamycin. Regarding the aminoglycosides, all strains were resistant to streptomycin, the majority to kanamycin and two to gentamycin. As for the antibiotics that inhibit the nucleic acids synthesis, all strains were resistant to trimethoprim/sulphametoxazole and none to ciprofloxacin. When multiple resistance is taken into account, two strains were resistant to two antibiotics, two to three, and two to four. Because of their long-time use in various food and feed products, LAB have been given the GRAS status [37, 38]; however, several studies have recently documented the presence and expression of antibiotic resistance genes in food-associated LAB including probiotics [39, 40], even though this trait is not commonly found in dairy LAB species [41]. Recently, the European Food Safety Authority (EFSA) has introduced the Qualified Presumption of Safety (QPS) concept, which is similar to the GRAS system in the United States and would allow microorganisms for which there are no special safety concerns to enter the market without extensive testing requirements [42]. The presence of acquired antibiotic resistance is considered by EFSA an important safety criterion for determining a strain's QPS status [43]. In this study, all strains analysed were generally resistant to aminoglycosides and trimethoprim/sulphametoxazole and sensitive to other clinically important antibiotics such as ampicillin, tetracycline, and vancomycin, in agreement with other findings [36, 44, 45], but in contrast with some reports where a high frequency of tetracycline resistance in L. lactis of probiotic and dairy origin was observed [46, 47], or a high percentage of L. lactis strains resistant to ciprofloxacin found [48]. As previously reported [48], different results may be explained by the lack of standardization in phenotypic antibiotic resistance testing in LAB food isolates, including differences in methods and media used.
Table 3.
Antibiotic resistance of the six bacteriocin-producing Lactococcus lactis subsp. lactis strains isolated from artisanal dairy products.
| Antibiotic tested | 9FS16 | 16FS16 | 9/20234 | 6LS5 | 3LC39 | 1LC18 |
|---|---|---|---|---|---|---|
| Ampicillin (10 μg) | S | S | S | S | S | S |
| Vancomycin (30 μg) | S | S | S | S | S | S |
| Streptomycin (10 μg) | R | R | R | R | R | R |
| Tetracycline (30 μg) | S | S | S | S | S | S |
| Gentamicin (10 μg) | R | R | S | S | S | S |
| Kanamycin (30 μg) | R | R | R | R | S | S |
| Erythromycin (15 μg) | S | S | S | S | S | S |
| Chloramphenicol (30 μg) | S | S | S | S | S | S |
| Clindamycin (2 μg) | S | S | S | S | S | S |
| Ciprofloxacin (5 μg) | S | S | S | S | S | S |
| Cotrimoxazole (25 μg) | R | R | R | R | R | R |
Resistance to some antibiotics such as aminoglycosides has been reported to be intrinsic for lactococci [44, 48, 49]; but particular attention should be paid to the presence of transferable resistance, since strains of L. lactis have been reported to harbor different plasmid-encoded resistance determinants [40, 46, 50].
4. Conclusions
The in situ production of bacteriocins by lactococcal strains in dairy foods provides a very attractive alternative to the use of purified bacteriocin, since many of them also generate specific aromas and flavors, but several issues including safety and adequate technological properties of the selected strains need to be addressed.
As the L. lactis strains tested in our study originated from artisanal goat and sheep dairy products and exhibited a strong inhibitory activity against L. monocytogenes, they may be useful in controlling the growth of this pathogen in dairy fermentation. The low level of antibiotic resistance observed in our strains could be of interest for a possible technological application since it has been demonstrated that L. lactis isolates displaying properties of technological interest generally exhibited a low-resistance phenotype (less than two antibiotics) [44].
Application of these bacteriocin-producing strains in food system studies is necessary to determine their effectiveness. The characterization of the bacteriocins and the technological properties of the strains are currently being investigated.
References
- 1.Paul Ross R, Morgan S, Hill C. Preservation and fermentation: past, present and future. International Journal of Food Microbiology. 2002;79(1-2):3–16. doi: 10.1016/s0168-1605(02)00174-5. [DOI] [PubMed] [Google Scholar]
- 2.Carr FJ, Chill D, Maida N. The lactic acid bacteria: a literature survey. Critical Reviews in Microbiology. 2002;28(4):281–370. doi: 10.1080/1040-840291046759. [DOI] [PubMed] [Google Scholar]
- 3.Gálvez A, Abriouel H, López RL, Omar NB. Bacteriocin-based strategies for food biopreservation. International Journal of Food Microbiology. 2007;120(1-2):51–70. doi: 10.1016/j.ijfoodmicro.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 4.Holzapfel WH. Biological preservation of foods with reference to protective cultures, bacteriocins and food-grade enzymes. International Journal of Food Microbiology. 1995;24(3):343–362. doi: 10.1016/0168-1605(94)00036-6. [DOI] [PubMed] [Google Scholar]
- 5.Cleveland J, Montville TJ, Nes IF, Chikindas ML. Bacteriocins: safe, natural antimicrobials for food preservation. International Journal of Food Microbiology. 2001;71(1):1–20. doi: 10.1016/s0168-1605(01)00560-8. [DOI] [PubMed] [Google Scholar]
- 6.De Vuyst L, Vandamme E. Antimicrobial potential of lactic acid bacteria. In: De Vuyst L, Vandamme E, editors. Bacteriocins of Lactic Acid Bacteria. London, UK: Blackie; 1994. pp. 91–142. [Google Scholar]
- 7.De Vuyst L, Leroy F. Bacteriocins from lactic acid bacteria: production, purification, and food applications. Journal of Molecular Microbiology and Biotechnology. 2007;13(4):194–199. doi: 10.1159/000104752. [DOI] [PubMed] [Google Scholar]
- 8.Allende A, Aguayo E, Artés F. Microbial and sensory quality of commercial fresh processed red lettuce throughout the production chain and shelf life. International Journal of Food Microbiology. 2004;91(2):109–117. doi: 10.1016/S0168-1605(03)00373-8. [DOI] [PubMed] [Google Scholar]
- 9.Ryan MP, Rea MC, Hill C, Ross RP. An application in cheddar cheese manufacture for a strain of Lactococcus lactis producing a novel broad-spectrum bacteriocin, lacticin 3147. Applied and Environmental Microbiology. 1996;62(2):612–619. doi: 10.1128/aem.62.2.612-619.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gandhi M, Chikindas ML. Listeria: a foodborne pathogen that knows how to survive. International Journal of Food Microbiology. 2007;113(1):1–15. doi: 10.1016/j.ijfoodmicro.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 11.Cole MB, Jones MV, Holyoak C. The effect of pH, salt concentration and temperature on the survival and growth of Listeria monocytogenes. Journal of Applied Bacteriology. 1990;69(1):63–72. doi: 10.1111/j.1365-2672.1990.tb02912.x. [DOI] [PubMed] [Google Scholar]
- 12.Mead PS, Dunne EF, Graves L, et al. Nationwide outbreak of listeriosis due to contaminated meat. Epidemiology and Infection. 2006;134(4):744–751. doi: 10.1017/S0950268805005376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McLauchlin J. The pathogenicity of Listeria monocytogenes: a public health perspective. Reviews in Medical Microbiology. 1997;8(1):1–14. [Google Scholar]
- 14.Rudolf M, Scherer S. High incidence of Listeria monocytogenes in European red smear cheese. International Journal of Food Microbiology. 2001;63(1-2):91–98. doi: 10.1016/s0168-1605(00)00413-x. [DOI] [PubMed] [Google Scholar]
- 15.Liu L, O’Conner P, Cotter PD, Hill C, Ross RP. Controlling Listeria monocytogenes in Cottage cheese through heterologous production of enterocin a by Lactococcus lactis. Journal of Applied Microbiology. 2008;104(4):1059–1066. doi: 10.1111/j.1365-2672.2007.03640.x. [DOI] [PubMed] [Google Scholar]
- 16.Samelis J, Kakouri A, Rogga KJ, Savvaidis IN, Kontominas MG. Nisin treatments to control Listeria monocytogenes post-processing contamination on Anthotyros, a traditional Greek whey cheese, stored at 4°C in vacuum packages. Food Microbiology. 2003;20(6):661–669. [Google Scholar]
- 17.Muriana PM. Bacteriocins for control of Listeria spp. in food. Journal of Food Protection. 1996;59(3):54–63. doi: 10.4315/0362-028X-59.13.54. [DOI] [PubMed] [Google Scholar]
- 18.Food and Drug Administration. Nisin preparation: affirmation of GRAS status as a direct human food ingredient. Federal Registry. 1998;53:11247–11251. [Google Scholar]
- 19.Delves-Broughton J. Nisin and its uses as a food preservative. Food Technology. 1990;44:100–117. [Google Scholar]
- 20.Trias R, Badosa E, Montesinos E, Bañeras L. Bioprotective Leuconostoc strains against Listeria monocytogenes in fresh fruits and vegetables. International Journal of Food Microbiology. 2008;127(1-2):91–98. doi: 10.1016/j.ijfoodmicro.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 21.Pisano MB, Fadda ME, Deplano M, Corda A, Cosentino S. Microbiological and chemical characterization of Fiore Sardo, a traditional Sardinian cheese made from ewe’s milk. International Journal of Dairy Technology. 2006;59(3):171–179. [Google Scholar]
- 22.Pu ZY, Dobos M, Limsowtin GKY, Powell IB. Integrated polymerase chain reaction-based procedures for the detection and identification of species and subspecies of the Gram-positive bacterial genus Lactococcus. Journal of Applied Microbiology. 2002;93(2):353–361. doi: 10.1046/j.1365-2672.2002.01688.x. [DOI] [PubMed] [Google Scholar]
- 23.Schillinger U, Lücke FK. Antibacterial activity of Lactobacillus sake isolated from meat. Applied and Environmental Microbiology. 1989;55(8):1901–1906. doi: 10.1128/aem.55.8.1901-1906.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bover-Cid S, Holzapfel WH. Improved screening procedure for biogenic amine production by lactic acid bacteria. International Journal of Food Microbiology. 1999;53(1):33–41. doi: 10.1016/s0168-1605(99)00152-x. [DOI] [PubMed] [Google Scholar]
- 25.Clinical and Laboratory Standard Institute. Performance Standards for Antimicrobial Disk Susceptibility Tests. 10th edition. Wayne, Ill, USA: Clinical and Laboratory Standard Institute; (Approved Standard). (CLSI document M02-A10), 2009. [Google Scholar]
- 26.Clinical and Laboratory Standard Institute. Performance Standards for Antimicrobial Disk Susceptibility Tests. Wayne, Ill, USA: Clinical and Laboratory Standard Institute; (12th Informational Supplement). (CLSI document M100-S20), 2010. [Google Scholar]
- 27.Ortolani MBT, Moraes PM, Perin LM, et al. Molecular identification of naturally occurring bacteriocinogenic and bacteriocinogenic-like lactic acid bacteria in raw milk and soft cheese. Journal of Dairy Science. 2010;93(7):2880–2886. doi: 10.3168/jds.2009-3000. [DOI] [PubMed] [Google Scholar]
- 28.Nero LA, Mattos MR, Beloti V, Barros MAF, Ortolani MBT, Franco BDGM. Autochthonous microbiota of raw milk with antagonistic activity against Listeria monocytogenes and salmonella enteritidis. Journal of Food Safety. 2009;29(2):261–270. [Google Scholar]
- 29.Coventry MJ, Gordon JB, Wilcock A, et al. Detection of bacteriocins of lactic acid bacteria isolated from foods and comparison with pediocin and nisin. Journal of Applied Microbiology. 1997;83(2):248–258. doi: 10.1046/j.1365-2672.1997.00216.x. [DOI] [PubMed] [Google Scholar]
- 30.Alegría A, Delgado S, Roces C, López B, Mayo B. Bacteriocins produced by wild Lactococcus lactis strains isolated from traditional, starter-free cheeses made of raw milk. International Journal of Food Microbiology. 2010;143(1-2):61–66. doi: 10.1016/j.ijfoodmicro.2010.07.029. [DOI] [PubMed] [Google Scholar]
- 31.Benkerroum N, Oubel H, Zahar M, Dlia S, Filali-Maltouf A. Isolation of a bacteriocin-producing Lactococcus lactis subsp. lactis and applicatin to control Listeria monocytogenes in Moroccan jben. Journal of Applied Microbiology. 2000;89(6):960–968. doi: 10.1046/j.1365-2672.2000.01199.x. [DOI] [PubMed] [Google Scholar]
- 32.Deepika Priyadarshani WM, Rakshit SK. Screening selected strains of probiotic lactic acid bacteria for their ability to produce biogenic amines (histamine and tyramine) International Journal of Food Science and Technology. 2011;46(10):2062–2069. [Google Scholar]
- 33.Novella-Rodríguez S, Veciana-Nogués MT, Roig-Sagués AX, Trujillo-Mesa AJ, Vidal-Carou MC. Influence of starter and nonstarter on the formation of biogenic amine in goat cheese during ripening. Journal of Dairy Science. 2002;85(10):2471–2478. doi: 10.3168/jds.S0022-0302(02)74329-4. [DOI] [PubMed] [Google Scholar]
- 34.Franz CMAP, Holzapfel WH, Stiles ME. Enterococci at the crossroads of food safety? International Journal of Food Microbiology. 1999;47(1-2):1–24. doi: 10.1016/s0168-1605(99)00007-0. [DOI] [PubMed] [Google Scholar]
- 35.Jett BD, Huycke MM, Gilmore MS. Virulence of Enterococci. Clinical Microbiology Reviews. 1994;7(4):462–478. doi: 10.1128/cmr.7.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maragkoudakis PA, Mountzouris KC, Psyrras D, et al. Functional properties of novel protective lactic acid bacteria and application in raw chicken meat against Listeria monocytogenes and Salmonella enteritidis. International Journal of Food Microbiology. 2009;130(3):219–226. doi: 10.1016/j.ijfoodmicro.2009.01.027. [DOI] [PubMed] [Google Scholar]
- 37.Borriello SP, Hammes WP, Holzapfel W, et al. Safety of probiotics that contain lactobacilli or bifidobacteria. Clinical Infectious Diseases. 2003;36(6):775–780. doi: 10.1086/368080. [DOI] [PubMed] [Google Scholar]
- 38.Salminen S, Von Wright A, Morelli L, et al. Demonstration of safety of probiotics—a review. International Journal of Food Microbiology. 1998;44(1-2):93–106. doi: 10.1016/s0168-1605(98)00128-7. [DOI] [PubMed] [Google Scholar]
- 39.Mathur S, Singh R. Antibiotic resistance in food lactic acid bacteria—a review. International Journal of Food Microbiology. 2005;105(3):281–295. doi: 10.1016/j.ijfoodmicro.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 40.Perreten V, Schwarz F, Cresta L, Boeglin M, Dasen G, Teuber M. Antibiotic resistance spread in food. Nature. 1997;389(6653):801–802. doi: 10.1038/39767. [DOI] [PubMed] [Google Scholar]
- 41.Ammor MS, Flórez AB, Van Hoek AHAM, et al. Molecular characterization of intrinsic and acquired antibiotic resistance in lactic acid bacteria and bifidobacteria. Journal of Molecular Microbiology and Biotechnology. 2008;14(1–3):6–15. doi: 10.1159/000106077. [DOI] [PubMed] [Google Scholar]
- 42.European Food Safety Authority. Brussels, Belgium: European Food Safety Authority; 2004. EFSA Scientific Colloquium Summary Report. QPS: qualified presumption of safety of microorganisms in food and feed. Tech. Rep. [Google Scholar]
- 43.European Food Safety Authority. The maintenance of the list of QPS microorganisms intentionally added to food or feed. European Food Safety Authority Journal. 2008;923:1–48. [Google Scholar]
- 44.Rodríguez-Alonso P, Fernández-Otero C, Centeno JA, Garabal JI. Antibiotic resistance in lactic acid bacteria and micrococcaceae/ staphylococcaceae isolates from artisanal raw milk cheeses, and potential implications on cheese making. Journal of Food Science. 2009;74(6):M284–M293. doi: 10.1111/j.1750-3841.2009.01217.x. [DOI] [PubMed] [Google Scholar]
- 45.Liu C, Zhang ZY, Dong K, Yuan JP, Guo XK. Antibiotic resistance of probiotic strains of lactic acid bacteria isolated from marketed foods and drugs. Biomedical and Environmental Sciences. 2009;22(5):401–412. doi: 10.1016/S0895-3988(10)60018-9. [DOI] [PubMed] [Google Scholar]
- 46.Devirgiliis C, Barile S, Caravelli A, Coppola D, Perozzi G. Identification of tetracycline- and erythromycin-resistant Gram-positive cocci within the fermenting microflora of an Italian dairy food product. Journal of Applied Microbiology. 2010;109(1):313–323. doi: 10.1111/j.1365-2672.2010.04661.x. [DOI] [PubMed] [Google Scholar]
- 47.Temmerman R, Pot B, Huys G, Swings J. Identification and antibiotic susceptibility of bacterial isolates from probiotic products. International Journal of Food Microbiology. 2003;81(1):1–10. doi: 10.1016/s0168-1605(02)00162-9. [DOI] [PubMed] [Google Scholar]
- 48.Hummel AS, Hertel C, Holzapfel WH, Franz CMAP. Antibiotic resistances of starter and probiotic strains of lactic acid bacteria. Applied and Environmental Microbiology. 2007;73(3):730–739. doi: 10.1128/AEM.02105-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ammor MS, Belén Flórez A, Mayo B. Antibiotic resistance in non-enterococcal lactic acid bacteria and bifidobacteria. Food Microbiology. 2007;24(6):559–570. doi: 10.1016/j.fm.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 50.Flórez AB, Ammor MS, Mayo B. Identification of tet(M) in two Lactococcus lactis strains isolated from a Spanish traditional starter-free cheese made of raw milk and conjugative transfer of tetracycline resistance to lactococci and enterococci. International Journal of Food Microbiology. 2008;121(2):189–194. doi: 10.1016/j.ijfoodmicro.2007.11.029. [DOI] [PubMed] [Google Scholar]


