Abstract
Two cases of primary plastid endosymbiosis are known. The first occurred ca. 1.6 billion years ago and putatively gave rise to the canonical plastid in algae and plants. The second is restricted to a genus of rhizarian amoebae that includes Paulinella chromatophora. Photosynthetic Paulinella species gained their plastid from an α-cyanobacterial source and are sister to plastid-lacking phagotrophs such as Paulinella ovalis that ingest cyanobacteria. To study the role of feeding behavior in plastid origin, we analyzed single-cell genome assemblies from six P. ovalis-like cells isolated from Chesapeake Bay, USA. Dozens of contigs in these cell assemblies were derived from prey DNA of α-cyanobacterial origin and associated cyanophages. We found two examples of horizontal gene transfer (HGT) in P. ovalis-like nuclear DNA from cyanobacterial sources. This work suggests the first evidence of a link between feeding behavior in wild-caught cells, HGT, and plastid primary endosymbiosis in the monophyletic Paulinella lineage.
The plastid in algae and plants almost certainly originated in the founding group of photosynthetic eukaryotes, the Plantae (or Archaeplastida1,2,3,4) and subsequently spread to all other major algal groups (e.g., diatoms, dinoflagellates, euglenids) through secondary and tertiary endosymbiosis5,6. Primary plastid acquisition occurred ca. 1.6 billion years ago7 putatively through the phagotrophic engulfment and permanent retention of a cyanobacterial endosymbiont1. Plastid evolution resulted in the endosymbiotic gene transfer (EGT) of hundreds of genes from the captured endosymbiont to the nucleus of the Plantae ancestor8,9.
The photosynthetic amoeba Paulinella chromatophora10 contains blue-green “chromatophores” (i.e., plastids) and was first described by Robert Lauterborn11. This genus has become a model for endosymbiosis research because it is widely accepted as a second case of cyanobacterial primary endosymbiosis12,13,14,15,16. Recent work shows many examples of EGT to the amoeba nuclear genome from the α-cyanobacterium-derived (e.g., Prochlorococcus and Synechococcus species) plastid16,17,18,19. To understand the processes that led to plastid origin in photosynthetic Paulinella we focused on its plastid-lacking sister taxa. Three heterotrophic Paulinella (P. ovalis, P. intermedia, and P. indentata) species are known20,21,22. P. ovalis feeds on cyanobacteria that have previously been identified in food vacuoles20. This suggests that the primary plastid in the monophyletic lineage of photosynthetic Paulinella14 is likely to be the outcome of permanent maintenance of captured cyanobacterial prey, as has been proposed for the origin of the Plantae plastid1,4. Given conservation in prey choice and the widespread abundance of α-Cyanobacteria in the oceans23,24, it also is possible that members of this prokaryote clade may be detected in the food vacuoles of heterotrophic Paulinella species. Because P. ovalis, although seasonally abundant in nature20, has not yet been successfully cultivated, it was until now not possible to generate genome data from this lineage to test for the presence of prey DNA or prey-derived HGT. This fundamental problem was recently solved with the development of single-cell genomic methods that allow the generation of draft genome data from cells collected in the natural environment25,26,27,28. These data not only provide insights into the genomes of the targeted cell but also identify the sources of foreign DNA present at the time of cell capture (e.g., from prey, pathogens, or symbionts28). Here we used single-cell genomics to generate draft assemblies from six P. ovalis-like cells isolated from Chesapeake Bay, USA. Specifically, we tested the idea that the source of the plastid in photosynthetic Paulinella reflects feeding behavior among its heterotrophic sister taxa.
Results
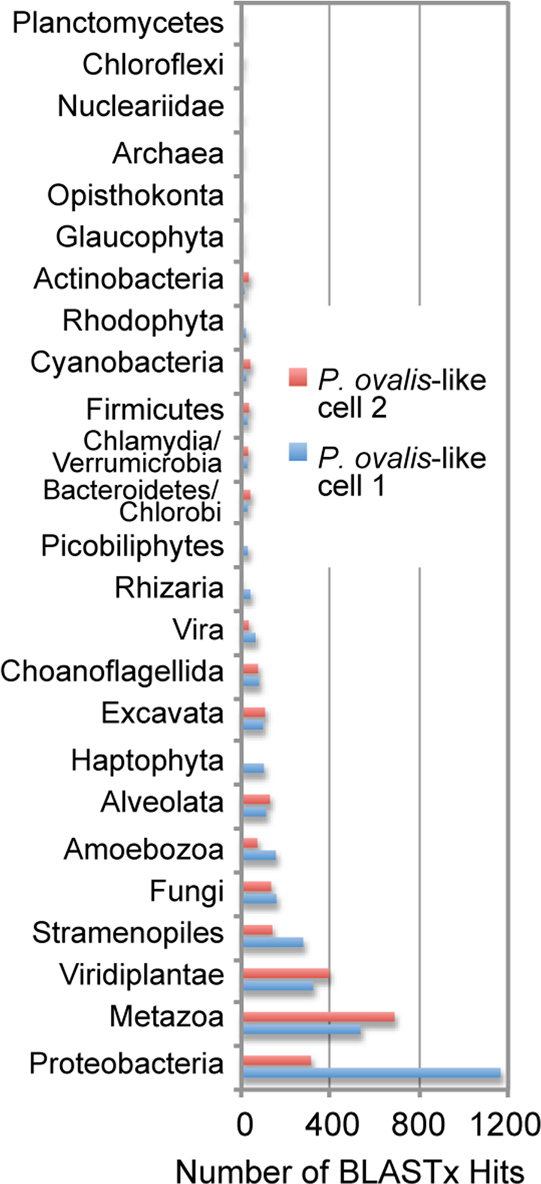
A water sample collected on May 30, 2009 from the dock of the Smithsonian Environmental Research Center, Edgewater, MD, USA, was used as input for flow cytometry. Single heterotrophic cells <10 μm in size that lacked chlorophyll autofluorescence were sorted. After whole genome amplification (WGA) of total DNA, the taxonomic identity of the single-cell amplified genomes (SAGs) was defined through analysis of the 18S rDNA sequence29. This showed that 10/48 SAGs were closely related to photosynthetic Paulinella lineages (referred to here as P. ovalis-like; Figs. 1A, 1B). Six of these SAGs that had identical small subunit rDNA sequences (P. ovalis-like cells 1–6 [Fig. 1B]) were chosen for draft genome sequencing using the Roche 454 GS-FLX system. This resulted in 180 – 308 Mbp of data from each of the cells that were used to generate individual genome assemblies (see Supplementary Table S1 online). Each assembly comprised several thousand contigs with the total number of assembled bases ranging from ca. 3.5 – 7.2 Mbp with the exception of the data-poor P. ovalis-like cell 6 that had a relatively small assembly of size 1.5 Mbp. All six assemblies were used in BLASTx sequence similarity searches against a comprehensive local database (see Methods and Supplementary Table S2) to identify top hits (e-value ≤ 10−5). The top hits were extracted and their numbers normalized (Supplementary Figs. S1, S2) to minimize the effect of uneven coverage bias introduced by multiple displacement amplification used in WGA28,30,31, resulting in the data shown in Figure 1C.
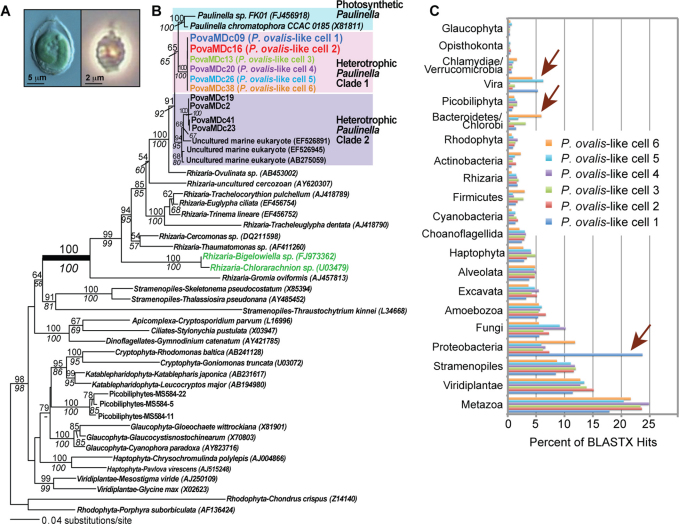
Figure 1. Evolutionary analyses of Paulinella ovalis-like SAGs.
(a) Light microscopy image of the photosynthetic Paulinella chromatophora (left) and its phagotrophic sister P. ovalis (right). (b) RAxML48 tree (GTR + Γ + I model) inferred from 18S rDNA showing the phylogenetic position of P. ovalis-like cells within Rhizaria. Single-cell sorting identified several P. ovalis-like cells that comprise two distinct heterotrophic Paulinella clades (Clade 1 and Clade 2) of which Clade 1 is most closely related to the photosynthetic P. chromatophora and Paulinella sp. FK0114, and is the subject of our study. RAxML and PhyML49 bootstrap values are shown above and below the branches, respectively (only those ≥ 60% are shown). The unit of branch length is the number of substitutions per site. The GenBank accession numbers (where available) are shown after each taxon name. (c) Taxonomic distribution of unique BLASTx hits (e-value ≤10−10) using the contigs from the six P. ovalis-like single cell SAGs for which we have 454 data. The percentage distribution of each phylum across all six SAGs is shown. The arrows indicate markedly different phyletic origins of DNA among the SAGs.
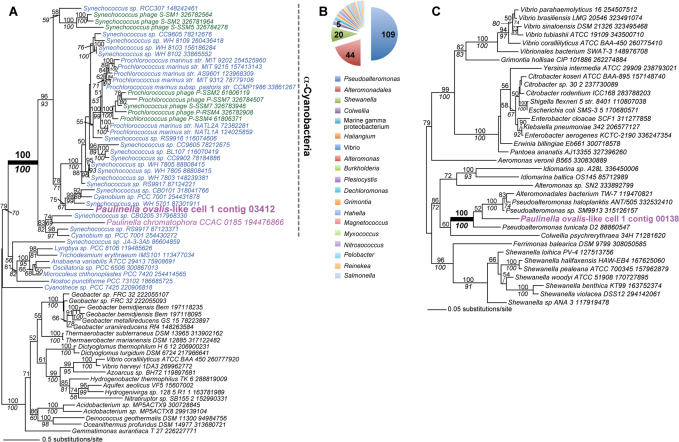
An example of a cyanobacterium-derived DNA fragment in the P. ovalis-like cell 1 assembly of the 454 data (contig 03412, length = 604 nt, 1449 reads) is shown in Figure 2A. This tree of a PstS phosphate ABC transporter shows that cell 1 contains DNA that is derived from α-Cyanobacteria (i.e., barring HGT of this gene into a non-cyanobacterial cell). Note that a homolog of the gene is present in the plastid (chromatophore) genome of the photosynthetic P. chromatophora CCAC 018515. Analysis of the proteobacterial DNA in cell 1 showed that the majority of contigs had top hits to the marine bacterial genus Pseudoalteromonas (i.e., Pseudoalteromonas sp. SM9913 [54 hits], P. tunicata D2 [30 hits], and P. haloplanktis [24 hits] Fig. 2B). One of the proteins encoded on contig 00138 (length = 4847 nt, 182 reads) that had a top hit to Pseudoalteromonas sp. SM9913 was used to infer a phylogeny. This protein encodes the highly conserved transcription elongation factor NusA (e-value 6.60 × 10−283) and demonstrates a strongly supported monophyletic group comprised of the P. ovalis-like cell 1 NusA sequence with Pseudoalteromonas/Alteromomonadales taxa (Fig. 2C). A second open reading frame on contig 00138 encodes the translation initiation factor IF-2 that is also most closely related to Pseudoalteromonas species. Despite this clear phylogenetic signal, given high rates of HGT among bacteria it is not assured that we have identified the true taxonomic source of the contig and whether single or multiple Proteobacteria are present in cell 1 DNA.
Figure 2. Bacterial DNA is present in the P. ovalis-like cell 1 SAG assembly.
(a) Maximum likelihood (RAxML, WAG + Γ + F model) phylogeny of PstS phosphate ABC transporter proteins. Cyanobacteria are in blue text, other Bacteria are in black text, the chromatophore (plastid) and the sequence encoded on P. ovalis-like cell 1 contig 03412 are in magenta text, and cyanophage sequences are in dark green. The well-supported clade that includes α-Cyanobacteria is identified with the dashed gray line. (b) Taxonomic distribution of BLASTx hits to Proteobacteria in the 454 assembly of P. ovalis-like cell 1. (c) Maximum likelihood (RAxML, WAG + Γ + F model) phylogeny of the transcription elongation factor NusA. P. ovalis-like cell 1 contig 00138 is shown in magenta text. RAxML and PhyML bootstrap values (100 replicates) in 2A and 2C are shown above and below the branches, respectively (only those ≥ 50% are shown). The unit of branch length is the number of substitutions per site. The NCBI “gi” numbers are shown after each taxon name.
To generate a more robust genome assembly from P. ovalis-like SAGs, we produced additional sequence data from cells 1 and 2 using an Illumina GAIIx instrument (see Methods). The Illumina data were co-assembled with the 454 reads and subjected to the BLASTx pipeline as described above. These results mirror the 454 data, with cell 1 showing a significantly larger number of proteobacterial hits (Fig. 3; Supplementary Fig. S2) than cell 2. To estimate the amount of coding DNA in the individual cell 1 and 2 combined (454 + Illumina) assemblies, we determined the number of nucleotides encoded on all contigs that had significant BLASTx hits. This showed that the cell 1 and cell 2 contigs contained 2.6 and 4.3 Mbp of eukaryote DNA, 1.8 and 0.9 Mbp of bacterial DNA, and 0.2 and 0.9 Mbp of viral DNA, respectively (Supplementary Fig. S3). The annotations (when present) for the top hits in the cell 1 and cell 2 contigs are found in Supplementary Tables S3 and S4, respectively.
Figure 3. Taxonomic distribution of BLASTx hit numbers using the contigs from P. ovalis-like cells 1 and 2 for which we have 454 + Illumina data.
Analysis of cyanobacterial and cyanophage gene fragments in the combined assembly
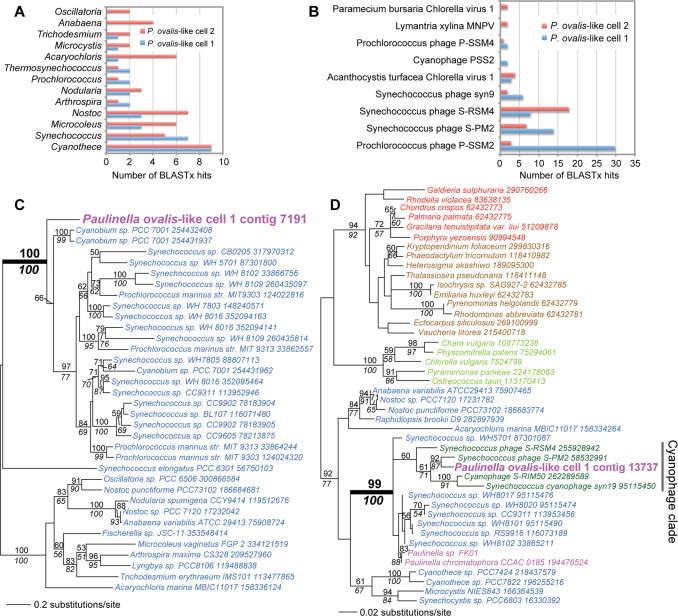
We searched for DNA fragments derived from cyanobacterial prey and associated phages in the combined assemblies. This BLASTx analysis turned up 35 and 62 hits for cell 1, and 53 and 31 hits for cell 2 to Cyanobacteria and cyanophages, respectively (see Supplementary Tables S3, S4 and Figs. 4A, 4B). A RAxML tree inferred from a protein (bacterial porin, OprB) encoded on one of the assembled fragments found in cell 1 is shown in Figure 4C and identifies prey DNA that is related to α-Cyanobacteria. The cyanobacterial fragment (contig 7191) is of length 11,565 nt and has an average coverage of 7,577x. Prediction of open reading frames using MAKER 2 (http://derringer.genetics.utah.edu/cgi-bin/MWAS/maker.cgi) revealed 8 putative proteins (see Supplementary Fig. S4) that encode porin, an ABC transporter subunit, a putative histidine kinase, a hypothetical protein, a putative p-pantothenate cysteine ligase, a HNH endonuclease family protein, a ribonucleotide-diphosphate reductase subunit beta, and a putative nicotinamide nucleotide transhydrogenase, all with cyanobacterial top hits. The absence of introns and gene richness suggest a prokaryotic origin of this contig.
Figure 4. Cyanobacterial and cyanophage DNA identified in the combined (454 + Illumina) assemblies of P. ovalis-like cells 1 and 2.
(a) Taxonomic distribution of BLASTx hits to Cyanobacteria, using the cell 1 and 2 contigs. (b) Taxonomic distribution of BLASTx hits to virus sequences using the cell 1 and 2 contigs. (c) Maximum likelihood (RAxML, WAG + Γ + F model) phylogeny of bacterial porin (OprB) proteins. (d) Maximum likelihood (RAxML, WAG + Γ + F model) phylogeny of photosystem II D2 (PsbD) proteins. In 4C and 4D, Cyanobacteria are in blue text, other Bacteria are in black text, the chromatophore (plastid) and the P. ovalis-like cell data are in magenta text, cyanophage sequences are in dark green, Viridiplantae is in light green text, red algae in red text, and chromalveolates in brown text. The well-supported clade that includes cyanophages is identified with the gray bar. RAxML and PhyML bootstrap values (100 replicates) are shown above and below the branches, respectively (only those ≥ 50% are shown). The unit of branch length is the number of substitutions per site. The NCBI “gi” numbers are shown after each taxon name.
The virus hits were studied to determine their putative taxonomic distribution. These data (Fig. 4B) show that the four most frequently recovered viral DNAs arise from cyanophages that infect Prochlorococcus and Synechococcus lineages. These phages are presumably associated with the different α-Cyanobacteria20 in the cells (Fig. 4A) or may be prey for P. ovalis-like cells. Cyanophage genomes encode genes of photosystems I and II that manipulate photosynthetic activity of the host to increase phage fitness32,33,34. Therefore we searched for contigs that encode these highly conserved genes in the assemblies. One of the contigs we found in cell 1 (contig 13737) is of length 16,010 nt and has an average coverage of 4,537x. Gene prediction using this contig (done as described above) identified 13 proteins that all encode cyanophage gene products such as a class II aldolase/adducin family protein (top hit, Synechococcus phage S-SM2), a 6-phosphogluconate dehydrogenase (top hit, Synechococcus phage S-RSM4), a photosystem II D1 protein (PsbA; top hit, Synechococcus phage S-PM2), a ferredoxin (top hit, Prochlorococcus phage Syn33), a virion structural protein (top hit, Synechococcus phage S-RSM4), a plastocyanin (top hit, Synechococcus phage S-SM2), and a photosystem II D2 protein (PsbD; top hit, Synechococcus phage S-PM2], among others (see Supplementary Fig. S5). The phylogeny of PsbD is shown in Figure 4D and demonstrates the close phylogenetic relationship between the protein encoded on contig 13737, cyanophage data available at NCBI (www.ncbi.nlm.nih.gov/), and the α-cyanobacterial sister clade that includes the plastid-encoded homologs in photosynthetic Paulinella species. These data provide a direct link between a phagotroph, its prey, phage that is associated with the prey (or is itself prey), and the source of the plastid in its sister group, the photosynthetic Paulinella clade14,17,18,19.
Have cyanobacterial genes been integrated into the nuclear genome of P. ovalis-like cells?
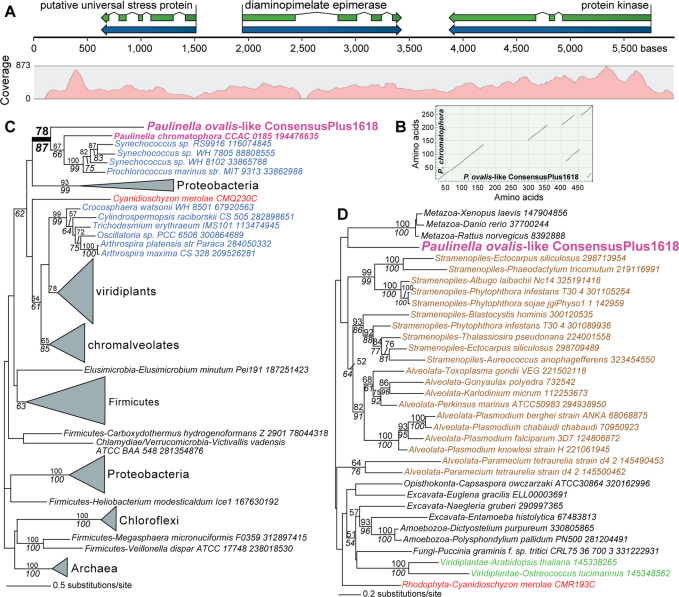
We analyzed manually each BLASTx hit of the combined assembly data listed in Supplementary Tables S3 and S4 to search for contigs that encode a conserved cyanobacterial protein that contains non-matching insertions, presumably resulting from nuclear introns. This search turned up one candidate in cell 1 (contig 4354, length = 1227 nt, average coverage = 26x) that encodes a diaminopimelate (DAP) epimerase gene containing a large insertion in the predicted gene. To extend this contig, we used BLASTn to identify regions with partial overlap in the 454 contigs from all six P. ovalis-like cell assemblies. This analysis identified two contigs (cell 1 contig 02238, length = 943 nt, 10 reads and cell 2 contig 00524, length = 2600 nt, 217 reads) that could be co-assembled with contig 4354 to generate a high quality consensus fragment (ConsensusPlus1618) of length 5970 nt, that when used to map all sequence reads had >100x coverage over most of the region (see Fig. 5A). The short regions of zero coverage in Figure 5A are due to repeated DNA that was masked by the assembler. Evidence that the Illumina paired-end reads span these repeat regions is shown in Supplementary Figure S6 demonstrating the contig is continuous. We also performed PCR using WGA-derived DNA from cell 1 and recovered fragments of expected size that span the length of contig ConsensusPlus1618, further validating the existence of this genomic region.
Figure 5. An example of α-cyanobacterial HGT found in the P. ovalis-like SAG data.
(a) Intron distribution and coverage of P. ovalis-like genome contig ConsensusPlus1618 that encodes three proteins. (b) Dot plot analysis of DAP epimerase from P. ovalis-like cells and the homolog that is encoded in the plastid genome of P. chromatophora CCAC 0185 showing the intron positions in the P. ovalis-like sequence. (c) Maximum likelihood (RAxML, WAG + Γ + F model) phylogeny of diaminopimelate epimerase (DapF) proteins. (d) Maximum likelihood (RAxML, WAG + Γ + F model) phylogeny of putative protein kinases. In 5B and 5C, Cyanobacteria are in blue text, other Bacteria are in black text, the chromatophore (plastid) and the P. ovalis-like cell data are in magenta text, Viridiplantae is in green text, red algae in red text, and chromalveolates in brown text. RAxML and PhyML bootstrap values (100 replicates) are shown above and below the branches, respectively (only those ≥ 50% are shown). The unit of branch length is the number of substitutions per site. The NCBI “gi” numbers (when available) are shown after each taxon name.
Protein prediction of this contig using AUGUSTUS (http://augustus.gobics.de/) and manual annotation identified three protein-coding regions that contained multiple spliceosomal introns (Fig. 5A, Supplementary Fig. S7). A dot plot analysis of the P. ovalis-like DAP epimerase when compared to the plastid-encoded homolog from P. chromatophora CCAC 0185 confirmed the presence of intervening sequences in the eukaryotic protein read-through product that correspond to the spliceosomal introns in this gene (Fig. 5B). Phylogenetic analysis of two of these proteins demonstrates that one (DAP epimerase [Fig. 5C]) originated via HGT from a α-cyanobacterial source, whereas the second (a protein kinase [Fig. 5D]) is of eukaryotic provenance. The third protein encoded on contig ConsensusPlus1618 is a putative universal stress protein that has a top BLASTp hit to a sequence from the human blood fluke Schistosoma japonicum (i.e., is eukaryotic in origin). To test the distribution of ConsensusPlus1618, we used the contig to map individual 454 reads from the six P. ovalis-like SAGs. This analysis showed that all cells had reads that mapped to this contig with data from some (e.g., cells 1, 2, and 4) nearly spanning the entire fragment (Supplementary Fig. S8). This suggests that the contig is likely to be present in all of the genomes. Our data therefore provide direct evidence for the integration of α-cyanobacterial DNA into the chromosome of P. ovalis-like cells.
We identified a second putative cyanobacterium-derived gene in P. ovalis-like cells that contains a large insertion when compared to prokaryote homologs. The encoded protein (leucyl-tRNA synthetase) is found on cell 2 contig 11624 (see Supplementary Fig. S9; length = 7,564 nt, avg. coverage = 299x) that also encodes a nuclear migration protein (nudC). Phylogenetic analysis demonstrates that P. ovalis-like leucyl-tRNA synthetase is sister to Cyanobacteria and monophyletic with oomycetes (Supplementary Fig. S10A). This is a more ancient HGT event that may have been shared by the ancestor of Rhizaria and stramenopiles (e.g., oomycetes), followed by widespread loss in other members of these lineages. Alternatively and more likely, based on the restricted distribution, these are independent HGTs from a cyanobacterial source. This contig in cell 2 has high sequence coverage (Supplementary Fig. S10B) and the neighboring gene that is a putative nudC homolog is clearly of eukaryotic provenance (Supplementary Fig. S10C).
Discussion
A key characteristic that has been postulated to underlie plastid endosymbiosis, and more generally genome evolution in eukaryotic microbes is long-term phagotrophy leading to HGT and ultimately plastid acquisition1,4,5,8,35. However, as appealing as these ideas may be they cannot be tested directly with Plantae whose plastid originated deep in the tree of Cyanobacteria36 about 1.6 billion years ago7. The Paulinella model therefore offers an opportunity to advance knowledge of plastid origin in a more recent, independent case of organelle origin in which the phagotrophic sister clade is available for study. Here we show that that P. ovalis-like SAG DNAs, although clearly of eukaryote provenance (i.e., containing identical rDNA sequences), harbor distinct pools of non-eukaryote sequence. These amoebae are heterotrophs based on the sorting procedure that excluded photosynthetic cells (see Methods) and the absence of plastid DNA in the assemblies. Therefore at the time of capture, the cells contained DNA from bacteria (and their associated phages) as prey28 in their food vacuoles or they ingested phage as prey. This hypothesis is in line with the observation that P. ovalis feeds on cyanobacteria20 and therefore likely ingests other bacteria and large phages as well. An alternative explanation is that the non-eukaryote hits derive from contamination associated with the cell surface and do not indicate intracellular DNA content. This interpretation is less favored for two reasons. First, the single cell approach has a low risk of DNA contamination from the sample matrix due to the small volume of fluorescence-activated cell sorting (FACS) microdroplets associated with each cell isolate; i.e., about 1–10 picoliter of the sample matrix37. Second, the different DNA compositions found in each SAG (in particular, from Proteobacteria, Bacteroidetes/Chlorobi, and viruses [Figs. 1C, 3, 4A, 4B]) is not consistent with the presence of a common cell surface contaminant shared by the captured cells. Nevertheless, we cannot exclude the possibility that some non-eukaryote DNAs may have originated from cells/virus particles externally attached to the sorted cells.
These results raise the possibility that given long-term phagotrophy in heterotrophic Paulinella species, prey DNA might have been integrated into the host nuclear genome35. Phagotrophy is widespread in “chromalveolates” and excavates and is widely regarded as an explanation for the increased rate of HGT in these taxa38,39,40. The key difference between HGT as a general phenomenon among protists and our study is that feeding behavior in Paulinella is tied to a fundamental change in lineage evolution, plastid primary endosymbiosis. In addition, EGT and HGT is known to be a major component of plastid establishment8,9,41,42, but does HGT occur from cyanobacterial prey prior to plastid endosymbiosis and could it play a role in this process? Although we cannot yet answer the second question with our data, we provide two examples of cyanobacterium-derived HGT in the P. ovalis-like SAGs. The case of DAP epimerase (DapF) is of particular interest because this gene is derived from α-Cyanobacteria. DapF carries out the second to last step in lysine biosynthesis in the DAP pathway. It is intriguing that plants that have a plastidial DAP pathway, encode a DapF gene of cyanobacterial origin, whereas all other genes in this pathway have proteobacterial or other affiliations43. The functional implication of a cyanobacterium-derived DapF gene in plastid-lacking P. ovalis-like cells is however unknown given incomplete knowledge of the DAP pathway in this lineage. Although the single cell genome approach does not provide expression data the presence of spliceosomal introns and high sequence conservation suggest a functional DapF in P. ovalis-like cells. Finally, we presume that the two cyanobacterium-derived genes we uncovered in the P. ovalis-like SAG genome data are not explained by a past photosynthetic history for these taxa. In the case that both the P. ovalis-like and photosynthetic Paulinella lineages once harbored a plastid, we would expect to find a more substantial imprint of EGT from alpha-cyanobacterial sources in the nuclear genome of the heterotrophic lineage16,19.
In summary, single-cell genome analysis provides several novel insights into phagotrophy and primary endosymbiosis in the Paulinella clade. Most important, we provide strong evidence that phagotrophic Paulinella feed on cyanobacterial prey derived from the same clade that gave rise to the plastid ca. 60 Mya15 in their photosynthetic sister group. The high abundance of α-Cyanobacteria in marine waters44 likely explains this conservation in prey choice that spans millions of years. Similar to what was found in the single cell genome analysis of wild-caught picobiliphyte cells28, P. ovalis-like cells isolated from the natural environment show distinct pools of non-eukaryote DNA, presumably derived from prey, symbionts, or pathogens. The wide variety of non-cyanobacterial prokaryote and viral DNA in the six cells also suggests that these (and likely most) phagotrophs have access to diverse prey DNAs that can be harnessed (e.g., via HGT35) to support an incipient endosymbiosis or other host functions. More generally, these data highlight the importance of analyzing single cells in their natural environment to understand protist-environment interactions.
Methods
Sample preparation
A surface water sample was collected on May 30, 2009 from the dock of the Smithsonian Environmental Research Center, Edgewater, MD, USA. Samples were kept in the dark at in situ temperature until processing. Subsamples (3 mL) were incubated for 10 min with Lysotracker Green DND-26 (75 nmol.L−1; Invitrogen), a pH-sensitive green fluorescing probe that stains food vacuoles in protists45. Target cells were identified and sorted using a MoFlo™ (Beckman-Coulter) flow cytometer equipped with a 488 nm laser for excitation. Prior to sorting, the cytometer was cleaned thoroughly with bleach. All tubes, plates, and buffers were UV-treated prior to use to remove any DNA contamination. A 1% NaCl solution (0.2 µm filtered and UV treated) was used as sheath fluid. The cleaning and preparation techniques were as previously described27,29.
Heterotrophic protists were identified by the presence of Lysotracker fluorescence and the absence of chlorophyll fluorescence (Fig. 6). Forward scatter was also used to select only the smaller protists that were ca. <10 µm in diameter. The sort criteria were optimized for a Lysotracker region that contained 5–10% heterotrophic Paulinella by positive microscopic identification, prior to single cell sorting. Individual target cells were deposited into 96 well plates, where some wells were dedicated for positive controls (10 cells/well) and negative controls (0 cells/well). All wells on the microplates contained 5 µL 1 x PBS or Lyse-N-Go (Pierce). The sorted microplates were centrifuged briefly and stored at −80°C.
Figure 6. Flow cytometric dot plot of the Lysotracker stained field sample.
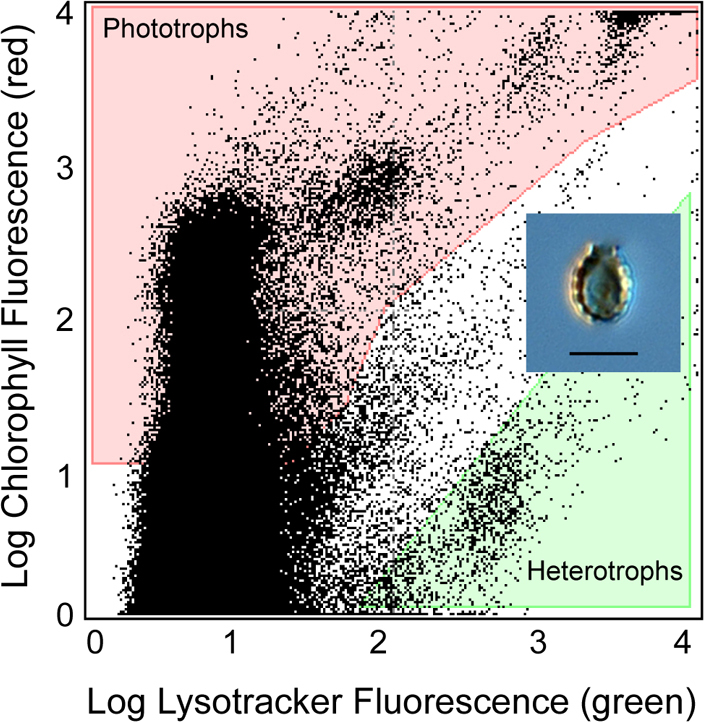
The heterotrophic protist sort region (shaded green) was identified as containing high relative green fluorescence (Lysotracker-stained food vacuoles) and low relative red fluorescence (indicative of chlorophyll). Phototrophs (shaded red) have both high chlorophyll fluorescence and Lysotracker fluorescence. A light microscopic image of a P. ovalis-like cell is shown in the inset (the scale bar indicates 5 μm).
Whole genome amplification
Cells deposited in PBS were lysed with cold KOH27. Cells deposited into Lyse-N-Go were lysed using a thermal cycle protocol provided by the manufacturer. Cell lysate genomic DNA was amplified using multiple displacement amplification (MDA46,47). All MDA reactions contained 2 U/µL Repliphi polymerase, 1 x reaction buffer, 0.4 mM dNTPs, 2 mM DTT (Epicentre), 1 µM SYTO-9 (Molecular Probes) and 50 nM random hexamer primers (IDT). Samples were incubated at 30°C for 6 h using a real-time thermal cycler with fluorescence measured at 6 min intervals. The Repliphi polymerase was inactivated by incubation for 3 min at 65°C, and the amplified DNA was stored at −80°C until further processing. After whole genome amplification, the SAGs were screened by PCR using conserved 18S rDNA primers to determine the phylogenetic origin of the nucleic acids. The genomic DNA of the six selected SAGs was re-amplified using the Repli-G midi kit (Qiagen) using the manufacturer's instructions. The products of the second MDA reaction were de-branched with S1 nuclease to reduce chimeric sequences during MDA25 and purified with a PCR purification kit (Qiagen).
Genome sequencing and assembly
About 5 μg of genomic DNA derived from each P. ovalis-like SAG with the A260/280 ratio of 1.85 was used for shotgun sequencing with the GS-FLX Titanium platform (Roche) at the DNA Facility at the University of Iowa (http://dna-9.int-med.uiowa.edu/). One-half of a picotitre plate was used to generate sequence data from each sample, resulting in over 600,000 reads per sample (Supplementary Table S1). All assemblies were generated with the native Roche Newbler Assembler, versions 2.3 and 2.5.3. The read depth/contig for the individual assemblies was determined by parsing the 454AlignmentInfo.tsv file, which is one of the output files generated by the Newbler assembler. The read depth is defined as the number of bases from all the reads used to assemble the contigs/Contig consensus length. All six assemblies were blasted (BLASTx) against RefSeq release 45 (http://www.ncbi.nlm.nih.gov/RefSeq/) and other publicly available sources (Supplementary Table S2). The top hits were extracted (leaving only 1 hit per contig). These were organized according to their phyletic grouping. This grouping was normalized such that, all P. ovalis-like SAG contigs with hits to the same target (overlapping and non-overlapping) were counted as one.
About 10 μg of WGA-derived DNA from P. ovalis-like cells 1 and 2 were each used to construct a library (i.e., sheared DNA fragments of size 500 bp) for 150 bp x 150 bp paired-end sequencing using an Illumina GAIIx instrument. Standard Illumina protocols (http://www.illumina.com/) were used to generate the library. For P. ovalis-like cell 1, a total of 46 million reads resulted in 4.7 Gbp of data that were assembled into 14,091 contigs with a N50 = 1.2 Kbp, totaling 11.1 Mbp. For P. ovalis-like cell 2, a total of 37 million reads resulted in 3.8 Gbp of data that were assembled into 17,793 contigs with a N50 = 994 bp, totaling 12.3 Mbp. The 454 + Illumina combined assemblies were done using the default settings and the CLC Genomics Workbench tools (http://www.clcbio.com/).
Author Contributions
HSY, DB, and RAA designed the research. NJP isolated the cells. HSY and ECY produced the amplified DNA. DB, DCP, and SPD performed all of the bioinformatic analyses and DCP contributed analytic tools. DB, DCP and HSY interpreted the data and DB wrote the paper.
Supplementary Material
Supplementary Information
Acknowledgments
This project was supported by NSF grants EF-0827023, DEB-0936884, DEB-0937975, OCE-0821374, and a grant from the Next-Generation BioGreen 21 Program (SSAC; 2011-PJ008177), Rural Development Administration, Korea to HSY. We thank Wayne Coats and Sunju Kim at the Smithsonian Environmental Research Center for assistance with sampling. Cell sorting services were provided by the J.J. MacIsaac Facility for Aquatic Cytometry at the Bigelow Laboratory for Ocean Sciences. The 10 novel rDNA sequences determined in this study from P. ovalis-like SAGs are available at GenBank under the accession numbers, JQ425612 – JQ425621. The 454 and Illumina sequence data used to assemble the P. ovalis-like SAG genomes are archived at the NCBI Sequence Read Archive (SRA) under Accession SRA049870.1. The 454 assemblies for each P. ovalis-like SAG, assembled 454+Illumina contigs for cells 1 and 2, and protein alignments used to generate the phylogenies in the paper are available at http://dbdata.rutgers.edu/data/ovalis/.
References
- Cavalier-Smith T. & Lee J. J. Protozoa as hosts for endosymbioses and the conversion of symbionts into organelles. J. Protozool. 32, 376–379 (1985). [Google Scholar]
- Rodríguez-Ezpeleta N. et al. Monophyly of primary photosynthetic eukaryotes: green plants, red algae, and glaucophytes. Curr. Biol. 15, 1325–1330 (2005). [DOI] [PubMed] [Google Scholar]
- Adl S. M. et al. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol. 52, 399–451(2005). [DOI] [PubMed] [Google Scholar]
- Chan C. X. et al. Red-and-green algal monophyly and extensive gene sharing found in a rich repertoire of red algal genes. Curr. Biol. 21, 328–333 (2011). [DOI] [PubMed] [Google Scholar]
- Archibald J. M. et al. Lateral gene transfer and the evolution of plastid-targeted proteins in the secondary plastid-containing alga Bigelowiella natans. Proc. Natl. Acad. Sci. USA 100, 7678–7683 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya D., Yoon H. S. & Hackett J. D. Photosynthetic eukaryotes unite: endosymbiosis connects the dots. BioEssays 26, 50–60 (2004). [DOI] [PubMed] [Google Scholar]
- Yoon H. S. et al. A molecular timeline for the origin of photosynthetic eukaryotes. Mol. Biol. Evol. 21, 809–818 (2004). [DOI] [PubMed] [Google Scholar]
- Martin W. et al. Gene transfer to the nucleus and the evolution of chloroplasts. Nature 393, 162–165 (1998). [DOI] [PubMed] [Google Scholar]
- Reyes-Prieto A. et al. Cyanobacterial contribution to algal nuclear genomes is primarily limited to plastid functions. Curr. Biol. 16, 2320–2325 (2006). [DOI] [PubMed] [Google Scholar]
- Bhattacharya D., Helmchen T. & Melkonian M. Molecular evolutionary analyses of nuclear-encoded small subunit ribosomal RNA identify an independent rhizopod lineage containing the Euglyphidae and the Chlorarachniophyta. J. Euk. Microbiol. 42, 65–69 (1995). [DOI] [PubMed] [Google Scholar]
- Lauterborn R. Protozoenstudien II. Paulinella chromatophora nov. gen., nov. spec., ein beschalter Rhizopode des Süsswassers mit blaugrünen chromatophorenartigen Einschlüssen. Z. Wiss. Zool. 59, 537–544 (1895). [Google Scholar]
- Yoon H. S. et al. Minimal plastid genome evolution in the Paulinella endosymbiont. Curr. Biol. 16, R670–R672 (2006). [DOI] [PubMed] [Google Scholar]
- Bodyl A., Mackiewicz P. & Stiller J. W. The intracellular cyanobacteria of Paulinella chromatophora: endosymbionts or organelles? Trends Microbiol. 15, 295–296 (2007). [DOI] [PubMed] [Google Scholar]
- Yoon H. S. et al. A single origin of the photosynthetic organelle in different Paulinella lineages. BMC Evol. Biol. 9, 98 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowack E. C. M., Melkonian M. & Glöckner G. Chromatophore genome sequence of Paulinella sheds light on acquisition of photosynthesis by eukaryotes. Curr. Biol. 18, 410–418 (2008). [DOI] [PubMed] [Google Scholar]
- Nowack E. C. et al. Endosymbiotic gene transfer and transcriptional regulation of transferred genes in Paulinella chromatophora. Mol. Biol. Evol. 28, 407–422 (2011). [DOI] [PubMed] [Google Scholar]
- Marin B. et al. The ancestor of the Paulinella chromatophore obtained a carboxysomal operon by horizontal gene transfer from a Nitrococcus-like γ-proteobacterium. BMC Evol. Biol. 7, 85 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T. & Ishida K. Another acquisition of a primary photosynthetic organelle is underway in Paulinella chromatophora. Curr. Biol. 19, R284–285 (2009). [DOI] [PubMed] [Google Scholar]
- Reyes-Prieto A. R. et al. Differential gene retention in plastids of common recent origin. Mol. Biol. Evol. 27, 1530–1537 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P. W., Hargraves P. E. & Sieburth J. M. Ultrastructure and ecology of Calycomonas ovalis Wulff, 1919, (Chrysophyceae) and its redescription as a testate rhizopod, Paulinella ovalis N. Comb. (Filosea: Euglyphina). J. Eukaryot. Microbiol. 35, 618–626 (1988). [Google Scholar]
- Vørs N. Marine heterotrophic amoebae, flagellates and Heliozoa from Belize (Central America) and Tenerife (Canary Islands), with descriptions of new species, Luffisphaera bulbochaete n. sp., L. longihastis n. sp., L. turriformis n. sp. and Paulinella intermedia n. sp. J. Eukaryot. Microbiol. 40, 272–287 (1993). [Google Scholar]
- Hannah F., Rogerson A. & Anderson O. R. A description of Paulinella indentata n. sp. (Filosea: Euglyphina) from subtidal coastal benthic sediments. J. Eukaryot. Microbiol. 43, 1–4 (1996). [Google Scholar]
- Zinser E. R. et al. Prochlorococcus ecotype abundances in the North Atlantic Ocean as revealed by an improved quantitative PCR method. Appl. Environ. Microbiol. 72, 723–732 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai V. & Palenik B. Temporal variation of Synechococcus clades at a coastal Pacific Ocean monitoring site. ISME J. 3, 903–915 (2009). [DOI] [PubMed] [Google Scholar]
- Zhang K. et al. Sequencing genomes from single cells by polymerase cloning. Nat. Biotechnol. 24, 680–686 (2006). [DOI] [PubMed] [Google Scholar]
- Marcy Y. et al. Dissecting biological "dark matter" with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc. Natl. Acad. Sci. USA 104, 11889–1194 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanauskas R. & Sieracki M. E. Matching phylogeny and metabolism in the uncultured marine bacteria, one cell at a time. Proc. Natl. Acad. Sci. USA 104, 9052–9057 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H. S. et al. Single cell genomics reveals trophic interactions and evolutionary history of uncultured protists. Science 332, 714–717 (2011). [DOI] [PubMed] [Google Scholar]
- Heywood J. L. et al. Capturing diversity of marine heterotrophic protists: one cell at a time. ISME J. 5, 674–684 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigue S. et al. Whole genome amplification and de novo assembly of single bacterial cells. PLoS ONE 4, e6864 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woyke T. et al. Assembling the marine metagenome, one cell at a time. PLoS ONE 4, e5299 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann N. H. et al. The Genome of S-PM2, a “photosynthetic” T4-type bacteriophage that infects marine Synechococcus strains. J. Bacteriol. 187, 3188–3200 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M. B. et al. Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol. 3, e144 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon I. et al. Photosystem I gene cassettes are present in marine virus genomes. Nature 461, 258–262 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle W. F. You are what you eat: a gene transfer ratchet could account for bacterial genes in eukaryotic nuclear genomes. Trends Genet. 14, 307–311 (1998). [DOI] [PubMed] [Google Scholar]
- Criscuolo A. & Gribaldo S. Large-scale phylogenomic analyses indicate a deep origin of primary plastids within cyanobacteria. Mol. Biol. Evol. 28, 3019–32 (2011). [DOI] [PubMed] [Google Scholar]
- Sieracki M., Poulton N. & Crosbie N. Automated isolation techniques for microalgae. In: Algal Culturing Techniques (ed R.A. Andersen) 101–116 (Elsevier Academic, New York, 2005) [Google Scholar]
- Andersson J. O. Horizontal gene transfer between microbial eukaryotes. Methods Mol. Biol. 532, 473–487 (2009). [DOI] [PubMed] [Google Scholar]
- Nosenko T. & Bhattacharya D. Horizontal gene transfer in chromalveolates. BMC Evol. Biol. 7, 173 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling P. J. & Palmer J. D. Horizontal gene transfer in eukaryotic evolution. Nat. Rev. Genet. 9, 605–618 (2008). [DOI] [PubMed] [Google Scholar]
- Timmis J. N. et al. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 5, 123–135 (2004). [DOI] [PubMed] [Google Scholar]
- Gross J. & Bhattacharya D. Mitochondrial and plastid evolution in eukaryotes: an outsider's perspective. Nat. Rev. Genet. 10, 495–505 (2009). [DOI] [PubMed] [Google Scholar]
- Sun G. & Huang J. Horizontally acquired DAP pathway as a unit of self-regulation. J. Evol. Biol. 24, 587–595 (2011). [DOI] [PubMed] [Google Scholar]
- Partensky F., Hess W. R. & Vaulot D. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol. Mol. Biol. Rev. 63, 106–127 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose J. M. et al. Counting heterotrophic nanoplanktonic protists in cultures and in aquatic communities by flow cytometry. Aquat. Microb. Ecol. 34, 263–277 (2004). [Google Scholar]
- Dean F. B. et al. Comprehensive human genome amplification using multiple displacement amplification. Proc. Natl. Acad. Sci. USA 99, 5261–5266 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunathan A. et al. Genomic DNA amplification from a single bacterium. Appl. Environ. Microbiol. 71, 3342–3347 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 (2006). [DOI] [PubMed] [Google Scholar]
- Guindon S. & Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information