Abstract
Catecholaminergic polymorphic ventricular tachycardia (CPVT) is an arrhythmogenic disease that manifests as syncope or sudden death during high adrenergic tone in the absence of structural heart defects. It is primarily caused by mutations in the cardiac ryanodine receptor (RyR2). The mechanism by which these mutations cause arrhythmia remains controversial, with discrepant findings related to the role of the RyR2 binding protein FKBP12.6. The purpose of this study was to characterize a novel RyR2 mutation identified in a kindred with clinically diagnosed CPVT.
Single-strand conformational polymorphism analysis and direct DNA sequencing were used to screen the RyR2 gene for mutations. Site-directed mutagenesis was employed to introduce the mutation into the mouse RyR2 cDNA. The impact of the mutation on the interaction between RyR2 and a 12.6 kDa FK506 binding protein (FKBP12.6) was determined by immunoprecipitation and immunoblotting and its effect on RyR2 function was characterized by single cell Ca2+ imaging and [3H]ryanodine binding.
A novel CPVT mutation, E189D, was identified. The E189D mutation does not alter the affinity of the channel for FKBP12.6, but it increases the propensity for store-overload-induced Ca2+ release (SOICR). Furthermore, the E189D mutation enhances the basal channel activity of RyR2 and its sensitivity to activation by caffeine.
The E189D RyR2 mutation is causative for CPVT and functionally increases the propensity for SOICR without altering the affinity for FKBP12.6. These observations strengthen the notion that enhanced SOICR, but not altered FKBP12.6 binding, is a common mechanism by which RyR2 mutations cause arrhythmias.
Key words: arrhythmia, calcium, death sudden, genetics, ion channels
Introduction
Catecholaminergic polymorphic ventricular tachycardia (CPVT) is an arrhythmogenic disease that manifests as recurrent syncope or sudden cardiac death, under conditions of exercise or emotional stress.1,2 CPVT is diagnosed by an index of suspicion and electrocardiographic demonstration of exercise-induced polymorphic ventricular arrhythmias or typical bidirectional ventricular tachycardia in a structurally normal heart.2 CPVT is predominantly caused by autosomal dominant mutations in the cardiac ryanodine receptor (RyR2), although seven recessive mutations causative of CPVT have also been identified in calsequestrin (CASQ2).3 To date, at least 143 CPVT-associated mutations in RyR2 have been identified and only a small portion have been functionally characterized.4 The disease-causing mutations in RyR2 are located in three distinct regions within the primary sequence of the channel; at the N-terminus, central region and C-terminus, a pattern that is shared by disease-causing mutations in the skeletal muscle ryanodine receptor (RyR1). In RyR1 these mutations give rise to the skeletal muscle diseases, malignant hyperthermia (MH) and central core disease (CCD).5
The cardiac ryanodine receptor (RyR2) is a large homotetrameric ion channel, with each subunit composed of a cytoplasmic and transmembrane domain. RyR2 is located in the membrane of the sarcoplasmic reticulum (SR) and is responsible for regulating SR Ca2+ release. It is well established that under normal conditions RyR2 is activated by Ca2+ influx through the L-type Ca2+ channels, which are triggered by membrane depolarization, in a process termed Ca2+-induced-Ca2+-release (CICR).6 It is also known that CPVT is caused by inappropriate openings of RyR2 during diastole, and that these inappropriate channel openings result in an increase in Na+/Ca2+ exchanger (NCX) activity, which depolarizes the cell membrane giving rise to delayed after depolariztions (DADs).7,8 It is these DADs that ultimately lead to cardiac arrhythmia.
However, the molecular mechanism that underlies the inappropriate opening of RyR2 is less well defined, and remains controversial. It has been proposed that CPVT RyR2 mutations alter the RyR2 macromolecular complex; more specifically, that these mutations reduce the affinity of RyR2 for the 12.6 kDa FK506-binding protein (FKBP12.6).9 It has been shown that a number of CPVT mutations within the central and C-terminal regions (S2245L, R2473S and R4496C) reduce the affinity of RyR2 for FKBP12.6 at rest, and that FKBP12.6-null mice have a phenotype that closely resembles that of CPVT mutant mice.10,11 Based on these observations, Marks and his colleagues have proposed a common model for the role of FKBP12.6 in CPVT. This model states that FKBP12.6 is required to stabilize RyR2 and that a reduced affinity for FKBP12.6 binding, as a result of CPVT RyR2 mutations, will therefore lead to a leaky channel.11
This mechanism has recently received additional support from high-resolution structural data for RyR1. Using sub-nanometer resolution electron cryomicroscopy and comparative modeling, Serysheva et al. mapped several MH and CCD mutations in the 3D structure of RyR1.12 They found that four mutations are located within a surface pocket that is predicted to be the binding site for FKBP12.0 (a RyR1 binding protein homologous to FKBP12.6), implicating a possible mechanism by which MH- and CCD-associated RyR1 mutations dissociate FKBP12.0 from RyR1.13,14 However, the role of this N-terminal region in FKBP binding remains controversial as other researchers have shown that CPVT mutant RyR2 channels display altered activity regardless of FKBP12.6 association. Multiple research groups have also been unable to repeat or extend the data showing that CPVT mutations reduce the affinity of RyR2 for FKBP12.6.15–18
As an alternative to the FKBP12.6 dissociation hypothesis, Jiang and Chen have proposed that CPVT mutations in RyR2 lead to alterations in intrinsic channel properties of RyR2.19 By measuring their responses to Ca2+ overload, they have shown that several CPVT mutations from all three mutation regions in RyR2 decrease the threshold for spontaneous Ca2+ release or store overload-induced Ca2+ release (SOICR).15,17 Interestingly, they have also identified a CPVT mutation that results in an increase in the threshold for SOICR, a suppression of function.20 Their data therefore suggest that CPVT may arise from either a gain of function or suppression of function in SOICR.
In this study we identified a kindred with recurrent, unexplained syncope and confirmed the diagnosis of CPVT. Using genetic screening we identified a novel mutation in RyR2, E189D. This mutation is the most N-terminal single mutation thus far characterized in CPVT and is located in close proximity to the proposed FKBP12.6 binding domain of RyR2. Using cellular Ca2+ measurements we show that the E189D mutation causes CPVT by increasing the propensity for SOICR. Further functional characterization of the E189D mutation shows that it increases the sensitivity of RyR2 to caffeine activation and enhances basal channel activity, similar to most of the other CPVT mutations characterized previously. Importantly, we found that the E189D mutation causes these changes in RyR2 without altering the affinity for FKBP12.6.
Results
Phenotypes associated with the RyR2 E189D mutation.
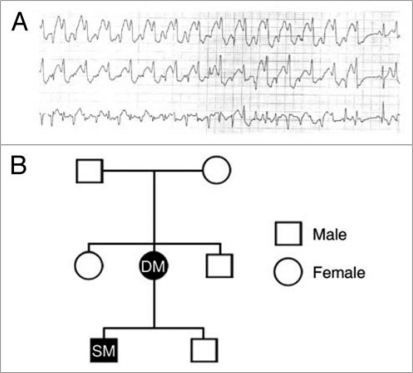
The index patient (DM) presented initially in 1978 (age 10) with recurrent profound syncope on exertion or emotional stress. Work-up failed to demonstrate any structural or reversible cause. An exercise treadmill test demonstrated a brief period of polymorphic ventricular tachycardia. The patient was started on beta-blockers with moderate activity restriction and had no further incidence of syncope. No formal diagnosis was made. Twenty six years later, her son (SM; age 7) also began to experience exertion-induced syncope. Following a complete investigation for structural aetiology, a Holter monitor was used to record the patient's electrocardiogram (Fig. 1A). During exercise, rapid bidirectional ventricular tachycardia was recorded and CPVT was diagnosed. This finding also led to a retrospective diagnosis of CPVT being made in the patient's mother, DM (Fig. 1B).
Figure 1.
Phenotype associated with E189D. (A) holter monitor recording capturing bidirectional ventricular tachycardia in patient SM. (B) Genealogy diagram. Males are denoted by squares, females by circles. Closed symbols indicate clinically affected individuals who are carriers for E189D. Open symbols denote normal phenotype and genotype.
Candidate gene approach reveals a novel RyR2 mutation E189D.
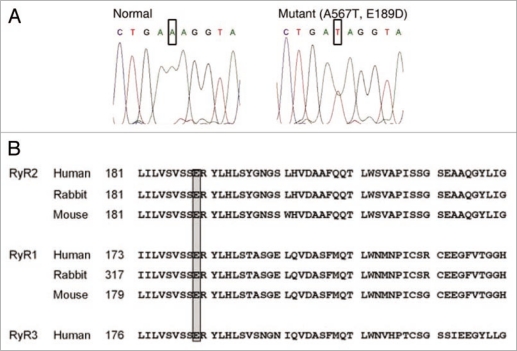
Sequencing analysis of both DM and SM revealed a single point mutation (base shift mutation A567T) in exon 8 of the RyR2 gene (Fig. 2A). This point mutation predicts a replacement of a glutamate residue for aspartate at position 189 (E189D). Residue E189 is located within a highly conserved region of RyR2 which is conserved between different species and receptor isoforms (Fig. 2B), suggesting an important role of this amino acid sequence in the structure/function of the channel. The remainder of the immediate family was tested and found negative for E189D. Thus, patient DM likely represents a de novo mutation. Alternatively, either parent may represent a germ line mosaic for the mutation or patient DM may not be related to her father.
Figure 2.
Molecular genetics and degree of conservation. (A) The mutation predicts the replacement of glutamic acid with aspartic acid at position 189 in exon 8 of the RyR2 gene. This residue is highly conserved among different species and receptor isoforms (B).
The E189D mutation does not alter the association of FKBP12.6 with RyR2.
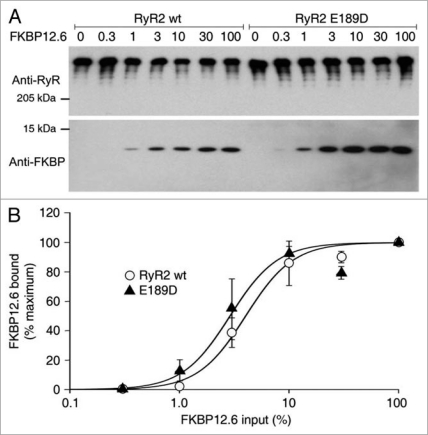
The molecular mechanism by which RyR2 mutations lead to arrhythmia is a contentious issue, with the field divided over the role of FKBP12.6. As the N-terminal location of E189D places it in close proximity to the proposed FKBP12.0 binding pocket of RyR1, we explored whether E189D can influence the association of RyR2 with FKBP12.6. An equal, fixed amount of cell lysate from HEK293 cells expressing RyR2 wt or E189D was used to co-immunoprecipitate varying input amounts of cell lysate from HEK293 cells expressing FKBP12.6 (0, 0.3, 1, 3, 10, 30, 100% of maximum input). It is important to note that HEK293 cells do not normally express FKBP12.6. Figure 3 shows that the concentration dependence of FKBP12.6 binding to E189D is not significantly different from that of FKBP12.6 binding to RyR2 wt (n = 3, p = 0.1–0.4) (Fig. 3B). These data indicate that the novel E189D mutation does not decrease the binding of FKBP12.6 to RyR2, and implies that other arrhythmogenic mechanisms are responsible.
Figure 3.
E189D does not alter the affinity of binding of FKBP12.6 to RyR2. (A) Western blots showing the interaction between RyR2 wt or E189D with various amounts of FKBP12.6 (values indicate % of maximal input). The RyR2-FKBP12.6 complex was co-immunoprecipitated using an anti-RyR antibody followed by immunoblotting with anti-RyR (upper part) and anti-FKBP12/12.6 (lower part) antibodies. Results shown are representative of three separate experiments. (B) Ratio of FKBP12.6 to RyR2, calculated using densitometry of the protein bands, normalized to maximal binding showed that there are no significant differences in the level of protein at any given FKBP12.6 concentration (p = 0.1–0.4).
E189D increases the propensity for spontaneous Ca2+ release events.
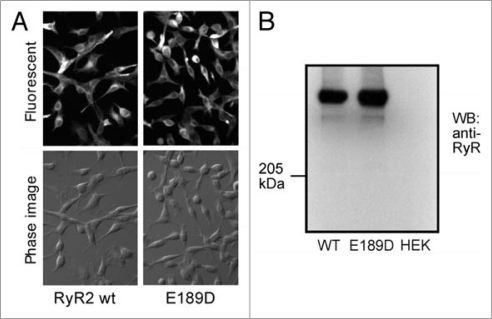
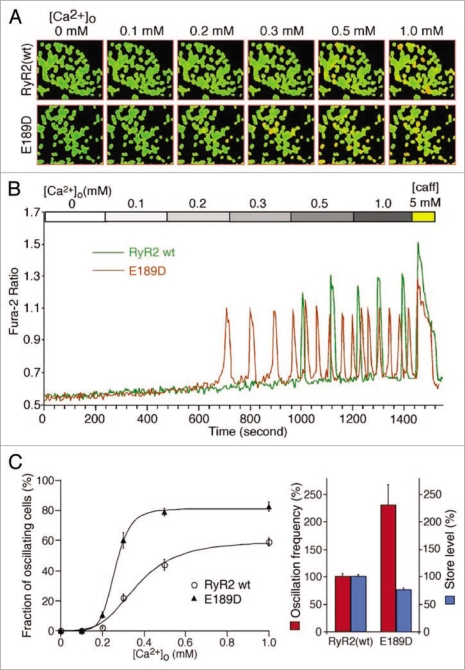
As E189D does not alter the association of RyR2 with FKBP12.6, we investigated whether it altered the propensity for store-overload-induced Ca2+ release (SOICR), another mechanism proposed to underlie RyR2-associated arrhythmias. We created stable-inducible HEK293 cell lines expressing either RyR2 wt or E189D, under the control of a tetracycline repressor. Although HEK293 cells expressing RyR2 wt or CPVT RyR2 mutants do not express a full complement of other cardiac proteins, such as triadin, junctin and calsequestrin, they have previously been shown to very closely recapitulate the SOICR behavior of adult cardiac myocytes from CPVT RyR2 transgenic animals.17,24 The distribution (Fig. 4A) and expression level (Fig. 4B) were determined by immunostaining and western blotting respectively, and found to be comparable between RyR2 wt and E189D. Figure 5 shows the SOICR activity of these cells, measured using the cytosolic Ca2+ indicator dye Fura-2AM, in response to increasing external Ca2+ concentrations (0–1 mM). Increasing the external Ca2+ concentration has previously been shown to be an effective way to induce Ca2+ store overload in HEK293 and cardiac myocytes.17 Figure 5A illustrates the response of a number of cells to this regime; red cells represent those cells displaying SOICR. Typical SOICR in a single cell is shown in Figure 5B with the pooled averaged data shown in Figure 5C. These data show that akin to previously characterized CPVT mutations, the E189D mutation results in an increased frequency, from 100 ± 4.8% for RyR2 wt (n = 16) to 230 ± 36% for E189D (n = 3) (p < 1.6 × 10−6) and a decreased amplitude (76.7 ± 2.7% for E189D, n = 3, compared to 100 ± 2.4% for RyR2 wt, n = 16, p < 0.005) of SOICR. Control wt HEK293 cells that do not express RyR2 have previously been shown to have no Ca2+ oscillations in response to increasing external Ca2+.17
Figure 4.
Expression of RyR2 wt and E189D in HEK293 cells. (A) Immunostaining of RyR2 wt and E189D using an anti-RyR antibody. (B) Western blot of equal amounts of cell lysate from RyR2 wt, E189D or HEK293 parental cells using an anti-RyR antibody.
Figure 5.
E189D increases the propensity for SOICR. Stable, inducible HEK293 cells expressing RyR2 wt or the E189D mutant were grown on glass coverslips. Cells were induced with 1 µg/ml tetracycline for 24 hours and loaded with 5 µM fura-2-AM in KRH buffer for 20 min at room temperature. Cells were perfused continuously with KRH buffer containing 0, 0.1, 0.2, 0.3, 0.5, 1.0 mM CaCl2 or 1.0 mM CaCl2 plus 5 mM caffeine. (A) Typical images of oscillating cells expressing RyR2 wt or E189D at various [Ca2+]. Oscillating cells are shown as red. (B) Typical recording of Fura-2 ratios from RyR2 wt (green trace) and E189D (red trace). (C) Percentage of cells expressing RyR2 wt (open circle) or E189D (closed triangle) oscillating at various [Ca2+]. Oscillation frequency of RyR2 wt and E189D at 1.0 mM CaCl2 and the store Ca2+ content, determined from the amplitude of caffeine (5 mM) induced Ca2+ release. Values are normalized to RyR2 wt level (100%). Data shown are mean ± seM from 3 or 16 separate experiments.
The E189D mutation in RyR2 causes [3H] ryanodine binding to be activated at a slightly lower Ca2+.
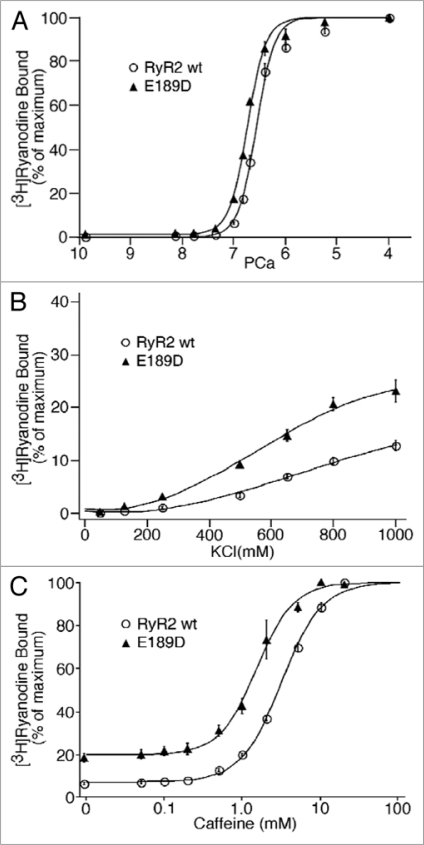
In order to determine the impact of E189D on the sensitivity of RyR2 to Ca2+ activation, we assessed the Ca2+ dependence of [3H]ryanodine binding. [3H]ryanodine binding is a good indicator of channel activity, as ryanodine only binds to the open state of the channel.16,17 Figure 6A shows the overall Ca2+ response of [3H]ryanodine binding to RyR2 wt and E189D. The EC50 value for Ca2+ activation of [3H]ryanodine binding to E189D is 0.18 ± 0.006 µM (n = 3), slightly lower than that for RyR2 wt (EC50 = 0.28 ± 0.017 µM, n = 15) (p < 0.03).
Figure 6.
The effect of E189D on [3H]Ryanodine binding to RyR2. [3H] ryanodine binding to cell lysate prepared from HEK293 cells transfected with RyR2 wt (open circle) or E189D (closed triangle) cDNA was performed at: (A) various Ca2+ concentrations (0.2 nM to 0.1 mM), 100 mM KCl and 5 nM [3H]ryanodine. (B) Various KCl concentrations (50 to 1,000 mM), 3 nM Ca2+ and 5 nM [3H]ryanodine. Amounts of [3H]ryanodine binding at various KCl concentrations were normalized to the maximal binding measured in the presence of 800 mM KCl and 100 µM Ca2+. (C) Various caffeine concentrations (0.01 to 20 mM), 43 nM Ca2+ and 100 mM KCl. Data points shown are mean ± SEM from 3 or 5 separate experiments.
E189D enhances the basal activity of RyR2.
Although the Ca2+ dependence of [3H]ryanodine binding to E189D and RyR2 wt is only slightly shifted, we have previously found that the basal activity of CPVT mutant channels is markedly increased.16,17 To examine whether this is also the case for E189D, we performed [3H]ryanodine binding in the near absence of Ca2+ and the presence of elevated KCl concentrations. Increasing the KCl concentration (50–1,000 mM) leads to an activation of RyR2 wt channels at very low Ca2+ concentrations (3 nM), by an unknown mechanism, but it likely reflects the destabilization of the closed state of the channel. Figure 6B shows that the E189D mutation enhances the basal activity of the channel. For instance, at 800 mM KCl, the level of [3H]ryanodine binding to E189D is 20.7 ± 1.2% (n = 3), significantly higher than the level of binding to RyR2 wt (10.1 ± 0.73%, n = 8) (p < 3.5 × 10−5). These data suggest that E189D de-stabilizes the closed state of RyR2.
E189D increases the sensitivity to caffeine.
Another property shared by CPVT causing RyR2 mutations is a sensitization to caffeine,17 which is also used to trigger CPVT in transgenic animals.25 To determine whether E189D also alters the sensitivity of the channel to caffeine, we performed [3H]ryanodine binding in the presence of a range of caffeine concentrations (0–20 mM). Figure 6C shows that E189D significantly increases the sensitivity of the channel to caffeine, resulting in a leftward shift in the EC50 for caffeine activation from 3.2 ± 0.08 mM (RyR2 wt, n = 4) to 1.6 ± 0.18 mM (E189D, n = 3) (p < 0.0003). Hence, this observation indicates that the E189D mutation sensitizes the channel to activation.
Discussion
CPVT is a relatively rare genetic disease caused by mutations in RyR2 or CASQ2.1,2 This study describes a novel CPVT RyR2 mutation, E189D. Molecular studies show that the E189D mutation results in an increase in SOICR, probably as a result of destabilization of the channel, without altering the affinity of RyR2 for FKBP12.6.
Role of FKBP12.6 in CPVT.
The role of FKBP12.6 in CPVT remains a controversial issue, with new evidence supporting or against its involvement published frequently. Much of the evidence that shows the importance of dissociation of FKBP12.6 comes from the laboratories of Marks and his colleagues. They have shown that several CPVT mutations reduce the binding affinity of RyR2 for FKBP12.6 at rest,10,11 and that FKBP12.6-null mice exhibit a CPVT like phenotype.11 This assertion has been challenged by George et al. who demonstrated that disease causing CPVT mutations (S2246L, N4104K and R4497C) enhance RyR2 function independent of FKBP12.6 dissociation when expressed in HL-1 cells.18 Chen and colleagues have performed similar studies and found that a number of CPVT mutations Q4201R, I4867M, S2246L, R2474S, R176Q/T2504M, L433P and V4653F have no significant effect on the RyR2-FKBP12.6 association when expressed in HEK293 cells.15–17 They also observed that FKBP12.6-null mice do not have an increased susceptibility for stress-induced arrhythmia compared to wt mice. Additionally, single RyR2 channels purified from FKBP12.6-null mice showed Ca2+-dependent activation, gating and single channel conductance similar to those of the wt mice. Lastly, it was demonstrated that direct dissociation of FKBP12.6 from native canine channels does not alter the gating and conductance of the channel.26 Recent structural modeling data for RyR1, however, has complemented the common model proposed by Marks, as it localizes a number of MH and CCD mutations (E161, R164, R402 and I404) to a proposed FKBP12.0 binding site,12 suggesting that these mutations may dissociate FKBP12.0 from RyR1. Although these data are from two different isoforms of RyRs, both the sequence identity and predicted structural homology are highly conserved between RyR1 and RyR2.27 However, the importance of the location of these residues in a binding site for FKBP is questionable, at least in RyR2, as a previous study has shown that deletion of this region (residues 0–305) does not markedly alter FKBP12.6 binding to RyR2.28 It is likely that the location of the FKBP binding site(s) on RyRs will remain controversial until a complete crystal structure is obtained. Nevertheless, in this study, we show that the novel CPVT mutation E189D does not decrease the binding of FKBP12.6 to RyR2. This observation adds to the growing common evidence that CPVT mutations do not alter the association of FKBP12.6 with RyR2.
How does E189D cause CPVT?
At least 143 CPVT RyR2 mutations have been identified, although a minority have been functionally characterized.4 These mutations are clustered in the N-terminus (aa residues 164–433), the central region (aa residues 2,246–2,504) and the C-terminus (aa residues 3,778–4,959).16 The C-terminal region is thought to comprise the channel pore,29,30 the site of ryanodine binding31,32 and the luminal portion of the protein. It is perhaps not surprising, therefore, that mutations within this region can destabilize the channel and lead to inappropriate channel openings.33–35 In contrast, the N-terminal and central regions are located within the large cytosolic portion of the protein, distal from the channel pore. Three dimensional modeling of RyR2 suggests that these regions lie in close proximity to one another.36,37 Therefore, alterations in these regions may destabilize the RyR2 channel by weakening inter-domain interactions, and thus facilitating conformational changes that activate the channel.35–37 The newly identified mutation, E189D, is located within the N-terminal region. Recent crystallography data from the N-terminus of RyR2 show that the side chain of E189D is orientated very similarly to that of V186 which can also cause CPVT when mutated to M186. The high resolution model suggests that the longer side chain of V186M bends and fills a deep surface exposed pocket in the N-terminus of RyR2 when mutated.38 The structure of V186M also shows that the bending of this side chain does not cause any changes in the overall structure or stability of the N-terminus. The authors, therefore, suggest that V186M must be located at an interface between domains and be important for domain-domain interactions.38 The proximity and orientation of the side chain of E189D to V186M suggest it may share a similar mechanism of destabilizing RyR2. In support of this data Amador et al. have created a homology model for the N-terminus of RyR2 based on high resolution structural data from RyR1. This model predicts that E189D would be located within β-sheet 10, which forms the cap of the β-trefoil structure.27 In this model the location of E189D is very close to R178C, a MH mutation in RyR1. Using nuclear magnetic resonance to determine the structure, they found that N-terminal fragments containing R178C do not display any significant chemical shift perturbations, suggesting that like V186M in RyR2, R178C does not cause any significant changes in the structure of the NH2-terminal domain itself.27 Combined these data suggest that E189D, V186M and R178C cause disease (CPVT or MH) by altering domain-domain interactions or by domain unzipping which have both previously been predicted to be mechanisms of disease.35–37 It is also interesting to note that the mutation itself, from glutamate to aspartate, is very conservative, retaining the charge and hydrophobicity. It is possible, therefore, that the size of the residue at position 189 is crucial for domain-domain interactions, perhaps via salt-bridge formation.
Clinical relevance.
As the role of RyR2 and SOICR in CPVT becomes better defined, it will allow for the development of more specific, targeted drugs. Importantly it has been shown that CPVT can result from either an increase or decrease in SOICR.16,20 These opposing findings, within the same disease phenotype, have profound implications for the novel treatment strategies in management of CPVT patients. Hence, it is imperative to fully characterize each newly identified mutation to determine the underlying mechanism, and to translate this to the management of CPVT in patients. Although crucial for CPVT, it still remains to be defined whether enhanced SOICR is common to other ventricular arrhythmias, as clinical and experimental evidence would indicate that these acquired conditions represent complex interactions between electrophysiological remodeling, structural alterations and possibly the myriad of RyR2 regulatory proteins.39,40
Summary.
This study identifies a novel CPVT RyR2 mutation, E189D. Characterization of this mutation serves to strengthen the notion that altered FKBP12.6 binding affinity is not a common mechanism underlying CPVT. It also reinforces the hypothesis that the majority of CPVT causing RyR2 mutations lead to enhanced channel activity and ultimately arrhythmia, by increasing the propensity for SOICR, conceivably as a result of intrinsic destabilization of the channel.
Materials and Methods
Human subjects.
The investigation conforms to the principles outlined in the Declaration of Helsinki. Informed consent was obtained from all participants in accordance with the University of Ottawa Ethics Committee. Participants were evaluated by medical history, physical examination, electrocardiogram, echocardiogram and exercise testing. Clinical assessment was performed prior to knowledge of genotype.
Human molecular genetics studies.
DNA was extracted using standard procedures. The coding exons of RyR2 gene were ampli- fied by PCR and subjected to mutation screening by single-strand conformational polymorphism (SSCP) analysis. Exons displaying abnormal conformational patterns were directly sequenced.
Site-directed mutagenesis.
The E189D mutation was inserted into the mouse RyR2 cDNA using the PCR-based overlap extension method.21 DNA sequencing was used to confirm the insertion and fidelity of the surrounding PCR-amplified region. RyR2 wt or E189D mutant RyR2 cDNA were then subcloned into the mammalian expression vector pcDNA3.
Generation of stable, inducible HEK293 cell lines.
Stable, inducible HEK293 cell lines expressing RyR2 wt or E189D were generated using the Flp-In T-REx Core Kit from Invitrogen as previously described.16 Briefly, the full-length cDNA encoding RyR2 wt or E189D were subcloned into the inducible expression vector pcDNA5/FRT/TO. Flp-In T-REx-293 cells were then co-transfected with pcDNA5/FRT/TO containing RyR2 wt or E189D and pOG44 vector encoding the Flp recombinase in 1:5 ratios using the Ca2+ phosphate precipitation method. Transfected cells were washed with PBS 1 day after transfection and allowed to grow for 1 more day in fresh medium. The cells were then washed again with PBS, harvested and plated onto new dishes. After the cells had attached (∼4 hrs), the growth medium was replaced with a selective medium containing 200 µg/ml hygromycin (Invitrogen). The selective medium was changed every 3–4 days until the desired number of cells was grown. The hygromycin-resistant cells were pooled, aliquoted and stored at −80°C. Both cell lines were tested for RyR2 expression using western blotting analysis and immunofluorescence staining (see below).
RyR2-FKBP12.6 co-immunoprecipitation assays and immunoblotting.
Western blotting analysis were performed as previously described.16 Briefly, HEK293 cells expressing RyR2wt or E189D with similar cell densities were harvested and lysed in lysis buffer (25 mM Tris/50 mM Hepes (pH 7.4), 137 mM NaCl, 1% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS), 0.5% soybean phosphatidylcholine, 5 mM MgCl2) containing a protease inhibitor mix (1 mM benzamidine, 2 µg/ml leupeptin, 2 µg/ml pepstatin A, 2 µg/ml aprotinin, 0.5 mM PMSF, 2.5 mM DTT). Equal amounts of cell lysate were incubated with glutathione-Sepharose, pre-bound with GST-FKBP12.6 (RyR2 alone, Fig. 4) or protein G-Sepharose pre-bound with anti-RyR antibody (34C) (for RyR2-FKBP12.6 binding assay, Fig. 3). For the RyR2-FKBP12.6 binding assay varying amounts of FKBP12.6 containing cell lysate (0–100%) were also added during the incubation. The resulting sepharose-precipitates were washed with ice-cold lysis buffer three times, each time for 2 min to remove unbound proteins. The proteins bound to the sepharose beads were then solubilized by the addition of 40 µl of Laemmli's sample buffer22 plus 5% (v/v) 2-mercaptoethanol and boiled for 5 min. The samples were then separated by SDS-PAGE (6% for RyR2 or 16% for FKBP12.6). The SDS-PAGE-resolved proteins were transferred to polyvinylidene fluoride membranes (PVDF) at 45 V for 18–20 h (for RyR2) or 100 V for 2 h (for FKBP12.6) at 4°C in the presence of 0.01% SDS according to the method of Towbin et al.23 The PVDF membranes containing the transferred proteins were blocked for 30 min with PBS containing 0.5% Tween 20 and 5% (w/v) skimmed milk powder. The blocked membranes were then incubated with anti-RyR (34C) or anti-FKBP antibodies (both 1:1,000) for 3 h and washed three times for 5 min in PBS containing 0.5% Tween 20. The membrane was then incubated with the appropriate horse-radish peroxidase-conjugated secondary antibody (1:20,000) for 30 min. After washing three times for 5 min in PBS containing 0.5% Tween 20, the RyR2 or FKBP12.6 proteins were detected by enhanced chemiluminescence (Pierce).
[3H]ryanodine binding.
Preparation of cell lysate and equilibrium [3H]ryanodine binding were carried out as described previously.16 Briefly, a binding mixture (300 µl) containing 30 µl of cell lysate expressing either RyR2 wt or E189D (3–5 mg/ml), 25 mM Tris/50 mM Hepes (pH 7.4), 5 nM [3H]ryanodine, a protease inhibitor mix and various concentrations of CaCl2, caffeine and KCl as indicated, was incubated at 37°C for 2.5–3.5 hrs. The binding mixture was diluted with 5 ml of ice-cold washing buffer containing 25 mM Tris (pH 8.0) and 250 mM KCl and immediately filtered through Whatman GF/B filters presoaked with 1% polyethylenimine. The filters were washed four times with 5 ml of ice-cold washing buffer and the radioactivity associated with the filters was determined by liquid scintillation counting. Nonspecific binding was determined by measuring [3H]ryanodine binding in the presence of 50 µM unlabeled ryanodine. Data shown are mean ± SEM for n experiments, and statistical significance was evaluated using the unpaired Student's t test.
Single cell Ca2+ imaging.
Intracellular Ca2+ transients in stable inducible HEK293 cells expressing the RyR2 wt or mutant channels were measured using single-cell Ca2+ imaging and the fluorescence Ca2+ indicator dye fura-2 acetoxymethyl ester (fura-2 AM) as described previously.16 Briefly, cells grown on glass coverslips for 24 hrs after induction by 1 µg/ml tetracycline (Sigma) were loaded with 5 µM fura-2 AM in Krebs-Ringer-Hepes (KRH) buffer (125 mM NaCl, 5 mM KCl, 1.2 mM KH2PO4, 6 mM glucose, 1.2 mM MgCl2, 25 mM HEPES, pH 7.4) plus 0.02% pluronic F-127 (Molecular Probes) and 0.1 mg/ml BSA for 20 min at room temperature. The coverslips were then mounted in a perfusion chamber (Warner Instruments, Hamden, CT) on a Nikon Ti-S microscope. Cells were perfused continuously with KRH buffer containing various concentrations of CaCl2 (0–1.0 mM) at room temperature. Fura-2 fluorescence was captured every 4 seconds through a Fluor x20 objective and a Chroma filter set using the C-imaging software. Data shown are mean ± SEM, and statistical significance was evaluated using the unpaired Student's t test.
Acknowledgements
We would like to thank our patients for their dedicated support and Jeff Bolstad for technical assistance. This work was supported by research grants from Alberta Heritage Foundation for Medical Research (Dawei Jiang, Peter P. Jones), Heart and Stroke Foundation of Ontario (Michael H. Gollob), the Heart and Stroke Foundation of Alberta, Northwest Territories and Nunavut (S.R. Wayne Chen, Peter P. Jones) and the National Institutes of Health (S.R. Wayne Chen). Michael H. Gollob is also supported by the Heart and Stroke Foundation of Ontario Clinician-Scientist Award.
Abbreviations
- CASQ2
cardiac calsequestrin
- CCD
central core disease
- CICR
Ca2+-induced-Ca2+-release
- CPVT
catecholaminergic polymorphic ventricular tachycardia
- DAD
delayed after depolarizations
- FKBP12.0
12.0 kDa FK506-binding protein
- FKBP12.6
12.6 kDa FK506-binding protein
- MH
malignant hyperthermia
- NCX
Na+/Ca2+ exchanger
- RyR1
skeletal muscle ryanodine receptor
- RyR2
cardiac muscle ryanodine receptor
- SOICR
store overload induced calcium release
- SSCP
single-strand conformational polymorphism
Footnotes
Previously published online: www.landesbioscience.com/journals/channels/article/12666
References
- 1.Reid DS, Tynan M, Braidwood L, Fitzgerald GR. Bidirectional tachycardia in a child. A study using his bundle electrography. Br Heart J. 1975;37:339–344. doi: 10.1136/hrt.37.3.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leenhardt A, Lucet V, Denjoy I, Grau F, Ngoc DD, Coumel P. Catecholaminergic polymorphic ventricular tachycardia in children. A 7-year follow-up of 21 patients. Circulation. 1995;91:1512–1159. doi: 10.1161/01.cir.91.5.1512. [DOI] [PubMed] [Google Scholar]
- 3.Kontula K, Laitinen PJ, Lehtonen A, Toivonen L, Viitasalo M, Swan H. Catecholaminergic polymorphic ventricular tachycardia: Recent mechanistic insights. Cardiovasc Res. 2005;67:379–387. doi: 10.1016/j.cardiores.2005.04.027. [DOI] [PubMed] [Google Scholar]
- 4.Medeiros-Domingo A, Bhuiyan ZA, Tester DJ, Hofman N, Bikker H, van Tintelen JP, et al. The RYR2-encoded ryanodine receptor/calcium release channel in patients diagnosed previously with either catecholaminergic polymorphic ventricular tachycardia or genotype negative, exercise-induced long QT syndrome: A comprehensive open reading frame mutational analysis. J Am Coll Cardiol. 2009;54:2065–2074. doi: 10.1016/j.jacc.2009.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Priori SG, Napolitano C. Cardiac and skeletal muscle disorders caused by mutations in the intracellular Ca2+ release channels. J Clin Invest. 2005;115:2033–2038. doi: 10.1172/JCI25664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bers DM. Cardiac excitation-contraction coupling. Nature. 2002;415:198–205. doi: 10.1038/415198a. [DOI] [PubMed] [Google Scholar]
- 7.Marban E. Cardiac channelopathies. Nature. 2002;415:213–218. doi: 10.1038/415213a. [DOI] [PubMed] [Google Scholar]
- 8.Pogwizd SM, Bers DM. Cellular basis of triggered arrhythmias in heart failure. Trends Cardiovasc Med. 2004;14:61–66. doi: 10.1016/j.tcm.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 9.Wehrens XH, Lehnart SE, Marks AR. Intracellular calcium release and cardiac disease. Annu Rev Physiol. 2005;67:69–98. doi: 10.1146/annurev.physiol.67.040403.114521. [DOI] [PubMed] [Google Scholar]
- 10.Lehnart SE, Wehrens XHT, Laitinen PJ, Reiken SR, Deng S, Cheng Z, et al. Sudden death in familial polymorphic ventricular tachycardia associated with calcium release channel (ryanodine receptor) leak. Circulation. 2004;109:3208–3214. doi: 10.1161/01.CIR.0000132472.98675.EC. [DOI] [PubMed] [Google Scholar]
- 11.Wehrens X, Lehnart S, Huang F, Vest J, Reiken S, Mohler P, et al. FKBP12.6 deficiency and defective calcium release channel (ryanodine receptor) function linked to exercise-induced sudden cardiac death. Cell. 2003;113:829–840. doi: 10.1016/s0092-8674(03)00434-3. [DOI] [PubMed] [Google Scholar]
- 12.Serysheva II, Ludtke SJ, Baker ML, Cong Y, Topf M, Eramian D, et al. Subnanometer-resolution electron cryomicroscopy-based domain models for the cytoplasmic region of skeletal muscle RyR channel. Proc Natl Acad Sci USA. 2008;105:9610–9615. doi: 10.1073/pnas.0803189105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samso M, Shen X, Allen PD. Structural characterization of the RyR1-FKBP12 interaction. J Mol Biol. 2006;356:917–927. doi: 10.1016/j.jmb.2005.12.023. [DOI] [PubMed] [Google Scholar]
- 14.Wagenknecht T, Radermacher M, Grassucci R, Berkowitz J, Xin HB, Fleischer S. Locations of calmodulin and FK506-binding protein on the three- dimensional architecture of the skeletal muscle ryanodine receptor. J Biol Chem. 1997;272:32463–32471. doi: 10.1074/jbc.272.51.32463. [DOI] [PubMed] [Google Scholar]
- 15.Jones PP, Jiang D, Bolstad J, Hunt DJ, Zhang L, Demaurex N, et al. Endoplasmic reticulum Ca2+ measurements reveal that the cardiac ryanodine receptor mutations linked to cardiac arrhythmia and sudden death alter the threshold for store-overload-induced Ca2+ release. Biochem J. 2008;412:171–178. doi: 10.1042/BJ20071287. [DOI] [PubMed] [Google Scholar]
- 16.Jiang D, Wang R, Xiao B, Kong H, Hunt DJ, Choi P, et al. Enhanced store overload-induced Ca2+ release and channel sensitivity to luminal Ca2+ activation are common defects of RyR2 mutations linked to ventricular tachycardia and sudden death. Circ Res. 2005;97:1173–1181. doi: 10.1161/01.RES.0000192146.85173.4b. [DOI] [PubMed] [Google Scholar]
- 17.Jiang D, Xiao B, Yang D, Wang R, Choi P, Zhang L, et al. RyR2 mutations linked to ventricular tachycardia and sudden death reduce the threshold for store-overload-induced Ca2+ release (SOICR) Proc Natl Acad Sci USA. 2004;101:13062–13067. doi: 10.1073/pnas.0402388101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.George CH, Higgs GV, Lai FA. Ryanodine receptor mutations associated with stress-induced ventricular tachycardia mediate increased calcium release in stimulated cardiomyocytes. Circ Res. 2003;93:531–540. doi: 10.1161/01.RES.0000091335.07574.86. [DOI] [PubMed] [Google Scholar]
- 19.Jiang D, Xiao B, Zhang L, Chen SR. Enhanced basal activity of a cardiac Ca2+ release channel (ryanodine receptor) mutant associated with ventricular tachycardia and sudden death. Circ Res. 2002;91:218–225. doi: 10.1161/01.res.0000028455.36940.5e. [DOI] [PubMed] [Google Scholar]
- 20.Jiang D, Chen W, Wang R, Zhang L, Chen SRW. Loss of luminal Ca2+ activation in the cardiac ryanodine receptor is associated with ventricular fibrillation and sudden death. Proc Natl Acad Sci USA. 2007;104:18309–18314. doi: 10.1073/pnas.0706573104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction [see comments] Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–665. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4434. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cerrone M, Napolitano C, Priori SG. Catecholaminergic polymorphic ventricular tachycardia: A paradigm to understand mechanisms of arrhythmias associated to impaired Ca2+ regulation. Heart Rhythm. 2009;6:1652–1659. doi: 10.1016/j.hrthm.2009.06.033. [DOI] [PubMed] [Google Scholar]
- 25.Liu N, Colombi B, Memmi M, Zissimopoulos S, Rizzi N, Negri S, et al. Arrhythmogenesis in catecholaminergic polymorphic ventricular tachycardia: Insights from a RyR2 R4496C knock-in mouse model. Circ Res. 2006;99:292–298. doi: 10.1161/01.RES.0000235869.50747.e1. [DOI] [PubMed] [Google Scholar]
- 26.Xiao J, Tian X, Jones PP, Bolstad J, Kong H, Wang R, et al. Removal of FKBP12.6 does not alter the conductance and activation of the cardiac ryanodine receptor or the susceptibility to stress-induced ventricular arrhythmias. J Biol Chem. 2007;282:34828–34838. doi: 10.1074/jbc.M707423200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amador FJ, Liu S, Ishiyama N, Plevin MJ, Wilson A, MacLennan DH, et al. Crystal structure of type I ryanodine receptor amino-terminal beta-trefoil domain reveals a disease-associated mutation “hot spot” loop. Proc Natl Acad Sci USA. 2009;106:11040–11044. doi: 10.1073/pnas.0905186106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masumiya H, Wang R, Zhang J, Xiao B, Chen SRW. Localization of the 12.6-kDa FK506-binding protein (FKBP12.6) binding site to the NH2-terminal domain of the cardiac Ca2+ release channel (ryanodine receptor) J Biol Chem. 2003;278:3786–3792. doi: 10.1074/jbc.M210962200. [DOI] [PubMed] [Google Scholar]
- 29.Bhat MB, Zhao J, Takeshima H, Ma J. Functional calcium release channel formed by the carboxyl-terminal portion of ryanodine receptor. Biophys J. 1997;73:1329–1336. doi: 10.1016/S0006-3495(97)78166-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu X, Bhat MB, Nishi M, Takeshima H, Ma J. Molecular cloning of cDNA encoding a drosophila ryanodine receptor and functional studies of the carboxyl-terminal calcium release channel. Biophys J. 2000;78:1270–1281. doi: 10.1016/S0006-3495(00)76683-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Witcher DR, McPherson PS, Kahl SD, Lewis T, Bentley P, et al. Photoaffinity labeling of the ryanodine receptor/Ca2+ release channel with an azido derivative of ryanodine. J Biol Chem. 1994;269:13076–13079. [PubMed] [Google Scholar]
- 32.Callaway C, Seryshev A, Wang JP, Slavik KJ, Needleman DH, Cantu C, 3rd, et al. Localization of the high and low affinity [3H]ryanodine binding sites on the skeletal muscle Ca2+ release channel. J Biol Chem. 1994;269:15876–15884. [PubMed] [Google Scholar]
- 33.Gao L, Tripathy A, Lu X, Meissner G. Evidence for a role of C-terminal amino acid residues in skeletal muscle Ca2+ release channel (ryanodine receptor) function. FEBS Lett. 1997;412:223–226. doi: 10.1016/s0014-5793(97)00781-3. [DOI] [PubMed] [Google Scholar]
- 34.Stewart R, Zissimopoulos S, Lai FA. Oligomerization of the cardiac ryanodine receptor C-terminal tail. Biochem J. 2003;376:795–799. doi: 10.1042/BJ20030597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.George CH, Jundi H, Thomas NL, Scoote M, Walters N, Williams AJ, et al. Ryanodine receptor regulation by intramolecular interaction between cytoplasmic and transmembrane domains 10.1091/mbc.E03-09-0688. Mol Biol Cell. 2004;15:2627–2638. doi: 10.1091/mbc.E03-09-0688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ikemoto N, Yamamoto T. Postulated role of interdomain interaction within the ryanodine receptor in Ca2+ channel regulation. Trends Cardiovasc Med. 2000;10:310–336. doi: 10.1016/s1050-1738(01)00067-6. [DOI] [PubMed] [Google Scholar]
- 37.Ikemoto N, Yamamoto T. Regulation of calcium release by interdomain interaction within ryanodine receptors. Front Biosci. 2002;7:671–683. doi: 10.2741/A803. [DOI] [PubMed] [Google Scholar]
- 38.Lobo PA, Van Petegem F. Crystal structures of the N-terminal domains of cardiac and skeletal muscle ryanodine receptors: Insights into disease mutations. Structure. 2009;17:1505–1514. doi: 10.1016/j.str.2009.08.016. [DOI] [PubMed] [Google Scholar]
- 39.Marks AR. A guide for the perplexed: Towards an understanding of the molecular basis of heart failure. Circulation. 2003;107:1456–1459. doi: 10.1161/01.cir.0000059745.95643.83. [DOI] [PubMed] [Google Scholar]
- 40.Bers DM, Eisner DA, Valdivia HH. Sarcoplasmic reticulum Ca2+ and heart failure: Roles of diastolic leak and Ca2+ transport. Circ Res. 2003;93:487–490. doi: 10.1161/01.RES.0000091871.54907.6B. [DOI] [PubMed] [Google Scholar]