Abstract
Sumoylation is an important posttranslational modification in which SUMO (small ubiquitin-related modifier) proteins are bonded covalently to their substrates. Studies on the roles of sumoylation in cell cycle regulation have been emerging in both mitosis from yeast to mammals and meiosis in budding yeast, but the functions of sumoylation in mammalian meiosis, especially in oocyte meiotic maturation are not well known. Here, we examined the localization and expression of SUMO-1 and SUMO-2/3, the two basic proteins in the sumoylation pathway and investigated their roles through overexpression of Senp2 during mouse oocyte maturation. Immunofluorescent staining revealed differential patterns of SUMO-1 and SUMO-2/3 localization: SUMO-1 was localized to the spindle poles in prometaphase I, MI and MII stages, around the separating homologues in anaphase I and telophase I stages of first meiosis, while SUMO-2/3 was mainly concentrated near centromeres during mouse oocyte maturation. Immunoblot analysis uncovered the different expression profiles of SUMO-1 and SUMO-2/3 modified proteins during mouse oocyte maturation. Overexpression of Senp2, a SUMO-specific isopeptidase, caused changes of SUMO-modified proteins and led to defects in MII spindle organization in mature eggs. These results suggest that the SUMO pathway may play an indispensable role during mouse oocyte meiotic maturation.
Key words: sumoylation, mouse oocyte maturation, overexpression, Senp2, MII spindle
Introduction
Sumoylation is an important posttranslational modification of proteins, which refers to SUMO (small ubiquitin-related modifier) proteins that are covalently attached to lysine residues in their substrate proteins in similar ways as in ubiquitination.1 To date, four SUMO proteins have been identified in mammals: SUMO-1,-2,-3 and -4. SUMO-2 and SUMO-3 share 96% sequence identity to each other and are referred to as SUMO-2/3, while they have only about 50% amino-acid sequence identity to SUMO-1. Sumoylation is ATP-dependent and requires E1-activating enzyme Aos1/Uba2, E2-conjugating enzyme Ubc9 responsible for the SUMO direct transfer, and diverse E3 ligases which can also function in SUMO transfer and may facilitate the sumoylation of specific substrates.2–4 Sumoylation can be reversed by a number of SUMO-specific proteases that have been discovered based on homology to yeast Ulp1, the first identified SUMO-specific protease. In mammals, at least seven Ulp1 homologs have been identified: Senp1 to -3 and Senp5 to -8, which share a conserved cysteine protease domain at their C termini.5 Sumoylation regulates diverse biological processes including gene expression, DNA repair and replication, signal transduction, nucleocytoplasmic trafficking as well as genomic and chromosomal stability and integrity.1,6–8
Accumulating evidence is showing that the SUMO pathway plays an essential role in regulation of mitotic cell cycle progression. Preliminary results are mainly obtained in studies on the budding yeast S. cerevisiae and fission yeast S. pombe. In S. cerevisia, impairment of components in the SUMO pathway results in significant defects in cell cycle progression.9–13 For fission yeast, defective chromosome segregation is seen in pmt3Δ strains14 and strains with mutations in S. pombe Aos1,15 or Ubc9,14 homologues. In yeast, a number of proteins modified by sumoylation have been identified. Pds5p is sumoylated in a cell cycle dependent manner peaking prior to the anaphase onset and sumoylation of Pds5p can disrupt Pds5p's interaction with the cohesion complex, leading to cohesion release from chromosomes.16–18 Top2p, the yeast homologue of Topoisomerase II, plays a critical role in centromeric cohesion and this function is downregulated by its sumoylation.13 Among clearly identified substrates of sumoylation are centromere- or kinetochore-associated proteins including Bir1p, Sli15p, Ipl1p, whose homologues in vertebrates are Survivin, INCENP and Aurora B, respectively; the three proteins are important components of the Chromosomal Passenger Complex (CPC), a key regulator of mitosis. Evidence for the involvement of sumoylation in the mitotic cell cycle in vertebrates including mammals is accumulating. Xenopus egg extracts fail to segregate sister chromatids when treated with a dominant-negative Ubc9 mutant. Ubc9-defecient mouse embryos are lethal in early embryonic stages because of chromosome defects in mitosis.19 The vertebrate SUMO E3 enzyme PIASy is indispensable for chromosome segregation in Xenopus egg extracts20 as well as in human tissue culture cells in which PIASy depletion results in the activation of the spindle assembly checkpoint and failure in sister-chromatid non-disjunction.21 Borealin and CENP-E, two kinetochorerelated proteins, are identified proteins of sumoylation during mitosis in mammals. Borealin is the fourth component of the CPC and its sumoylation is dynamically regulated during mitotic progression, peaking in early mitosis.22 Global inhibition of sumoylation in Hela cells leads to prometaphase arrest during the mitotic cell cycle through impairment of CENP-E targeting to kinetochores.23 Although CENP-E is identified as a substrate of SUMO-2/3, the recruitment of CENP-E to kinetochores is dependent on its binding by polySUMO-2/3.
Meiosis shares similarities with mitosis but it also displays significant differences. As for mitosis, pioneering studies used budding yeast to investigate the roles of sumoylation in meiosis. Mutant UBC9 gene in budding yeast displays defects in Zip1 polymerization along homologous chromosomes, causing structural damage of the synaptonemal complex (SC).24 Zip3 involved in the initiation of SC formation is identified as a SUMO E3 ligase.25,26 The functions of sumoylation in higher organisms have been explored in studies on spermatogenesis; it has been shown that sumoylation plays crucial roles in several processes during spermatogenesis, including XY body formation, microtubule nucleation and nuclear restructuring.27,28 Further investigations revealed expression of sumoylation pathway genes and proteins, implying their functions in meiosis and spermiogenesis.29 Although several reports refer to the SUMO pathway in oogenesis in Drosophila,30–32 no direct evidence exists to link sumoylation to the meiotic cell cycle in Drosophila. A recent study on sumoylation in mouse oocyte development suggested that sumoylation may play a role in regulating gene expression by modulating transcription and RNA processing.33 However, the role of the SUMO pathway in mouse oocyte maturation is not clear.
Oocyte maturation is an important cell cycle and development process during which immature oocytes grow to mature MII eggs awaiting fertilization. In the present study, we demonstrated the subcellular localization of SUMO-1 and SUMO-2/3 during mouse oocyte meiotic maturation and performed immunoblot analysis to show the profiles of SUMO-1 and SUMO-2/3 modified proteins. To identify the roles of the SUMO pathway during mouse oocyte maturation, we overexpressed Senp2 protein, a SUMO-specific isopeptidase, to disturb sumoylation in mouse oocytes and showed that impairment of sumoylation led to defects in MII spindle organization in mature eggs.
Results
Spatial and temporal subcellular localization of SUMO-1 and SUMO-2/3 during mouse oocyte meiotic maturation.
To examine the spatial and temporal subcellular localization of SUMO-1 and SUMO-2/3 during mouse oocyte meiotic maturation, mouse oocytes at different stages of maturation were fixed for immunofluorescent staining. As shown in Figure 1, SUMO-1 was only shown in the Germinal Vesicle (GV) of oocytes at the GV stage displaying a punctate staining pattern, but previously reported33 nuclear membrane localization of SUMO-1 was not detected. As oocytes progressed to prometaphase I and the condensed chromosomes began to migrate to the equator of the spindle, SUMO-1 gradually translocated to the spindle poles. By metaphase I, chromosomes aligned at the equatorial plate and SUMO-1 was concentrated at the spindle poles. At anaphase I to/and at telophase I during separation of the homologous chromosomes and polar body emission, SUMO-1 was detected as cloud-like formation around the separating homologues; then, part of SUMO-1 became separated into the polar body, leaving the remaining SUMO-1 in the vicinity of the chromosomes. At metaphase II, SUMO-1 again translocated to the spindle poles.
Figure 1.
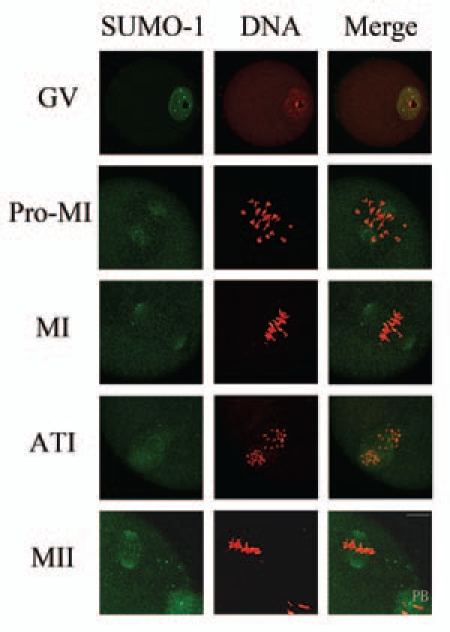
Subcellular localization of SUMO-1 during mouse oocyte maturation. Oocytes were fixed at 0 h, 4 h, 8 h, 9.5 h and 12 h of culture and then stained with a mouse monoclonal antibody specific for SUMO-1 and counterstained with the fluorescent dye propidium Iodide (PI) to visualize DNA. Green, SUMO-1; red, DNA; GV, oocytes at germinal vesicle stage; Pro-MI, oocytes at first prometaphase; MI, oocytes at first metaphase; ATI, oocytes at first anaphase; MII, oocytes at second metaphase. PB, first polar body; Bar = 20 µm. Experiments were repeated at least three times and representative images are shown.
SUMO-2/3 showed a localization pattern distinct from-SUMO-1 during mouse oocyte maturation except in the oocytes at the GV stage. Similar to SUMO-1, at the GV stage SUMO-2/3 was mainly localized as concentrated dots in the GV at the GV stage. Shortly after GVBD, SUMO-2/3 became localized to foci on condensed chromosomes. By metaphase I, SUMO-2/3 was localized near the centromeres. When oocytes proceeded to anaphase I, the localization of SUMO-2/3 did not change. In MII eggs SUMO-2/3 was clearly detected near the centromeres (Fig. 2).
Figure 2.
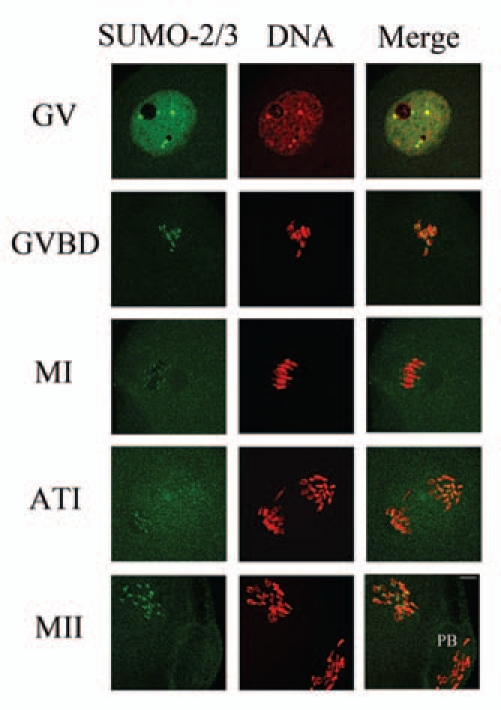
Subcellular localization of SUMO-2/3 during mouse oocyte maturation. Oocytes at different stages (0 h, 2 h, 8 h, 9.5 h and 12 h of culture, respectively) were immunolabeled with a mouse monoclonal antibody specific to SUMO-2/3 (green). GV, oocytes at germinal vesicle stage; GVBD, oocytes after germinal vesicle breakdown; MI, oocytes at first metaphase; ATI, oocytes at first anaphase; MII, oocytes at second metaphase. Each sample was counterstained with PI (red) to visualize DNA. PB, first polar body; Bar = 20 µm. Experiments were repeated at least three times and representative images are shown.
Expression profiles of SUMO-1 and SUMO-2/3 modified proteins during mouse oocyte meiotic maturation.
To investigate the expression profiles of SUMO-1 and SUMO-2/3 modified proteins during mouse oocyte maturation, we performed immunoblot analysis with samples collected after mouse oocytes had been cultured for 0 h, 4 h, 8 h, 9.5 h and 12 h, corresponding to the GV, premetaphase I, metaphase I, anaphase I and MII stages respectively, as reported by us previously.34–36 Distinct profiles of modified proteins were recognized by anti-SUMO-1 and anti-SUMO-2/3 monoclonal antibodies. We detected only two bands (up, band A; low, band B) which were seen between the 72 kDa and 55 kDa markers, using the anti-SUMO-1 antibody (Fig. 3). Both bands were only faintly detected at the GV stage and then showed different expression patterns at later stages: band A remained stable from prometaphase I to MII, but band B peaked at prometaphase I, dramatically decreased in metaphase I and then remained in low amount (Fig. S1A and B). The anti-SUMO-2/3 antibody mainly recognized three bands: two bands (up, band C; low, band D) were seen above 170 kDa and the third band (band E) was seen slightly below 72 kDa (Fig. 3). Bands C and D shared almost the same expression pattern: only small amounts could be detected in the GV stage, then increased to a higher amount in prometaphase I and remained stable until MII stage (Fig. S1C and D). Band E was little detectable in the GV stage, increased to a stable amount in later stages except for a decrease in anaphase I (Fig. S1E).
Figure 3.
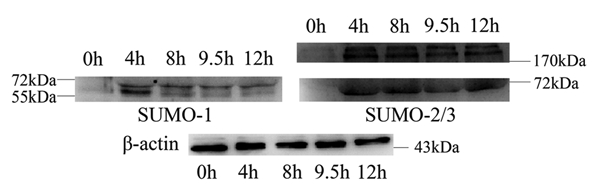
Expression of SUMO-1 and SUMO-2/3 modified proteins during mouse oocyte maturation. Samples were collected after oocyte culture for 0, 4, 8, 9.5 and 12 h, corresponding to the GV, prometaphase I, metaphase I, anaphase I, metaphase II stages, respectively. Proteins from a total of 250 oocytes were loaded for each sample. Experiments were repeated three times.
Overexpression of Senp2 caused inhibition or accumulation of SUMO modified proteins and resulted in defects in MII spindle formation in mature eggs.
Following the localizations of SUMO-1 and SUMO-2/3 during mouse oocyte maturation, we investigated the roles of sumoylation. Sumoylation of most substrates is dynamic, controlled by the relative rates of conjugation and isopeptidase-mediated deconjugation. It was recently shown in Hela cells that overexpression of Senp2, a SUMO-specific isopeptidase, inhibited accumulation of sumoylated proteins and caused prometaphase arrest.23 Myc-tagged Senp2 mRNA produced in vitro was microinjected into GV oocytes and the effects on expression of SUMO-1 and SUMO-2/3 modified proteins were examined by immunoblot analysis. Figure 4A shows a clear band agreeing with the predicted molecular weight of Myc-Senp2, indicating that Myc-Senp2 protein was expressed in mouse oocytes. As expected, Senp2 overexpression inhibited expression of SUMO modified proteins including proteins indicated by band B and band C (Figs. 4B, and S2B and C). Surprisingly, overexpression of Senp2 also caused accumulation of SUMO modified proteins including proteins revealed by band A and band E (Figs. 4B, and S2A and E).
Figure 4.

Overexpression of Senp2 resulted in changes of SUMO modified proteins. (A) Myc-Senp2 protein was expressed in mouse oocytes. Myc-Senp2 mRNA microinjected GV oocytes were incubated in M16 medium containing 2.5 µM milrinone for 5 h and collected immediately; the lysates were then analyzed by immunoblot with mouse monoclonal anti-myc antibody. Equal numbers of oocytes microinjected with Myc mRNA was analyzed as a control. (B) Germinal vesicle oocytes were microinjected with the control Myc mRNA and Myc-Senp2 mRNA, respectively. After microinjection, oocytes were incubated in M16 medium with 2.5 µM milrinone for 5 h, then transferred to milrinone-free M16 medium for 10 h and collected, followed by immunoblot analysis with SUMO-1 or SUMO-2/3 specific mouse monoclonal antibodies. Three independent experiments were performed.
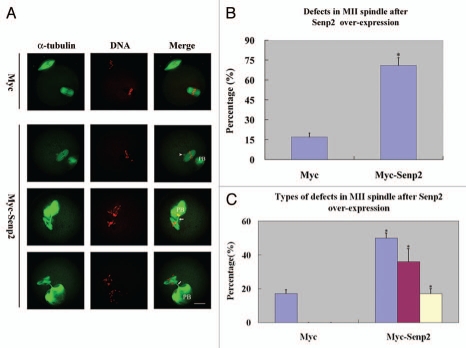
An obvious aberrance in Myc-Senp2 mRNA microinjected oocytes was observed in the MII spindle organization of mature eggs. At 14 hours of culture, most mouse oocytes had emitted the first polar body and formed normal MII spindles and only 17.3 ± 2.9% of oocytes displayed defects in MII spindle organization in the control group (Fig. 5B). However, in the Myc-Senp2 mRNA microinjection group, a large amount of MII spindles exhibited various abnormalities and defects (69.6 ± 5.8%; p < 0.05; Fig. 5B). These defects could be divided into three types: MII spindles of defective morphology, MII spindles with branched bundles of microtubules connecting to the first polar body, MII spindles with both the above defects (Fig. 5A). The percentages of the defective oocytes within the total number of oocytes were 50.4 ± 2.3%, 36.3 ± 7.6%, 17.1 ± 3.1%, respectively, while the defects in the control group all displayed the first type (Fig. 5C).
Figure 5.
Overexpression of Senp2 impaired spindle organization in MII mature eggs. (A) MII spindle morphologies in Myc mRNA and Myc-Senp2 mRNA microinjection groups. After microinjection, GV oocytes were incubated in M16 medium with 2.5 µM milrinone for 5 h, then transferred to milrinone-free M16 medium for 14 h to fully mature into MII eggs and fixed immediately, followed by immunoflurescent analysis with anti-α-tubulin-FITC antibody (green) and PI (red). In the Myc mRNA microinjection group, normal MII spindles formed. In the Myc-Senp2 mRNA microinjection group, several forms of morphologically aberrant MII spindles were seen in mature eggs. Arrow head, multipolar spindle; arrows, microtubule bundles connecting MII spindles and the first polar bodies. PB, first polar body, Bar = 20 µm. Three independent experiments were performed and representative images are shown. (B) The rate of abnormal MII spindles was recorded in the Myc mRNA microinjection group and the Myc-Senp2 mRNA microinjection group. Data are presented as mean percentage (mean ± SEM) from three independent experiments *p < 0.05. (C) The rates of the main forms of defects in MII spindles were recorded in the Myc mRNA microinjection group and the Myc-Senp2 mRNA microinjection group. Blue: MII spindles with defective morphology; purple: MII spindles with branched microtubule bundles connecting the first polar body; Yellow: MII spindles with both of the above defects. Data are presented as mean percentage (mean ± SEM) from three independent experiments *p < 0.05.
Discussion
In this study, we investigated the localization and expression of SUMO-1 and SUMO-2/3 modified proteins during mouse oocyte maturation using antibodies to endogenous proteins and found that SUMO-1 and SUMO-2/3 were conjugated to differential subsets of proteins. Overexpression of Senp2, a SUMO-specific isopeptidase, led to changes of both SUMO-1 and SUMO-2/3 modified proteins and resulted in defects in MII spindle formation in mature eggs.
Differential pathways through SUMO-1 and SUMO-2/3.
Our results showed that SUMO-1 and SUMO-2/3 were localized to distinct subcellular structures during mouse oocyte maturation, indicating that they were conjugated to differential groups of proteins regulating unique cellular functions. In mitosis, SUMO-1 is localized to the mitotic spindles in cells progressing to metaphase,23,37,38 then translocating to the spindle midzone in anaphase and telophase, and to the cleavage furrow during cell division; SUMO-1 reappears at the reforming nuclear envelope during late telophase. During mouse oocyte maturation, SUMO-1 was mainly concentrated at the MI and MII spindle poles and there was no evident signal at the spindle midzone although a SUMO-1 cloud-like pattern was seen around the separating homologues and the spindle (Fig. 1). In mitosis, RanGAP1 is the major SUMO-1 modified protein associated with mitotic spindles23 and this modification is required for the association of RanGAP1 with the spindle.38 The two bands detected by anti-SUMO-1 antibody in immunoblots (Fig. 3, band A and band B) did not correspond in molecular mass to sumoylated RanGAP1, and the main substrates of SUMO-1 during mouse oocyte maturation remain to be further determined.
In our study, SUMO-2/3 was shown to localize near centromeres during mouse oocyte maturation (Fig. 2). A similar localization of SUMO-2/3 in metaphase I spermatocytes was reported recently.29 The most widely studied substrate of SUMO-2/3 is Topoisomerase II. Topoisomerase II is localized to and associates with the mitotic spindles and/or centromeres in several systems including budding yeast,39 Xenopus egg extract20,40 and human cells.23,41 Topoisomerase II is critical for the separation of sister chromatids in mitosis and is associated with the transient modification of Topoisomerase IIa by SUMO-2/3 at the metaphaseanaphase transition.40 In mammalian cells, SUMO-2/3 modified proteins concentrating at centromeres and kinetochores include CENP-E, BubR1 and Nuf2,23 besides Topoisomerase IIa. The proteins indicated by the three bands (bands C, D and E) recognized by anti-SUMO-2/3 antibody need further exploration. The identification of the substrates for SUMO-1 and SUMO-2/3 during mouse oocyte maturation would be important to determine the underlying mechanisms for the defects in MII spindles caused by overexpression of Senp2.
Senp2 overexpression caused unexpected accumulation of SUMO modified proteins and led to defects in MII spindle organization.
The SUMO-specific proteases Senps are responsible for removing SUMO from SUMO modified proteins. Overexpression of Senp may result in deconjugation of SUMO modified proteins, and the overexpression of Senp2 in Hela cells brought about global inhibition of sumoylation.23 Unexpectedly, overexpression of Senp2 in mouse oocytes not only caused inhibition but also accumulation of SUMO modified proteins. Besides deconjugation functions, Senps are also responsible for initial processing of SUMO precursors to form a C-terminal diglycine motif required for sumoylation or conjugation.1,5 Whether the accumulation of SUMO modified proteins was caused by the processing activity of overexpressed Senp2 needs further investigation.
Sumoylation by SUMO-1 and SUMO-2/3 require the only conjugating enzyme Ubc9, so this protein might be an ideal candidate to study the role of sumoylation in mouse oocyte maturation. A recent study showed that sumoylation regulated gene expression by coordinating transcription and RNA processing in mouse oocyte development. In this report, overexpression of Ubc9 stimulated transcription, but the function was independent of its catalytic activity because the potentially dominant-negative version of Ubc9 (Ubc9 C93S) also increased transcriptional activity.33 This report did not refer to any effect on oocyte maturation by dominant-negative Ubc9, and we did not discover apparent phenotypes by injection of Ubc9 siRNA (data not shown) during mouse oocyte maturation. Inhibition of Ubc9 might not be an appropriate method to investigate the roles of sumoylation in mouse oocyte maturation. Alternative approaches to disrupt sumoylation occurs through deconjugation by Senps. Overexpression of Senp2 causes both inhibition and accumulation of SUMO modified proteins suggests that the method of Senps overexpression requires cautionary approaches.
There are two main types of defects in MII spindles resulting from Senp2 overexpression: MII spindles with defective morphology and MII spindles with branched microtubule bundles connecting the first polar body. In mature MII eggs, the existence of branched microtubule bundles connecting MII spindles with the fist polar body might be recognized as a cell division failure. Knockdown of Senp5, another Senp protein in Hela cells resulted in cells with more than one nucleus or impaired nuclear structure correlated with defects in mitosis and cytokinesis.1 The relationship between Senp protein and cytokinesis needs further more detailed studies. The small GTPase Ran vplays crucial roles in the control of spindle organization during the M phase42 and MII spindle assembly is strictly dependent on RanGTP levels in mouse oocytes.43 The production of GTPase depends on the activity of the regular of chromosome condensation (RCC1), Ran's nucleotide exchange factor. The Ran pathway and SUMO pathway share a number of proteins. The Ran-binding protein RanBP2 functions as an E3 ligase for smoylation by SUMO-1,44,45 and SUMO-2/3.22 The Ran GTPase activating protein RanGAP1 is a substrate of SUMO-1 and this modification is required for the target of RanGAP1 to mitotic spindles. Taken together, overexpression of Senp2 may affect components in the Ran pathway, leading to MII spindle disorganization in mature eggs.
Materials and Methods
Antibodies.
Mouse monoclonal anti-SUMO-1 antibody, clone 21C7, was purchased from ZYMED Laboratories, Inc., (Carlsbad, CA); mouse monoclonal anti-SUMO-2/3 antibody, clone 8A2, was a gift kindly provided by Dr. Xiang-Dong Zhang and Dr. Michael J. Matunis, Johns Hopkins University; mouse monoclonal anti-α-tubulin-fluorescein isothiocyanate (FITC) antibody was obtained from Sigma-Aldrich Co., (St. Louis, MO); mouse monoclonal anti-β-actin antibody was purchased from Proteintech Group Inc., (Chicago, IL); mouse monoclonal anti-myc antibody was obtained from Invitrogen (Carlsbad, CA).
Oocyte collection and culture.
Animal care and use were carried out in accordance with the Animal Research Committee guidelines of the Institute of Zoology, Chinese Academy of Sciences.
Oocytes were collected from ovaries of 8-week-old female CD-1 mice in M2 medium (Sigma, St. Louis, MO). Only the immature oocytes arrested at prophase of meiosis I, showing a germinal vesicle (GV) were selected and cultured in M16 medium under liquid paraffin oil at 37°C in an atmosphere of 5% CO2 in air. The oocyte samples were collected for immunofluorescent staining, microinjection or immunoblot analysis at specific times of culture.
Plasmid construction, RNA synthesis and microinjection.
A total of 150 GV-stage mouse oocytes were used to extract the total RNA using the RNeasy micro purification kit (Qiagen), and the first strand cDNA generation was performed following the cDNA synthesis kit instructions (Takara). The full length of Senp2 cDNA was cloned by PCR using the following two pairs of nested primers. F1: ACA GCT CTG GGG TTC GCG T, R1: TGA GAG CTC AGT ACC TGT GAG; F2: TCA GGC CGG CCG ATG TAC AGA TGG CTG GCT AAG GT, R2: GTT GGC GCG CCT CAC AGC AAC TGC TGG TGA AGG A. For in vitro transcription reaction, the Senp2 cDNA was subcloned into the pCS2+ Myc6 vector.
The pCS2+-Myc6-Senp2 plasmid was linearized by Sal I and purified using a gel extraction kit (Qiagen). SP6 Message Machine kit (Ambion) was used to produce the capped Myc-Senp2 mRNA and the mRNA was purified using the RNeasy cleanup kit (Qiagen). Control Myc mRNA was obtained from pCS2+ Myc6 vector following the same procedure.
Microinjection of Myc-Senp2 mRNA or Myc mRNA was performed using an Eppendorf microinjector (Hamburg, Germany) and completed within 30 minutes. For Myc-Senp2 expression, Myc-Senp2 mRNA was microinjected into the cytoplasm of GV oocytes. Oocytes were arrested at the GV stage in M16 medium with 2.5 µM milrinone for 5 hours. The same amount of Myc mRNA was microinjected as control.
Immunofluorescence and confocal microscopy.
For single staining of SUMO-1, SUMO-2/3 and α-tubulin, oocytes were fixed in 4% paraformaldehyde in PBS (pH 7.4) for at least 30 min at room temperature. After being permeabilized with 0.5% Triton X-100 at room temperature for 20 min, oocytes were blocked in 1% BSA-supplemented PBS for 1 h and incubated overnight at 4°C with 1:50 mouse anti-SUMO-1 antibody, 1:200 mouse anti-SUMO-2/3 antibody and 1:200 anti-α-tubulin-FITC antibody, respectively. After three washes in PBS containing 0.1% Tween-20 and 0.01% Triton X-100 for 5 min each, the oocytes were labeled with 1:100 FITC-conjugated goat anti-mouse IgG for 1 h at room temperature (for staining of α-tubulin, this step was omitted). After three washes in PBS containing 0.1% Tween-20 and 0.01% Triton X-100, the oocytes were stained with propidium iodide (PI; 10 µg/ml in PBS). Then, the oocytes were mounted on glass slides and examined with a confocal laser scanning microscope (Zeiss LSM 510 META, Germany).
Immunoblot analysis.
The samples each containing 250 mouse oocytes were collected in SDS sample buffer at the appropriate stage of meiotic maturation and heated for 5 min at 100°C. Immunoblot analysis was conducted following the procedures reported by us previously.46 The proteins were separated by SDS-PAGE and then electrically transferred to polyvinylidene fluoride membranes. After transfer, the membranes were blocked in TBST buffer (TBS containing 0.1% Tween-20) containing 5% skimmed milk for 2 h, followed by incubation overnight at 4°C with 1:500 mouse monoclonal anti-SUMO-1 antibody, 1:750 mouse monoclonal anti-SUMO-2/3 antibody, 1:500 mouse monoclonal anti-myc antibody and 1:1,000 mouse monoclonal anti-β-actin antibody. After three washes for 10 min each in TBST, the membranes were incubated for 1 h at 37°C with 1:1,000 horseradish peroxidase-conjugated goat antimouse IgG. Finally, the membranes were processed using the enhanced chemiluminescence detection system (Amersham, Piscataway, NJ).
Statistics.
All percentages from at least three repeated experiments were expressed as mean ± SEM. Data were analyzed by paired-samples t-test. p < 0.05 was considered statistically significant.
Acknowledgements
We are grateful to Dr. Michael J. Matunis and Dr. Xiang-Dong Zhang for generously providing the mouse monoclonal anti-SUMO2/3 antibody. We also wish to thank Shi-Wen Li for her technical assistance. This work was supported by the National Basic Research Program of China (2006CB944001, 2006CB504004) and National Natural Science Foundation of China (No. 30930065).
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/12120
Supplementary Material
References
- 1.Di Bacco A, Ouyang J, Lee HY, Catic A, Ploegh H, Gill G. The SUMO-specific protease SENP5 is required for cell division. Mol Cell Biol. 2006;26:4489–4498. doi: 10.1128/MCB.02301-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 3.Zhao J. Sumoylation regulates diverse biological processes. Cell Mol Life Sci. 2007;64:3017–3033. doi: 10.1007/s00018-007-7137-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meulmeester E, Melchior F. Cell biology: SUMO. Nature. 2008;452:709–711. doi: 10.1038/452709a. [DOI] [PubMed] [Google Scholar]
- 5.Mukhopadhyay D, Dasso M. Modification in reverse: the SUMO proteases. Trends Biochem Sci. 2007;32:286–295. doi: 10.1016/j.tibs.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 6.Bossis G, Melchior F. SUMO: regulating the regulator. Cell Div. 2006;1:13–13. doi: 10.1186/1747-1028-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hay RT. SUMO: a history of modification. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 8.Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 9.Seufert W, Futcher B, Jentsch S. Role of a ubiquitin-conjugating enzyme in degradation of S- and M-phase cyclins. Nature. 1995;373:78–81. doi: 10.1038/373078a0. [DOI] [PubMed] [Google Scholar]
- 10.Dieckhoff P, Bolte M, Sancak Y, Braus GH, Irniger S. Smt3/SUMO and Ubc9 are required for efficient APC/C-mediated proteolysis in budding yeast. Mol Microbiol. 2004;51:1375–1387. doi: 10.1046/j.1365-2958.2003.03910.x. [DOI] [PubMed] [Google Scholar]
- 11.Jacquiau HR, van Waardenburg RC, Reid RJ, Woo MH, Guo H, Johnson ES, et al. Defects in SUMO (small ubiquitin-related modifier) conjugation and deconjugation alter cell sensitivity to DNA topoisomerase I-induced DNA damage. J Biol Chem. 2005;280:23566–23575. doi: 10.1074/jbc.M500947200. [DOI] [PubMed] [Google Scholar]
- 12.Li SJ, Hochstrasser M. A new protease required for cell cycle progression in yeast. Nature. 1999;398:246–251. doi: 10.1038/18457. [DOI] [PubMed] [Google Scholar]
- 13.Bachant J, Alcasabas A, Blat Y, Kleckner N, Elledge SJ. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol Cell. 2002;9:1169–1182. doi: 10.1016/s1097-2765(02)00543-9. [DOI] [PubMed] [Google Scholar]
- 14.Tanaka K, Nishide J, Okazaki K, Kato H, Niwa O, Nakagawa T, et al. Characterization of a fission yeast SUMO-1 homologue, pmt3p, required for multiple nuclear events, including the control of telomere length and chromosome segregation. Mol Cell Biol. 1999;19:8660–8672. doi: 10.1128/mcb.19.12.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shayeghi M, Doe CL, Tavassoli M, Watts FZ. Characterisation of Schizosaccharomyces pombe rad31, a UBA-related gene required for DNA damage tolerance. Nucleic Acids Res. 1997;25:1162–1169. doi: 10.1093/nar/25.6.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stead K, Aguilar C, Hartman T, Drexel M, Meluh P, Guacci V. Pds5p regulates the maintenance of sister chromatid cohesion and is sumoylated to promote the dissolution of cohesion. J Cell Biol. 2003;163:729–741. doi: 10.1083/jcb.200305080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baldwin ML, Julius JA, Tang X, Wang Y, Bachant J. The yeast SUMO isopeptidase Smt4/Ulp2 and the polo kinase Cdc5 act in an opposing fashion to regulate sumoylation in mitosis and cohesion at centromeres. Cell Cycle. 2009;8:3406–3419. doi: 10.4161/cc.8.20.9911. [DOI] [PubMed] [Google Scholar]
- 18.Guacci VA. The yeast SUMO isopeptidase Smt4/Ulp2 and the polo kinase Cdc5 act in an opposing fashion to regulate sumoylation in mitosis and cohesion at centromeres. Cell Cycle. 2009;8:3811–3812. [PubMed] [Google Scholar]
- 19.Nacerddine K, Lehembre F, Bhaumik M, Artus J, Cohen-Tannoudji M, Babinet C, et al. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev Cell. 2005;9:769–779. doi: 10.1016/j.devcel.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 20.Azuma Y, Arnaoutov A, Anan T, Dasso M. PIASy mediates SUMO-2 conjugation of Topoisomerase-II on mitotic chromosomes. EMBO J. 2005;24:2172–2182. doi: 10.1038/sj.emboj.7600700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diaz-Martinez LA, Gimenez-Abian JF, Azuma Y, Guacci V, Gimenez-Martin G, Lanier LM, et al. PIASgamma is required for faithful chromosome segregation in human cells. PLoS One. 2006;1:53–53. doi: 10.1371/journal.pone.0000053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klein UR, Haindl M, Nigg EA, Muller S. RanBP2 and SENP3 function in a mitotic SUMO2/3 conjugation-deconjugation cycle on Borealin. Mol Biol Cell. 2009;20:410–418. doi: 10.1091/mbc.E08-05-0511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang XD, Goeres J, Zhang H, Yen TJ, Porter AC, Matunis MJ. SUMO-2/3 modification and binding regulate the association of CENP-E with kinetochores and progression through mitosis. Mol Cell. 2008;29:729–741. doi: 10.1016/j.molcel.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hooker GW, Roeder GS. A Role for SUMO in meiotic chromosome synapsis. Curr Biol. 2006;16:1238–1243. doi: 10.1016/j.cub.2006.04.045. [DOI] [PubMed] [Google Scholar]
- 25.Cheng CH, Lo YH, Liang SS, Ti SC, Lin FM, Yeh CH, et al. SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae. Genes Dev. 2006;20:2067–2081. doi: 10.1101/gad.1430406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Carvalho CE, Colaiacovo MP. SUMO-mediated regulation of synaptonemal complex formation during meiosis. Genes Dev. 2006;20:1986–1992. doi: 10.1101/gad.1457806. [DOI] [PubMed] [Google Scholar]
- 27.Rogers RS, Inselman A, Handel MA, Matunis MJ. SUMO modified proteins localize to the XY body of pachytene spermatocytes. Chromosoma. 2004;113:233–243. doi: 10.1007/s00412-004-0311-7. [DOI] [PubMed] [Google Scholar]
- 28.Vigodner M, Morris PL. Testicular expression of small ubiquitin-related modifier-1 (SUMO-1) supports multiple roles in spermatogenesis: silencing of sex chromosomes in spermatocytes, spermatid microtubule nucleation and nuclear reshaping. Dev Biol. 2005;282:480–492. doi: 10.1016/j.ydbio.2005.03.034. [DOI] [PubMed] [Google Scholar]
- 29.La Salle S, Sun F, Zhang XD, Matunis MJ, Handel MA. Developmental control of sumoylation pathway proteins in mouse male germ cells. Dev Biol. 2008;321:227–237. doi: 10.1016/j.ydbio.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Apionishev S, Malhotra D, Raghavachari S, Tanda S, Rasooly RS. The Drosophila UBC9 homologue lesswright mediates the disjunction of homologues in meiosis I. Genes Cells. 2001;6:215–224. doi: 10.1046/j.1365-2443.2001.00413.x. [DOI] [PubMed] [Google Scholar]
- 31.Hashiyama K, Shigenobu S, Kobayashi S. Expression of genes involved in sumoylation in the Drosophila germline. Gene Expr Patterns. 2009;9:50–53. doi: 10.1016/j.gep.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 32.Nie M, Xie Y, Loo JA, Courey AJ. Genetic and proteomic evidence for roles of Drosophila SUMO in cell cycle control, Ras signaling and early pattern formation. PLoS One. 2009;4:5905–5905. doi: 10.1371/journal.pone.0005905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ihara M, Stein P, Schultz RM. UBE2I (UBC9), a SUMO-conjugating enzyme, localizes to nuclear speckles and stimulates transcription in mouse oocytes. Biol Reprod. 2008;79:906–913. doi: 10.1095/biolreprod.108.070474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun SC, Wei L, Li M, Lin SL, Xu BZ, Liang XW, et al. Perturbation of survivin expression affects chromosome alignment and spindle checkpoint in mouse oocyte meiotic maturation. Cell Cycle. 2009;8:3365–3372. doi: 10.4161/cc.8.20.9855. [DOI] [PubMed] [Google Scholar]
- 35.Yuan J, Li M, Wei L, Yin S, Xiong B, Li S, et al. Astrin regulates meiotic spindle organization, spindle pole tethering and cell cycle progression in mouse oocytes. Cell Cycle. 2009;8:3384–3395. doi: 10.4161/cc.8.20.9885. [DOI] [PubMed] [Google Scholar]
- 36.Wei L, Liang XW, Zhang QH, Li M, Yuan J, Li S, et al. BubR1 is a spindle assembly checkpoint protein regulating meiotic cell cycle progression of mouse oocyte. Cell Cycle. 2010;9:1112–1121. doi: 10.4161/cc.9.6.10957. [DOI] [PubMed] [Google Scholar]
- 37.Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joseph J, Tan SH, Karpova TS, McNally JG, Dasso M. SUMO-1 targets RanGAP1 to kinetochores and mitotic spindles. J Cell Biol. 2002;156:595–602. doi: 10.1083/jcb.200110109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takahashi Y, Yong-Gonzalez V, Kikuchi Y, Strunnikov A. SIZ1/SIZ2 control of chromosome transmission fidelity is mediated by the sumoylation of topoisomerase II. Genetics. 2006;172:783–794. doi: 10.1534/genetics.105.047167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Azuma Y, Arnaoutov A, Dasso M. SUMO-2/3 regulates topoisomerase II in mitosis. J Cell Biol. 2003;163:477–487. doi: 10.1083/jcb.200304088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dawlaty MM, Malureanu L, Jeganathan KB, Kao E, Sustmann C, Tahk S, et al. Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIalpha. Cell. 2008;133:103–115. doi: 10.1016/j.cell.2008.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng Y. G protein control of microtubule assembly. Annu Rev Cell Dev Biol. 2004;20:867–894. doi: 10.1146/annurev.cellbio.20.012103.094648. [DOI] [PubMed] [Google Scholar]
- 43.Dumont J, Petri S, Pellegrin F, Terret ME, Bohnsack MT, Rassinier P, et al. A centriole- and RanGTP-independent spindle assembly pathway in meiosis I of vertebrate oocytes. J Cell Biol. 2007;176:295–305. doi: 10.1083/jcb.200605199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pichler A, Gast A, Seeler JS, Dejean A, Melchior F. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell. 2002;108:109–120. doi: 10.1016/s0092-8674(01)00633-x. [DOI] [PubMed] [Google Scholar]
- 45.Azuma Y, Dasso M. A new clue at the nuclear pore: RanBP2 is an E3 enzyme for SUMO1. Dev Cell. 2002;2:130–131. doi: 10.1016/s1534-5807(02)00124-7. [DOI] [PubMed] [Google Scholar]
- 46.Zhang D, Ma W, Li YH, Hou Y, Li SW, Meng XQ, et al. Intra-oocyte localization of MAD2 and its relationship with kinetochores, microtubules and chromosomes in rat oocytes during meiosis. Biol Reprod. 2004;71:740–748. doi: 10.1095/biolreprod.104.028282. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.