Abstract
Translocation of RNA polymerase on DNA is thought to involve oscillations between pretranslocated and posttranslocated states that are rectified by nucleotide addition or pyrophosphorolysis. The pretranslocated register is also a precursor to transcriptional pause states that mediate regulation of transcript elongation. However, the determinants of bias between the pretranslocated and posttranslocated states are incompletely understood. To investigate translocation bias in multisubunit RNA polymerases, we measured rates of pyrophosphorolysis, which occurs in the pretranslocated register, in minimal elongation complexes containing T. thermophilus or E. coli RNA polymerase. Our results suggest that the identity of RNA:DNA nucleotides in the active site are strong determinants of susceptibility to pyrophosphorolysis, and thus translocation bias, with the 3′ RNA nucleotide favoring the pretranslocated state in the order U > C > A > G. The preference of 3′ U vs G for the pretranslocated register appeared to be universal among both bacterial and eukaryotic RNA polymerases and was confirmed by exonuclease III footprinting of defined elongation complexes. However, the relationship of pyrophosphate concentration to the rate of pyrophosphorolysis of 3′ U- versus 3′ G-containing elongation complexes did not match predictions of a simple mechanism in which 3′-RNA seqeunce affects only translocation bias and pyrophosphate (PPi) binds only to the pretranslocated state.
Cellular gene expression in all free-living organisms relies on evolutionarily conserved, highly regulated, multi-subunit RNA polymerases (RNAPs). During transcription, synthesis of the RNA transcript requires stepping the DNA template strand through the RNAP active site one nucleotide (nt) at a time in a process called translocation (recently reviewed in refs 1, 2). Translocation is one of four steps in a nucleotide addition cycle (NAC; Figure 1) that also includes, NTP binding, catalysis, and pyrophosphate (PPi) release. At the beginning of the NAC, the RNA 3′ nt is in the i+1 subsite of RNAP, corresponding to the pretranslocated register of the elongating complex (EC). Translocation of DNA through RNAP generates the posttranslocated register, positions the RNA 3′ nt into the product (P) or i subsite, and opens the i+1 (A) subsite to bind the incoming cognate NTP complexed with Mg2+II. Catalysis occurs by alignment of the phosphate of the bound NTP with the RNA 3′ OH upon formation of a three-helix bundle in the active site consisting of the trigger helices (TH; which form from the trigger loop; TL) and the bridge helix (BH). TH-NTP contacts facilitate an SN2 nucleophilic reaction in which a trigonal bipyramidyl transition state is stabilized by two catalytic Mg2+ ions (3). This reaction extends the RNA transcript by one nt and generates PPi, the release of which leaves the EC in the pretranslocated register ready for the next round of nucleotide addition. The NAC is reversible at high PPi concentration. Although the basic features of the NAC are established, the order in which translocation occurs relative to NTP binding and PPi release, which step is rate-limiting, and the path of NTP loading remains uncertain and under study (1, 2).
FIGURE 1.
Nucleotide addition and pyrophosphorolysis cycle. Template DNA is shown in black, non-template DNA in blue, RNA in red, incoming nucleotide triphosphate and pyrophosphate in green, and Mg2+ in yellow. One template/nontemplate position is colored orange to illustrate translocation. The EC alternates between pre and posttranslocated states with the RNA 3′ nt in the i or i+1 subsites, prior to NTP binding (step 1). NTPs enter the active site when the EC is posttranslocated (step 2). Catalysis (step 3) requires a rate-limiting conformational change in which the trigger loop folds into the trigger helices (not shown). Release of PPi (step 4) completes the nucleotide addition cycle. Pyrophosphorolysis is the reverse reaction of nucleotide addition. Since catalysis itself is reversible, net pyrophosphorolysis requires that the thermodynamic driving force of PPi conversion to NTP+RNA−1 exceed the conversion of NTP to PPi+RNA+1. Pyrophosphorolysis requires the EC be in the pretranslocated register, but whether PPi binding/release and translocation occur with obligate order is not established.
Translocation is thought to bear features of a thermal ratchet mechanism in which RNAP can oscillate between the pre and posttranslocated registers, with the bias between registers determined by their relative stabilities and the net motion of DNA through RNAP resulting from nucleotide addition or pyrophosphorolysis (2). However, the elemental rates of forward and reverse translocation, the effects of scaffold structure (e.g., RNA:DNA hybrid length) on translocation bias and the sequence-specific effects of interactions between nucleic acid bases and RNAP side chains on translocation bias are unknown (2). In the simplest form of this mechanism (Figure 1), translocation bias should affect the apparent binding affinities of NTP and PPi by dictating the fraction of time their binding sites are available. It also should determine susceptibility of the EC to transcriptional pausing, which plays many roles in the regulation of transcript elongation by multisubunit RNAPs. Although the mechanism of pausing remains under study, current proposals all identify the pretranslocated EC as the starting point for either (i) a structural isomerization in the EC that can be followed by reverse translocation (backtracking) or by other events that stabilize the paused EC (7, 8), (ii) direct conversion to backtracked (paused) states (9–11), or (iii) inhibition of conversion to the posttranslocated state by thermodynamic stability of the pretranslocated state (12). The energetics of translocation bias during backtracking are better understood and are governed by the relative stabilities of the RNA:DNA hybrid and the transcription bubble in different translocation registers (9, 13, 14). Translocation bias may also affect susceptibility to termination, which is thought to occur via either hypertranslocation without RNA synthesis or hybrid shearing (15, 16), both of which should be easier from the posttranslocated state (although alternative, non-translocational termination models also have been proposed; Refs. (17, 18). Thus, understanding the nature of translocation bias is important not only to gain insight in the NAC but also to understand regulatory events like pausing and termination and the mechanisms of regulators like NusA and NusG that are proposed to alter translocational bias (19, 20).
We have investigated the contribution of hybrid length and scaffold sequence to translocation bias using sensitivity to pyrophosphorolysis, which can occur only from the pretranslocated register (21). Initially, we eliminated contributions of the transcription bubble, upstream DNA, and upstream RNA using minimal scaffolds containing only the hybrid and downstream DNA (Figure 2A). We then extended the experiments to use full scaffolds, test contributions to translocation bias by exonuclease III footprinting, and remove the effect of the reverse reaction (nucleotide addition) by using apyrase to destroy NTPs generated by pyrophosphorolysis. Our findings suggest that the 3′ RNA dinucleotide sequence is a primary determinant of translocation bias, and that the PPi concentration-dependence of the pyrophosphorolysis reaction is inconsistent with the simple thermal ratchet model of the reaction.
FIGURE 2.
Pyrophosphorolysis by TthRNAP on a minimal scaffold. (A) A representative example of a minimal nucleic acid scaffold used in this study (yields EC8AU). *, 5′ 32P. The color scheme for RNA and DNA is the same as in Figure 1A. The posttranslocated state is resistant to pyrophosphorolysis; the pretranslocated state is competent for pyrophosphorolysis, yielding UTP in this case. (B) ECs containing TthRNAP were assembled on four minimal nucleic acid scaffolds that differ only in the sequence shown above the gel panels. The time course of pyrophosphorolysis (0.5 mM PPi) at 60°C is shown, with the sizes of RNAs indicated.
Materials and Methods
Materials
All DNA and RNA oligonucleotides (Table S1) were obtained from IDT (Corvalville, IA) and purified by denaturing PAGE before use. [γ-32P]ATP was from PerkinElmer Life Sciences and NTPs were from GE Healthcare (Piscataway, NJ). Exonuclease III (100,000 U/mL), T4 polynucleotide kinase, and apyrase (50 units/mL) were obtained from New England Biolabs (Ipswich, MA).
Proteins
Core E. coli and T. thermophilus RNAPs were purified as described previously (22, 23). Calf thymus RNAP was purified as described previously (24). Yeast (Saccharomyces cerevisiae) RNA polymerase II was a generous gift from Dr. Yuichiro Takagi (Indiana University, Indianapolis).
In Vitro EC Reconstitution
Nucleic acid scaffolds for reconstituting ECs were assembled in reconstitution buffer (RB; 10 mM Tris·HCl, pH 7.9, 40 mM KCl, 5 mM MgCl2) by heating 5′-32P-labeled RNAs (500 nM; Table S1), tDNAs (1 μM; Table S1), and ntDNAs (1 μM; Table S1) to 95 °C for 2 min, rapidly cooling to 45 °C, and then cooling to room temperature in 2 °C/2 min steps as described previously (25). Reconstitution of ECs was performed by incubating core RNAPs with the nucleic acid scaffold (2:1 RNAP:nucleic acid scaffold) in elongation buffer (EB; 25 mM HEPES-KOH, pH 8.0, 130 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol, 0.15 mM EDTA, 5% glycerol, and 25 μg of acetylated bovine serum albumin/ml) for 15 min at 37 °C as described previously (25). When fully complementary DNA strands were used, RNA and tDNA were preannealed and incubated with RNAP at 37 °C followed by incubation with ntDNA for an additional 10 min at 37 °C.
Pyrophosphorolysis
Pyrophosphorolysis was performed by incubating reconstituted ECs (~50 nM) with different concentrations of PPi in EB at 37 °C for EcoRNAP, 60 °C for TthRNAP, and 30 °C for SceRNAPII and BtaRNAPII. Aliquots were removed at the indicated times and quenched with an equal volume of 2X stop buffer (8 M urea, 50 mM EDTA, 90 mM Tris-borate buffer, pH 8.3, 0.02% bromphenol blue, and 0.02% xylene cyanol) and analyzed by denaturing (8 M urea) 25% polyacrylamide (19:1) gel electrophoresis (45 mM Tris, pH 8.3, 1.25 mM EDTA, 45 mM Boric acid). To remove the NTP accumulated from pyrophosphorolysis, 1 μL of apyrase (50 milliunits) was added to 100 μL of reaction mixture containing 50 nM EC.
Exonuclease III Footprinting
Exonuclease III digestion was performed as described previously (22). ECs were assembled with TthRNAP by annealing appropriate oligonucleotides as described above. Either the template or nontemplate DNA strand was 5′-end labeled with 32P. The non-labeled DNA strand in the scaffold contained phosphothioate linkage at the 3′ penultimate position to inhibit cleavage of that strand by ExoIII. ExoIII digestion was initiated by addition of 100 U of ExoIII to ECs (44 μL) at 37 °C. Aliquots were removed at the indicated times and stopped with the addition of an equal volume of 2X stop buffer. Products of the reaction and G+A nucleotide-sequencing reaction (26) of the same DNA fragment were analyzed by denaturing 15% polyacrylamide gel electrophoresis.
Intrinsic transcript cleavage assay
ECs were reconstituted in 20 mM Tris·HCl, pH 9.0, 20 mM NaCl, 0.1mM EDTA by incubating TthRNAP with nucleic acid scaffolds at 37 °C for 15 min as described above. The cleavage reaction was initiated by the addition of MgCl2 to 20 mM and incubated at 37 °C. At the indicated times, aliquots were removed, and samples were processed as for the pyrophosphorolysis assay.
Data Quantitation and Analysis
Gels were exposed to phosphorimager screens, scanned using a Typhoon PhosphorImager, and quantitated using the ImageQuant Software (GE Healthcare). The 9 nt RNA present in each lane was quantitated as a fraction of the total RNA in each lane (Figure 3) and corrected for the fraction remaining in the chase lane. The rate of pyrophosphorolysis was then determined as described below.
FIGURE 3.
Identity of RNA 3′ terminal nucleotide affects translocation register of TthRNAP. (A) Quantitative analysis of pyrophosphorolysis of EC9CU, which favors the pretranslocated state. TthEC9CU was reconstituted from tDNA #6059, ntDNA #5848, and RNA #6042 (Table S1). ECs (50 nM) were incubated with 0.5 mM pyrophosphate at 60°C and disappearance of the 9 nt RNA measured at the times indicated in the inset. +NTP, UTP (1mM) was added at the end of the time course to confirm that the unreacted EC9CU remained active and could extend the RNA 1 nt by UMP incorporation. Errors are SD from three independent experiments. Data were fit to a simple reversible mechanism of pyrophosphorolysis (equation 3; see Materials and Methods). A value for k1 that approximated the initial rate of pyrophosphorolysis (0.29 ± 0.04 min-1) and a final fraction of pyrophosphorolysed transcript (PPi-sensitive fraction, remaining 9-nt RNA/total RNA = 0.74 ± 0.03) were determined by nonlinear regression. (B) Quantitative analysis of pyrophosphorolysis of EC9CG, which favors the posttranslcated state. The experiment was performed identically to that shown in panel A except that TthEC9CG was reconstituted from tDNA #6062, ntDNA #5848, and RNA #6045 (Table S1). (C) A plot of fraction of EC susceptible to pyrophosphorolysis (PPi-sensitive fraction) with 0.5 mM PPi versus the rate of pyrophosphorolysis for 16 combinations of 3′-proximal dinucleotide sequence.
Estimation of Rates and Kinetic Modeling
To calculate the apparent rate of pyrophosphorolysis of 16 different variants in table 1, we determined rapid decay of 9 nt RNA by fitting the fraction of 9 nt (Figure 3) using the equation (3). A simple reversible mechanism of pyrophosphorolysis was used in the fitting for simplicity.
| (1) |
The overall rate of pyrophosphorolysis (equation 1) is given by
| (2) |
Integration between t = 0 and t = t at concentrations [EC9]o and [EC9] gives
| (3) |
The values of k1 and k−1 for 16 ECs are summarized in Table 1 and the rate constant (k1) of 16 ECs was used to generate a plot in Figure 3C.
To characterize the [PPi]-dependence of pyrophosphorolysis in ECs with 3′ U or 3′ G transcripts, we assayed pyrophosphorolysis of EcoEC9GU and EcoEC9UG at different [PPi]. The fraction remaining EC9 was plotted as a function of time (Figure S4). Since we included apyrase (Apy) in these reactions, we modified reaction 1 and included the rate of apyrase action (reaction 4).
 |
(4) |
We then estimated initial rates of pyrophosphorolysis by fitting the [EC9] versus time at different [PPi] to mechanism 4 using the program KinTek Global Kinetic Explorer (27, 28), with diffusion limited NTP binding (k−2 = 6000 μM−1 min−1) a dissociation rate to give a KNTP of ~55 μM (k2 =330,000 min−1) (29). At the resulting NTP concentrations, reported kinetic values for apyrase were consistent with NTP degradation becoming rate limiting for overall pyrophosphorolysis (diffusion limited k3 = 6000 μM−1 min−1; k−3 = 132,000 μM−1 min−1; k4 = 190 min−1; Ref. 30). The concentration of apyrase, which varied somewhat due to the error in small-volume pipetting, was allowed to float. The initial rate (k1) of pyrophosphorolysis of EC9GU was obtained using these simulations and plotted versus [PPi] (Figure 8B). Because it was too slow for the apyrase reaction to become rate limiting, the pyrophosphorolysis rate of EC9UG was determined by nonlinear regression assuming a pseudo-first order reaction and plotted versus [PPi] (Figure 8B).
FIGURE 8.
PPi-concentration dependence of pyrophosphorolysis in EcoEC9GU and EcoEC9UG. (A) A predicted relationship for the rate of pyrophosphorolysis vs. [PPi] for the ordered translocation/PPi binding mechanism shown in Figure 6A and Figure 1. The difference in apparent KPPi (~75x) between EC9GU and EC9UG and in rate at 0.5 mM PPi concentration are indicated on the plot. (B) Kinetic analysis of [PPi]-dependence of pyrophosphorolysis in EC9GU and EC9UG. Rate of pyrophosphorolysis for EcoEC9GU and EcoEC9UG over a wide range of PPi concentrations. (C) Approximate, apparent kinetic constants of EC9GU and EC9UG are calculated from the plot shown in Figure 8B (see Materials and Methods).
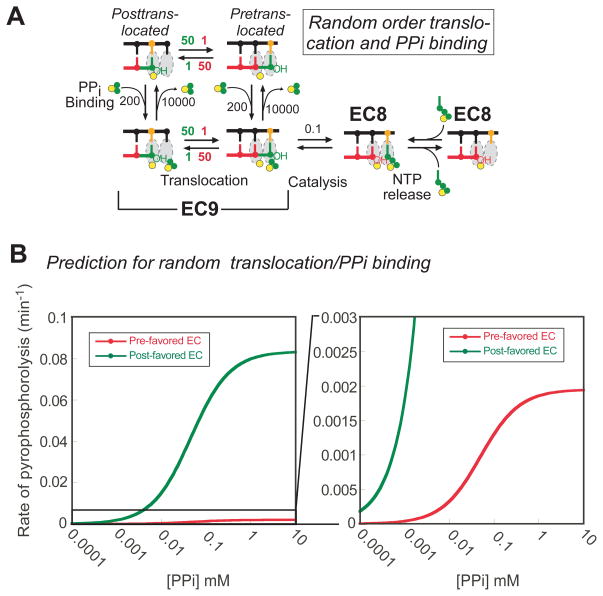
To obtain the kinetic simulation graph depicted in Figure 9B, the arbitrary kinetic parameters shown in Figure 9A were used. The synthetic rate of pyrophosphorolysis for pre-favored EC and that of post-favored EC over a wide range of PPi concentrations were generated using KinTek Global Kinetic Explorer. The predicted values were used to construct complete curves by nonlinear regression to a hyperbola (Figures. 9B).
FIGURE 9.
A random-order of translocation of PPi binding/release can explain differences in pyrophosphorolysis of EC9UG and EC9GU. (A) A random-order translocation and PPi binding/release mechanism. With the arbitrarily chosen rate constants shown in the figure, the mechanism can account for a Vmax difference with little effect on apparent KPPi. Translocation rates were assigned based on translocation bias of a particular EC. Forward and backward translocation rate constants for pre-favored ECs are shown in green (50 and 1 s-1, respectively), whereas those for post-favored ECs are in red (1 s-1; forward rate constant and 50 s-1; reverse rate constant). (B) A kinetic simulation graph of pretranslocation-favoring EC9GU (green) and posttranslocation favoring EC9UG (red) ECs was generated using the reaction scheme shown in Figure 9A using the program KinTek Global Kinetic Explorer (see Materials and Methods).
RESULTS
RNA 3′ sequence, not an 8-bp vs. 9-bp hybrid, primarily controls translocation bias
We first investigated the relationship between 8- vs. 9 bp hybrids, transcript sequence, and translocation register using minimal scaffolds that eliminate effects of transcription bubble energetics on translocation bias, and sequences that led Kashkina et al. (31) to suggest that the pre or posttranslocated registers were favored by 8-bp or 9-bp hybrids, respectively. We reconstituted ECs using T. thermophilus RNAP (TthRNAP) on scaffolds that differed either by hybrid length or by 3′ trinucleotide sequence, and then assessed translocational bias by measuring the rates of pyrophosphorolysis for each scaffold (Figure 2). We observed the same difference reported by Kashkina et al. (31) between the sensitive 8-bp hybrid scaffold EC8GAU (superscript indicates the 3′-proximal RNA sequence in the EC) and the resistant 9-bp hybrid scaffold EC9AUA (Figure 2B). However, when we examined pyrophosphorolysis of ECs in which the 3′ RNA sequences of 8-bp and 9-bp hybrid were swapped, we found that EC8AUA was resistant to pyrophosphorolysis whereas EC9GAU was sensitive to pyrophsophorolysis. In other words, both EC8GAU and EC9GAU appear biased toward the pretranslocated state whereas both EC8AUA and EC9AUA appear biased toward the posttranslocated state. This result strongly suggests that the RNA 3′ sequence in the sensitive 3′-proximal GAU and resistant 3′-proximal AUA is the dominant determinant of translocation bias relative to any effects of 8- vs. 9-bp hybrids.
RNA 3′-dinucleotide dictates translocation bias
Given the dominant effect of the 3′-proximal sequence on pyrophosphorolysis, we next sought to determine which RNA 3′-dinucleotides are most and least sensitive to pyrophosphorolysis (presumably corresponding to the most and least biased toward the pretranslocated register). To accomplish this test, we generated 16 different ECs in which the 3′ dinucleotide sequence was varied to create all possible combinations, again using minimal scaffolds to avoid effects of transcription bubble energetics. When we tested pyrophosphorolysis in the 16 different ECs, we noticed that they exhibited differences not only in the rate of pyrophosphorolysis but also in the fraction of ECs in which the RNA that was shortened (e.g., compare EC9CG and EC9CU; Figures 3A & B). Incomplete RNA shortening was not due to incomplete reconstitution of ECs because addition of cognate NTP resulted in extension of all 9mer RNA (+NTP lane; Figures 3A & 3B). Rather, we suspected (and later confirmed; see below) that incomplete pyrophosphorolysis reflected equilibration of pyrophorphorolysis with the reverse reaction of nucleotide addition as the concentration of NTP produced by pyrophorphorolysis increased. Therefore, we fit the reaction progress curves to a simple reversible mechanism (reaction 1: see Materials and Methods).
This yielded a value for k1 that approximated the initial rate of pyrophosphorolysis and allowed us to rank the pyrophosphorolysis sensitivities of the 16 different ECs by assigning both an observed rate of pyrophosphorolysis and an equilibrium fraction ([EC8]eq/[EC9]eq) of pyrophosphorolysed transcript (Table 1 and Figure 3C). For any given penultimate RNA nucleotide, both the rate of pyrophosphorolysis and the equilibrium fraction yielded the same order of sensitivity to pyrophosphorolysis for the 3′-terminal nucleotide: U > C > A > G. Interestingly, 3′-penultimate nucleotide gave the inverse order of effects on sensitivity to pyrophosphorolysis: G > A > C > U. In general, faster pyrophosphorolysis rate correlated with a larger sensitive EC fraction (Figure 3C). These data are consistent with preferential interaction of G > A > C > U in the product (i) subsite and U > C > A > G in the NTP-binding (i+1) subunit (see Discussion).
RNA 3′ dinucleotide effects on pyrophosphorolysis are evolutionarily conserved
To investigate whether similar effects of RNA 3′ dinucleotides would be observed in ECs reconstituted with other multisubunit RNAPs, we compared ECs reconstituted on the most and least sensitive scaffolds (EC9GU and EC9UG, respectively) with Thermus thermophilus RNAP (TthRNAP), Escherichia coli RNAP (EcoRNAP), Saccharomyces cerevisiae RNAPII (SceRNAPII), and calf thymus (Bos taurus) RNAPII (BtaRNAPII). For all four sources of RNAP, EC9GU was sensitive to pyrophosphorolysis and EC9UG was resistant (Table 2; Figure S1). We also confirmed that incomplete pyrophosphorolysis was not due to incomplete reconstitution of ECs by the addition of cognate NTP, which resulted in extension of all 9mer RNA (+NTP lane; Figures 3A & 3B; C lane; Figure S1A & S1B) Thus, the same RNA 3′ dinucleotide characteristics appear to control translocation bias for evolutionarily diverse multisubunit RNAPs.
NTP accumulation causes incomplete pyrophosphorolysis
We assumed that incomplete pyrophosphorolysis (e.g., Figure 3) could be explained by reverse-reaction (nucleotide addition) with NTPs generated during pyrophosphorolysis because the KNTP is low and nucleotide addition is fast (kcat≈103 s−1; Refs. 29, 32). To test this hypothesis, we examined pyrophosphorolysis of EC9GU (Table S1) in the presence of apyrase. Apyrase rapidly hydrolyzes NTPs to NDPs (30). We used EcoRNAP instead of TthRNAP for these experiments because the optimal temperature for apyrase is 30 °C, a temperature at which TthRNAP is relatively inactive. EcoEC9GU exhibited sensitivity to pyrophosphorolysis similar to TthEC9GU, but with a higher unreacted fraction (~65% vs ~30%; compare Figure 4A to Figure 3C). If the 65% unreacted fraction was due to reverse-reaction with NTP, addition of apyrase should shift the reaction toward completion. Consistent with our hypothesis, addition of apyrase at the beginning of the reaction eliminated the plateau at partial completion and gave a pseudo-first-order rate of 0.016 ± 0.001 min−1 for the most of reaction (Figure 4A). Addition of apyrase after the plateau had been achieved (at 60 min) caused resumption of pyrophosphorolysis, unambiguously establishing that incomplete reaction reflected accumulation of NTP. Since initial concentration of EcoEC9GU is 50 nM and 65% of it remains unreactive when the reaction reaches the equilibrium, we infer that the net forward flux of 0.5 mM PPi + ~30 nM EcoEC9GU is equal to the reverse flux from 20 nM UTP + 20 nM EcoEC8G. We noted, however, that at later times in the reaction (after ~2 hours) the equilibrium in the absence of apyrase was replaced by further pyrophorphorolysis and the rate of pyrophosphorolysis in the presence of apyrase slowed, most likely because one or more reaction components degraded upon prolonged incubation.
FIGURE 4.
Incomplete pyrophosphorolysis by EcoRNAP is caused by NTP accumulation. (A) Rates of pyrophosphorolysis of EcoEC9GU. EcoEC9GU reconstituted using E. coli RNAP and the pre-translocation favoring scaffold (#6063, #5848, #6046; Table S1). EcoEC9GU (50 nM) was incubated at 37°C with 0.5 mM PPi alone (red), in the presence of 0.5 U apyrase/ml (blue), or with addition of apyrase to 0.5 U/ml 60 min after the reaction was initiated (green) for the time indicated. Samples were removed at the times indicated and the fraction EC9GU remaining was plotted as a function of time. The error bars represent standard deviations obtained from 4 different experiments. The data from reaction without apyrase (red) are fit to a simple reversible mechanism of pyrophosphorolysis (equation 3; see Materials and Methods), whereas data from reaction with apyrase (blue) are fit to a single exponential for a pseudo-first-order reaction. (B) The fraction of EcoEC9GU and EcoEC9UG resistant to pyrophosphorolysis at 0.5 mM PPi in the absence (gray) or presence (black) of apyrase.
We next tested whether the resistance of EcoEC9UG to pyrophosphorolysis could possibly be explained by an exceptionally low KGTP for the EcoEC8U + GTP reaction. To test this possibility, we performed pyrophosphorolysis of EcoEC9UG in the presence of apyrase in parallel to reactions in which apyrase made pyrophosphorolysis of EcoEC9GU go to completion. Although we observed the expected shift in EcoEC9GU completion, no reaction of EcoEC9UG was observed even in the presence of apyrase (Figure 4B and Figure S2A). We verified that apyrase effectively destroyed NTPs by preincubating 200 nM UTP with apyrase under the same conditions used for pyrophophorolysis, then adding EcoEC9UG, and observing that elongation occurred only when apyrase was omitted (Figure S2B). We concluded that the resistance of EcoEC9UG to pyrophosphorolysis is an inherent property of EcoEC9UG, most likely a strong preference for the posttranslocated register, and not to a strong reverse-reaction of EcoEC8U with GTP.
Effects of RNA 3′-dinucleotide sequence on pyrophosphorolysis were preserved on a complete nucleic acid scaffold
We used minimal nucleic-acid scaffolds in initial experiments to avoid complications from transcription bubble energetics. To test whether the strong effects of RNA 3′ dinucleotide sequence persisted on complete scaffolds containing fully complementary DNA strands, a transcription bubble, and upstream and downstream duplexes, we tested complete-scaffold versions of the PPi-resistant EC9UG and the PPi-sensitive EC9GU reconstituted ECs with TthRNAP or EcoRNAP (Figure 5A and Table S1). Similar to the minimal scaffolds, EcoEC9UG (<0.002 min−1) and TthEC9UG (0.12 ± 0.04 min−1) were less sensitive to pyrophosphorolysis than EcoEC9GU (0.034 ± 0.004 min−1) and TthEC9GU (1.7 ± 0.1 min−1) (Figure 5B). In contrast to the minimal scaffolds, however, both EcoEC9UG and TthEC9UG exhibited measurable rates of pyrophosphorolysis. We also tested the effects of apyrase on the complete scaffold version of EcoEC9GU and verified that apyrase addition caused the 40% unreactive EcoEC9GU to complete pyrophosphorolysis (Figure S3). We drew two conclusions from these results. First, pyrophosphorolysis is faster in ECs reconstituted with complete scaffolds than in those with minimal scaffolds. This could reflect a greater bias of complete vs. minimal scaffolds toward the pretranslocated register for these specific complexes or a lower activation barrier to formation of the trigger helices in the presence of an intact fork junction (trigger helices formation strongly stimulates the rate of pyrophosphorolysis (32, 35). Second, faster pyrophosphorolysis and presumably greater bias toward the pretranslocated register for the RNA 3′dinucleotide GU vs. UG occurs on both minimal and complete scaffolds.
FIGURE 5.
ExoIII footprints of TthEC9GU (pretranslocated) and TthEC9UG (posttranslocated). (A) Schematic of the complete scaffold used for exoIII footprinting experiments. The sizes of the template DNA (upstream) and the nontemplate DNA (downstream) fragments protected by RNAP from digestion by ExoIII are illustrated. For the nontemplate strand assay, the nontemplate strand contained a 5′ 32P label and the template strand contained a 3′ phosphorothioate bond. For template strand assay, the template strand contained a 5′ 32P label and the nontemplate strand containing a 3′ phosphorothioate bond. (B) Pyrophosphorolysis results of EC9GU and EC9UG on complete scaffolds used for ExoIII footprinting experiments. The RNA and template DNA (GU: #6046, #6355, and UG: #6053, #6357; Table S1) were first annealed, then mixed with TthRNAP, then annealed to the nontemplate strand (GU: #6354 and UG: #6356; Table S1; see Materials and Methods). ECs (50 nM) were incubated with 0.5 mM PPi at 60 C. The initial rate of pyrophosphorolysis (k1) and the fraction resistant to pyrophosphorolysis were determined as described in Materials and Methods. (C) Upstream ExoIII footprinting (left) and downstream ExoIII footprinting (right) of EC9GU and EC9UG. Plot on the left depicts the appearance of the nontemplate DNA 14 band, which should be faster when the pretranslocated register is favored. Plot on the right depicts the appearance of the template DNA +12 band, which should be faster when the posttranslocated register is favored.
Direct ExoIII footprinting confirmed translocation bias due to RNA 3′ dinucleotide sequence
To verify that the differences in pyrophosphorolysis sensitivity were due to differences in translocation register and not to inherent differences in the chemical reactivities of RNA 3′ dinucleotides with PPi or to sequence-specific effects of the RNAP catalytic center on pyrophosphorolysis, we assayed translocation register using the independent criterion of exonuclease III (ExoIII) footprinting (31, 33, 34). ExoIII is a double-strand-specific deoxyribonuclease that processively digests a single-strand of DNA from the 3′ end and detects the boundary of RNAP on DNA when bound RNAP inhibits access of the ExoIII active site to the DNA phosphodiester backbone. Because RNAP may be in rapid oscillation between pre and posttranslocated registers and because ExoIII exhibits some sequence specificity in cleavage rates, assessment of translocation register with ExoIII requires measuring the rates of ExoIII digestion rather than arbitrary reaction endpoints (34).
We determined the upstream and downstream ExoIII boundaries for TthEC9GU and TthEC9UG reconstituted on complete scaffolds (Figure 5A). On this scaffold, an EC with greater pretranlocation bias will exhibit a greater barrier to ExoIII digestion of −15 to −14 on the template strand and a weaker barrier to ExoIII digestion of +13 to +12 on the nontemplate strand (Figure 5A). The upstream footprint slowly shifts one bp downstream in both TthEC9GU and TthEC9UG but the TthEC9GU presents a stronger barrier that results in an initial lag in the rate of the shift and a difference in the final distribution of DNA fragments (Figure 5C). This is consistent with the hypothesis that TthEC9GU was more biased in the pretranslocated direction than TthEC9UG. A different effect was observed during ExoIII digestion of downstream DNA, with the initial rates of digestion being similar, but TthEC9UG posing a stronger barrier as the reaction progressed. This result also is consistent with TthEC9GU being more pretranslocated than TthEC9UG. These ExoIII footprinting assays are less clear-cut than one might hope, but the consistent pattern of effects strongly supports the interpretation that RNA 3′ GU more favors the pretranslocated register than RNA 3′ UG.
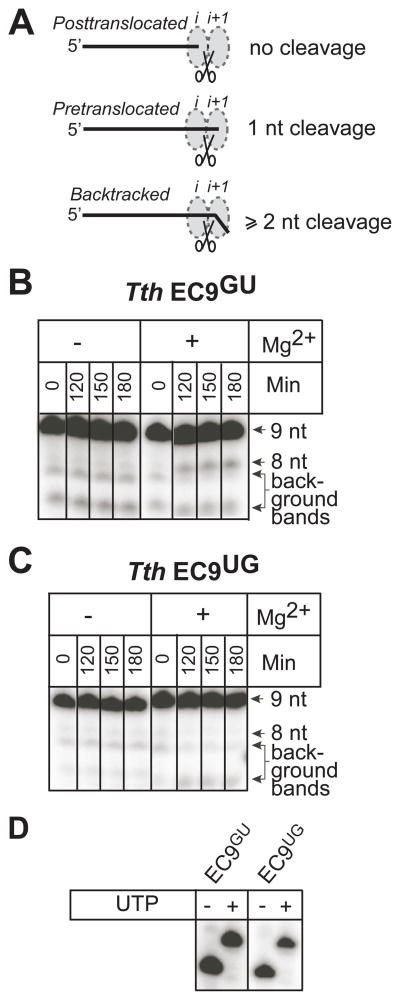
EC9UG resistance to pyrophosphorolysis was not caused by backtracking
Although our results strongly favored the idea that EC9UG is biased toward the posttranslocated register, we wished to rule out the alternative possibility that EC9UG is backtracked and therefore resistant to pyrophosphorolysis. To this end, we performed hydrolytic transcript cleavage in TthEC9UG reconstituted on a minimal scaffold. The transcript hydrolysis (i.e., intrinsic cleavage) reaction cleaves nascent RNA at the phosphodiester bond located in the RNAP active site so that a pretranslocated EC yields a one nt 3′ cleavage product and a backtracked EC yield a larger 3′ cleavage product (Figure 6A); the hydrolysis reaction is faster at elevated pH and high Mg2+ concentration (3, 35–37). We first generated TthEC9UG and TthEC9GU in transcription buffer at pH 9 lacking Mg2+ and then initiated transcript cleavage by addition of Mg2+ to 20 mM. No cleavage products were observed for TthEC9UG, whereas TthEC9GU generated the one-nt cleavage product expected for a pretranslocated EC (Figures 6B and C). We confirmed that these ECs were fully reconstituted and active by extending the transcripts from 9 to 10 nt by incubation with UTP (Figure 6D). Further, cleavage products were not observed when RNAP was omitted (no enzyme control; Figure S5). The absence of cleavage products >1 nt is consistent with strong preference of the RNA 3′ GU dinucleotide for the pretranslocated register relative to the GG dinucleotide that would occupy the active site if the EC were to backtrack by 1 bp. We concluded that TthEC9UG is not backtracked and that the simplest explanation for its resistance to pyrophosphorolysis is bias toward the posttranslocated register.
FIGURE 6.
The pyrophosphorolysis-resistant TthEC9UG is not backtracked. (A) Predicted outcomes for intrinsic cleavage reactions of pretranslocated, posttranslocated, and backtracked ECs. Posttranslocated ECs does not generate cleavage products, whereas pretranslocated and backtracked complexes produce 1-nt cleavage products, and 2 or more cleavage products, respectively. (B) Intrinsic cleavage reaction of TthEC9GU. TthEC9GU was reconstituted using TthRNAP and the pretranslocation-favoring scaffold (#6063, #5848, #6046; Table S1) containing 5′ end 32P-labeled RNA at pH 9 without Mg2+. Intrinsic cleavage reaction was initiated by the addition of 20 mM Mg2+ and samples were removed and separated by electrophoresis at the times indicated (see Materials and Methods). (C) Intrinsic cleavage reaction of TthEC9UG. TthEC9UG was reconstituted and assayed as in panel A except with the postranslocation-favoring scaffold (#6070, #5848, #6053: Table S1). (D) EC9GU and EC9UG were incubated with 100 ?M UTP to extend their respective RNAs by one nucleotide, thus showing the ECs were active and not arrested.
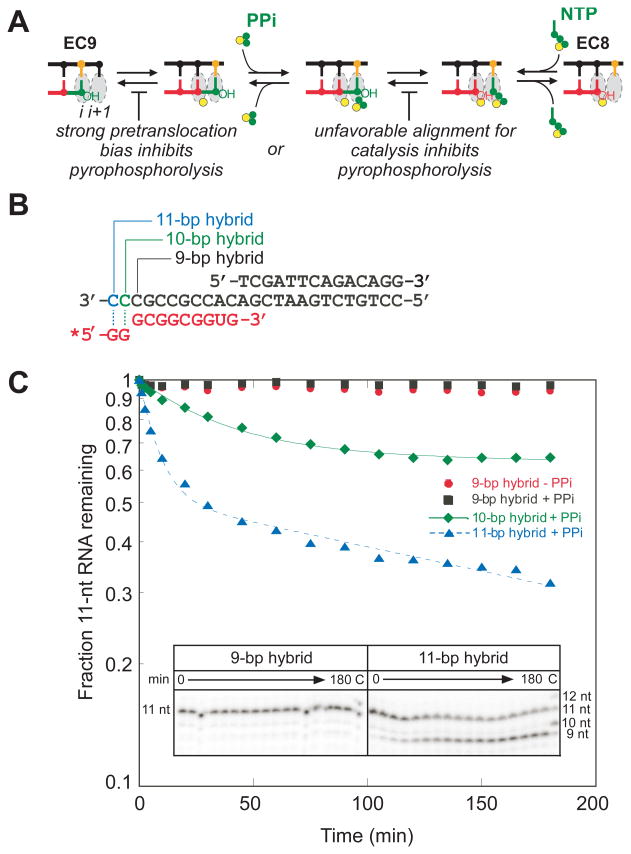
Inhibiting forward translocation with extended hybrids makes EC9UG PPi-sensitive
We reasoned that if bias toward the posttranslocated register in EC9UG rather than an inherently slow catalytic reaction explained its pyrophosphorolysis resistance (Figure 7A), then it might be possible to increase the rate of pyrophosphorolysis by increasing the length of the RNA:DNA past 9 bp. A longer hybrid, especially one with GC-bp at the upstream end, should be harder to melt and thus be shifted toward pretranslocated register to fit the longer hybrid within the RNAP main channel. To test this prediction, we used EcoRNAP to allow use of apyrase (to reduce potential interference from reverse nucleotide addition) and measured pyrophosphorolysis of EcoEC9UG, EcoEC10UG, and EcoEC11UG reconstituted on minimal scaffolds (Figure 7B). These scaffolds use the same 11-nt, 3′-UG RNA and differ only in potential hybrid length due to changes in the 3′ portion of the template strands, which are 3′-GCC··· for EC9UG, 3′-CGCC··· for EC10UG, and 3′-CCGCC··· for EC11UG. In agreement with the idea that pyrophosphorolysis resistance of EC9UG reflects translocation bias, EC10UG exhibited increased sensitivity to PPi and EC11UG exhibited a greater increase in sensitivity to PPi (Figure 7C). To confirm that longer hybrids inhibit forward translocation, we also performed hydrolytic transcript cleavage in EcoEC9UG and EcoEC11UG. As expected, EcoEC11UG generated a two-nt 3′ cleavage product, which indicated that forward translocation is hindered in this EC, whereas EcoEC9UG gave little cleavage product (Figure S5). These results are inconsistent with inherence resistance of the 3′-UG dinucleotide to pyrophosphorolysis but consistent with the view that posttranslocation bias confers PPi-resistance on EC9UG. Pyrophosphorolysis of EC11UG was noticeably biphasic. This could either reflect structural heterogeneity in the elongation complexes or an inability of apyrase to hydrolyze NTPs rapidly at the concentrations generated in this experiment.
FIGURE 7.
The RNA 3′ dinucleotide UG is not intrinsically resistant to pyrophosphorolysis by EcoRNAP. (A) translocation bias towards posttranslocated state or a slow catalysis could account for the insensitivity of EC9UG. (B) Nucleic acid scaffolds with different potential RNA:DNA hybrid length. RNA is shown in red, template and nontemplate DNA strands are in black. The 9-bp, 10-bp, and 11-bp potential RNA:DNA hybrid are created by varying the length of template DNA without changing the length of the 11-nt RNA (the template DNAs in 9-bp, 10-bp, and 11-bp hybrids end at the positions marked with a black line, a green, or a blue line, respectively. RNA was labeled with 32P at its 5′ end (*). (C) Plot of pyrophosphorolysis of ECs reconstituted with EcoRNAP and different scaffolds shown in Figure 6B. ECs were incubated with 0.5 mM PPi, 0.5 U apyrase/ml at 37°C for the times indicated. Representative gel panels for EC9UG (9-bp hybrid) and EC11UG (11-bp hybrid) are shown in the inset. “C”, chase lane in which ECs were chased with 1 mM UTP at the end of pyrophosphorolysis time course (after 3 hours).
We note that the 2-nt cleavage interval observed for EcoEC9UG and EcoEC11UG (Figure S5) differs from the 1-nt hydrolytic cleavage interval observed for TthEC9GU (Figure 6). This could reflect, at least in part, differences in Eco and Tth RNAPs. However, it is notable that our analysis of RNA 3′ dinucleotide effects on pyrophosphorolysis (Fig. 3C and Table 1) suggested that the UG dinucleotide disfavors the pretranslocated register. Upon a 1 nt backtrack, the active site of EcoEC9UG and EcoEC11UG would be occupied by the GU dinucleotide that appears to favor the pretranslocated register. Thus, the observed difference in hydrolytic cleavage intervals is consistent with favorable interactions of the GU dinucleotide and unfavorable interactions of the UG dinucleotide in the pretranslocated active site.
PPi concentration-dependence of pyrophosphorolysis is inconsistent with ordered translocation and PPi binding and effects of 3′ dinucleotide only on translocation bias
A simple model in which rapid translocation equilibrium is linked to PPi binding to the pretranslocated EC (Figure 7A) predicts that increasing PPi concentration should reduce occupancy of the posttranslocated state. Indeed, studies of the single-subunit polymerases HIV-1 reverse transcriptase and T7 RNAP show that binding of PPi or the PPi-analog foscarnet does shift the translocation bias towards the pretranslocated state, suggesting these single-subunit polymerases adhere to the predictions of this simple model (38, 39). If such a model operates for multisubunit RNAPs and if all the effects of RNA 3′ dinucleotide sequence are on translocation bias, then a greater posttranslocation bias should increase apparent KPPi by reducing the fraction of time the pretranslocated state is available to bind PPi. Thus, in this scenario, EC9UG should undergo pyrophosphorolysis if incubated at a high [PPi] and it should exhibit a higher apparent KPPi than EC9GU (e.g., Figure 8A). To investigate this prediction, we sought to measure the PPi-concentration dependence of pyrorophosphorolysis for EC9UG and EC9GU using EcoRNAP and complete nucleic acid scaffolds (Figure 5A and Figure S4). Although we included apyrase in these reactions, we found it was not possible to obtain accurate kinetic data at low or high PPi concentrations. At low PPi concentration, the background rate of intrinsic transcript hydrolysis prevented detection of slow pyrophosphorolysis (Figure S4). At high PPi concentation, the reactions of EC9GU became biphasic (Figure S4). The biphasic kinetics could be explained by structural heterogeneity of the reconstituted ECs or by an inability of apyrase to degrade NTPs rapidly at very low NTP concentration (or the combination of both; Figure S4). We favor the latter view, but were unable to establish it conclusively.
Since complete kinetic profiling of pyrophosphorolysis proved difficult, we instead estimated initial rates of pyrophosphorolysis by fitting the relative [EC9] at different [PPi] using the mechanism 4 in the program KinTek Global Kinetic Explorer (See Materials and Methods). These calculations yielded estimates of initial rates that saturated at ~0.08 min−1 for EC9GU and ~0.003 min−1 for EC9UG (Figures. 8B and 8C). Although these data could suggest that the intrinsic rate of EC9UG is inherently slow even at saturated PPi, such a conclusion is inconsistent with the observation that EC11UG reacts rapidly with even 0.5 mM PPi (>0.04 min−1, Figure 7C, vs. <0.002 min−1 for EC9UG, Table 2). Further, the apparent half-maximal concentration of PPi (KPPiapp) was actually lower for EC9UG (0.1 ± 0.02 mM) than for EC9GU (0.5 ± 0.1 mM) (Figures. 8B and 8C). Taken together, these results are inconsistent with a simple model in which a rapid equilibrium of pre and posttranslocated states is coupled to PPi binding to the pretranslocated EC and in which posttranslocation bias completely explains the pyrophosphorolysis of EC9UG. Such a model (e.g., Figure 7A) predicts a roughly 75-fold greater KPPiapp for EC9UG vs. EC9GU to account for the 30-fold faster observed rate of pyrophosphorolysis at 0.5 mM PPi (Figure 8A).
These results led us to ask if it is possible to explain the observed rates of pyrophosphorolysis for EC9UG and EC9GU if translocation and PPi binding were not ordered events; that is, if PPi could bind to either the pre or posttranslocated EC. Although PPi release has been postulated to cause translocation at least in single-subunit RNAPs (40), we are unaware of data establishing the order of PPi release and translocation for multisubunit RNAPs. To evaluate the consequences of a random order for translocation and PPi binding/release, we considered the hypothetical reaction scheme in which PPi can bind to pre- and posttranslocated ECs (Figure 9A). We considered only the simplest version of such a mechanism in which pre- and posttranslocated ECs bind PPi with equal affinity and in which the translocation equilibrium is unaffected by PPi binding. Even with these constraints, we found that the random-order mechanism yielded predictions that were remarkably similar to our observed data by assuming a 50x pretranslocation bias for EC9GU and a 50x posttranslocation bias for EC9UG (Figures 9B and 9C). We draw no conclusion about the actual translocation equilibria in EC9UG and EC9GU from this prediction, given the arbitrary constraints introduced to simplify modeling and the lack of comprehensive kinetic data. Rather, we conclude that a mechanism involving random order of PPi binding and translocation can, in principle, generate rates of pyrophosphorolysis that match those we observed.
DISCUSSION
Our investigation of the mechanistic basis of translocation bias uncovered basic effects of RNA:DNA hybrid sequence that appear universal among multisubunit RNAPs. As such, they provide insight into how RNAP interacts with the nucleic acid scaffold in an EC that will be generally applicable to understanding the regulation of transcript elongation. Additionally, our findings uncovered preliminary evidence that highlights the need for experiments that establish the order of translocation and PPi release/binding in the nucleotide addition cycle.
RNA 3′ dinucleotide sequence strongly influences translocation bias
Our central findings are that the RNA 3′ dinucleotide sequence has a major effect on bias of ECs between the pre and posttranslocated registers, and that this effect is conserved from bacterial to mammalian RNAPs. This translocation bias was evident in both the initial rate of pyrophosphorolysis and the point at which product inhibition (build up of NTP) caused the EC9 and EC8 species to equilibrate. The rate-limiting step in pyrophosphorolysis appears to be folding of the trigger loop into the trigger helices, as proline substitutions that block folding decrease the rate of pyrophosphorolysis by a factor of ~200 (35). Thus, we also needed to consider the possibility that different 3′ dinucleotides could affect trigger loop folding and give rise to the differences in the rates pyrophosphorolysis rather than being attributable to translocation bias. This concern was heightened by our finding that the rates of pyrophosphorolysis differed between EC9GU and EC9UG even at saturating PPi (Figure 8).
Although we cannot exclude differences in kcat as contributing factors in the observed differences in pyrophosphorolysis rate, three arguments favor a dominant contribution of translocation bias. First, the strong inhibition of pyrophosphorolysis observed in EC9UG was dramatically lessened when the RNA:DNA hybrid was overextended to 11 bp, presumably because the longer hybrid favored the pretranslocated register through steric clash between the upstream end on the hybrid and the wall of the hybrid-binding cleft (Figure 7). Second, an independent measure of translocation bias, exoIII footprinting, also detected a pretranslocation bias of EC9GU vs EC9UG (Figure 5). Third, an effect of RNA sequence on pyrophosphorolysis rate was also observed for the 3′-penultimate position of the RNA (G > A > C > U). It is not obvious how the 3′-penultimate base would directly affect folding of the trigger loop during catalysis (unlike for the 3′ base, which contacts the trigger helices), whereas these effects are easily rationalized by differences in contacts to active-site side chains that could favor or disfavor positioning nucleotide in the i site. It is notable in this regard that the order of effects of the penultimate nucleotide is the reverse of the order of effects of the 3′-nucleotide on pyrophosphorolysis rate (U > C > A > G). This inverse relationship is consistent with a preference for binding of G > A > C > U in the i site and a preference for binding of U > C > A > G in the i+1 site.
Although our results favor a dominant contribution of 3′-dinucleotide sequence to pyrophosphorolysis, we do not mean to suggest that hybrid length makes no contribution. Indeed, we observed a strong effect of increasing pretranslocation bias of lengthening the hybrid past 9 bp. Our results disfavor the idea that an 8-bp hybrid necessarily favors the pretranslocated register (31), but we note that a 8-bp hybrid may not be a state ordinarily observed in ECs. Crystal structures of both yeast and bacterial RNAP in a posttranslocated EC detect 9-bp hybrids (41–43), whereas paused ECs thought to be in the pretranslocated register appear to contain a 10-bp hybrid (25). A pretranslocated yeast RNAPII EC resolved only 9 nt of nascent RNA, leaving the hybrid length uncertain but consistent with 10-bp in the pretranslocated state (44).
We note that assays of translocation bias employed in our study are suboptimal in that they are all indirect. More robust assays based on cleavage of DNA from locally generated free radicals have been used to detect translocation states in T7 RNAP and in reverse transcriptase (38, 45), but have not yet been applied to multisubunit RNAPs. A direct assay of sequence length on translocation bias, for instance using fluorescence quenching (46, 47), would also be desirable. In general, the paucity of good methods to assay translocation register/bias limits our understanding. Although pre- and posttranlocated registers are thought to equilibrate in halted ECs, we lack measurements of the rates of interconversion. Development of such methods for multisubunit RNAPs would significantly advance study of the regulation of transcript elongation.
PPi binding may not be tightly coupled to translocation register
The relationship between PPi release (or PPi binding during pyrophosphorolysis) and translocation is one of the least well understood aspects of the nucleotide addition cycle catalyzed by multisubunit RNAPs. No structure of a multisubunit RNAP with bound PPi has been reported. Our attempts to reconcile pretranslocation bias of RNA 3′-GU relative to 3′UG with the apparently greater KPPi of 3′-GU for initial pyrophosphorolysis rate suggests that another possibility should be considered, specifically that PPi release (or binding) might occur in either a pre or posttranslocated EC. There is no obvious structural impediment to PPi binding to a posttranslocated EC, unlike the obvious steric clash that would result from NTP binding to a pretranslocated EC. Even a highly constrained version of a model that allows random order of translocation vs. PPi binding/release appears able to explain a lack of effect of 3′-dinucleotide sequence on apparent KPPi even while the sequence has a large effect on translocation bias. We suggest that this idea merits further investigation.
Translocation bias may contribute to transcriptional pausing
Transcriptional pausing is thought to arise initially by a structural rearrangement in the active site of a pretranslocated EC (7, 8). Although the sequence and structure contributions to transcriptional pausing are complex and include structures in the exiting RNA, the sequence of the RNA:DNA hybrid, the bases in the active site, and the sequence of the downstream DNA duplex (48–50), the RNA 3′ nucleotide has among the strongest effects. Pausing is favored by an RNA 3′ U or C (48, 51). Thus, our finding that an RNA 3′ U or C exhibits a pretranslocation bias relative to an RNA 3′ A or G suggests a possible mechanistic basis for at least one component of the contribution of the 3′ nucleotide to pause proclivity. By increasing the fraction of time that ECs spend in the pretranslocated register prior to NTP binding, an RNA 3′ U or C may increase the probability that the EC isomerizes to a paused state and conversely decrease the probability of NTP binding. An increased probability of pausing at templates positions that favor the pretranslocated register is distinct from the proposed existence of “pretranslocated pauses” (12). In the mechanism we propose, the pretranslocated register increases the probability of an isomerization to an off-line paused state in a branched mechanism such that a fraction of RNAPs pause. The proposed “pretranslocated pause” is an on-line state in which the energetic barrier to formation of the posttraslocated state is so high that all RNAPs are delayed at the pause position.
The effect of RNA 3′ dinucleotide sequence on translocation bias may synergize with a G at −10 that also could favor the pretranslocated state by inhibiting hybrid melting at the upstream end (25, 52). Thus, an important component of pausing may be a hybrid sequence that favors the pretranslocated state, thereby allowing isomerization into the paused state, and one way that regulators may influence pausing could be to affect translocation bias.
Supplementary Material
Table S1. Oligonucleotides used in this study
Figure S1. RNA 3′ dinucleotide effects on pyrophosphorolysis are evolutionarily conserved
Figure S2. Effect of apyrase on pyrophosphorolysis of EC9UG
Figure S3. Incomplete pyrophosphorolysis caused by NTP accumulation using a complete scaffold
Figure S4. PPi-concentration dependence of pyrophosphorolysis of EcoEC9GU reconstituted on a complete nucleic acid scaffold
Figure S5. Extension of the RNA:DNA hybrid past 9bp shift translocation bias
Acknowledgments
We are grateful to Dr. Yuichiro Takagi for providing Yeast (Saccharomyces cerevisiae) RNA polymerase II. We thank the members of the Landick laboratory for critical reading and comments on the manuscript.
Abbreviations
- RNAP
RNA polymerase
- EC
elongation complex
- nt
nucleotide
- Eco
Escherichia coli
- Tth
Thermus thermophilus
- SceRNAPII
Saccharomyces cerevisiae RNA polymerase II
- BtaRNAPII
calf thymus (Bos taurus) RNA polymerase II
- ExoIII
exonuclease III
- TL
trigger loop
- BH
Bridge helix
- PPi
pyrophosphate
- NTP
nucleoside triphosphate
- Apy
apyrase
- NAC
nucleotide addition cycle
Footnotes
This work was funded by Grant GM38660 from the National Institute of General Medical Sciences.
Supporting Information Available
This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Zhang JaLR. Substrate loading, nucleotide addition, and translocation by RNA Polymerase. In: Buc HaST., editor. RNA Polymerase as Molecular Motors. 1. Royal Society of Chemistry; Cambridge, UK: 2009. pp. 206–235. [Google Scholar]
- 2.Kireeva M, Kashlev M, Burton ZF. Translocation by multi-subunit RNA polymerases. Biochim Biophys Acta. 2010 doi: 10.1016/j.bbagrm.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 3.Sosunov V, Sosunova E, Mustaev A, Bass I, Nikiforov V, Goldfarb A. Unified two-metal mechanism of RNA synthesis and degradation by RNA polymerase. Embo J. 2003;22:2234–2244. doi: 10.1093/emboj/cdg193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guajardo R, Sousa R. A model for the mechanism of polymerase translocation. J Mol Biol. 1997;265:8–19. doi: 10.1006/jmbi.1996.0707. [DOI] [PubMed] [Google Scholar]
- 5.Abbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, Block SM. Direct observation of base-pair stepping by RNA polymerase. Nature. 2005;438:460–465. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bai L, Fulbright RM, Wang MD. Mechanochemical kinetics of transcription elongation. Phys Rev Lett. 2007;98:068103. doi: 10.1103/PhysRevLett.98.068103. [DOI] [PubMed] [Google Scholar]
- 7.Landick R. The regulatory roles and mechanism of transcriptional pausing. Biochem Soc Trans. 2006;34:1062–1066. doi: 10.1042/BST0341062. [DOI] [PubMed] [Google Scholar]
- 8.Landick R. Transcriptional pausing without backtracking. Proc Natl Acad Sci U S A. 2009;106:8797–8798. doi: 10.1073/pnas.0904373106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galburt EA, Grill SW, Wiedmann A, Lubkowska L, Choy J, Nogales E, Kashlev M, Bustamante C. Backtracking determines the force sensitivity of RNAP II in a factor-dependent manner. Nature. 2007;446:820–823. doi: 10.1038/nature05701. [DOI] [PubMed] [Google Scholar]
- 10.Mejia YX, Mao H, Forde NR, Bustamante C. Thermal probing of E. coli RNA polymerase off-pathway mechanisms. J Mol Biol. 2008;382:628–637. doi: 10.1016/j.jmb.2008.06.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Depken M, Galburt EA, Grill SW. The origin of short transcriptional pauses. Biophys J. 2009;96:2189–2193. doi: 10.1016/j.bpj.2008.12.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bai L, Shundrovsky A, Wang MD. Sequence-dependent kinetic model for transcription elongation by RNA polymerase. J Mol Biol. 2004;344:335–349. doi: 10.1016/j.jmb.2004.08.107. [DOI] [PubMed] [Google Scholar]
- 13.Komissarova N, Kashlev M. Transcriptional arrest: Escherichia coli RNA polymerase translocates backward, leaving the 3′ end of the RNA intact and extruded. Proc Natl Acad Sci U S A. 1997;94:1755–1760. doi: 10.1073/pnas.94.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. The RNA:DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell. 1997;89:33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- 15.Larson MH, Greenleaf WJ, Landick R, Block SM. Applied force reveals mechanistic and energetic details of transcription termination. Cell. 2008;132:971–982. doi: 10.1016/j.cell.2008.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santangelo TJ, Roberts JW. Forward translocation is the natural pathway of RNA release at an intrinsic terminator. Mol Cell. 2004;14:117–126. doi: 10.1016/s1097-2765(04)00154-6. [DOI] [PubMed] [Google Scholar]
- 17.Epshtein V, Cardinale CJ, Ruckenstein AE, Borukhov S, Nudler E. An allosteric path to transcription termination. Mol Cell. 2007;28:991–1001. doi: 10.1016/j.molcel.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 18.Epshtein V, Dutta D, Wade J, Nudler E. An allosteric mechanism of Rho-dependent transcription termination. Nature. 2010;463:245–249. doi: 10.1038/nature08669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bar-Nahum G, Epshtein V, Ruckenstein A, Rafikov R, Mustaev A, Nudler E. A ratchet mechanism of transcription elongation and its control. Cell. 2005;120:183–193. doi: 10.1016/j.cell.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 20.Pasman Z, von Hippel PH. Regulation of rho-dependent transcription termination by NusG is specific to the Escherichia coli elongation complex. Biochemistry. 2000;39:5573–5585. doi: 10.1021/bi992658z. [DOI] [PubMed] [Google Scholar]
- 21.Rozovskaya TA, Chenchik AA, Beabealashvilli R. Processive pyrophosphorolysis of RNA by Escherichia coli RNA polymerase. FEBS Lett. 1982;137:100–104. doi: 10.1016/0014-5793(82)80323-2. [DOI] [PubMed] [Google Scholar]
- 22.Toulokhonov I, Zhang J, Palangat M, Landick R. A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Mol Cell. 2007;27:406–419. doi: 10.1016/j.molcel.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 23.Vassylyeva MN, Lee J, Sekine SI, Laptenko O, Kuramitsu S, Shibata T, Inoue Y, Borukhov S, Vassylyev DG, Yokoyama S. Purification, crystallization and initial crystallographic analysis of RNA polymerase holoenzyme from Thermus thermophilus. Acta Crystallogr D Biol Crystallogr. 2002;58:1497–1500. doi: 10.1107/S0907444902011770. [DOI] [PubMed] [Google Scholar]
- 24.Hu X, Malik S, Negroiu CC, Hubbard K, Velalar CN, Hampton B, Grosu D, Catalano J, Roeder RG, Gnatt A. A Mediator-responsive form of metazoan RNA polymerase II. Proc Natl Acad Sci U S A. 2006;103:9506–9511. doi: 10.1073/pnas.0603702103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kyzer S, Ha KS, Landick R, Palangat M. Direct versus limited-step reconstitution reveals key features of an RNA hairpin-stabilized paused transcription complex. J Biol Chem. 2007;282:19020–19028. doi: 10.1074/jbc.M701483200. [DOI] [PubMed] [Google Scholar]
- 26.Maxam AM, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 27.Johnson KA, Simpson ZB, Blom T. Global kinetic explorer: a new computer program for dynamic simulation and fitting of kinetic data. Anal Biochem. 2009;387:20–29. doi: 10.1016/j.ab.2008.12.024. [DOI] [PubMed] [Google Scholar]
- 28.Johnson KA, Simpson ZB, Blom T. FitSpace explorer: an algorithm to evaluate multidimensional parameter space in fitting kinetic data. Anal Biochem. 2009;387:30–41. doi: 10.1016/j.ab.2008.12.025. [DOI] [PubMed] [Google Scholar]
- 29.Foster JE, Holmes SF, Erie DA. Allosteric binding of nucleoside triphosphates to RNA polymerase regulates transcription elongation. Cell. 2001;106:243–252. doi: 10.1016/s0092-8674(01)00420-2. [DOI] [PubMed] [Google Scholar]
- 30.Molnar J, Lorand L. Studies on apyrases. Arch Biochem Biophys. 1961;93:353–363. doi: 10.1016/0003-9861(61)90278-8. [DOI] [PubMed] [Google Scholar]
- 31.Kashkina E, Anikin M, Tahirov TH, Kochetkov SN, Vassylyev DG, Temiakov D. Elongation complexes of Thermus thermophilus RNA polymerase that possess distinct translocation conformations. Nucleic Acids Res. 2006;34:4036–4045. doi: 10.1093/nar/gkl559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vassylyev D, Vassylyeva M, Zhang J, Palangat M, Artsimovitch I, Landick R. Structural basis for substrate loading in bacterial RNA. olymerase. Nature. 2007;448:163–168. doi: 10.1038/nature05931. [DOI] [PubMed] [Google Scholar]
- 33.Landick R, Yanofsky C. Isolation and structural analysis of the Escherichia coli trp leader paused transcription complex. J Mol Biol. 1987;196:363–377. doi: 10.1016/0022-2836(87)90697-8. [DOI] [PubMed] [Google Scholar]
- 34.Kireeva ML, Nedialkov YA, Cremona GH, Purtov YA, Lubkowska L, Malagon F, Burton ZF, Strathern JN, Kashlev M. Transient reversal of RNA polymerase II active site closing controls fidelity of transcription elongation. Mol Cell. 2008;30:557–566. doi: 10.1016/j.molcel.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J, Palangat M, Landick R. Role of the RNA polymerase trigger loop in catalysis and pausing. Nat Struct Mol Biol. 2010;17:99–104. doi: 10.1038/nsmb.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orlova M, Newlands J, Das A, Goldfarb A, Borukhov S. Intrinsic transcript cleavage activity of RNA polymerase. Proc Natl Acad Sci USA. 1995;92:4596–4600. doi: 10.1073/pnas.92.10.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Surratt CK, Milan SC, Chamberlin MJ. Spontaneous cleavage of RNA in ternary complexes of Escherichia coli RNA polymerase and its significance for the mechanism of transcription. Proc Natl Acad Sci USA. 1991;88:7983–7987. doi: 10.1073/pnas.88.18.7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo Q, Sousa R. Translocation by T7 RNA polymerase: a sensitively poised Brownian ratchet. J Mol Biol. 2006;358:241–254. doi: 10.1016/j.jmb.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 39.Marchand B, Tchesnokov EP, Gotte M. The pyrophosphate analogue foscarnet traps the pre-translocational state of HIV-1 reverse transcriptase in a Brownian ratchet model of polymerase translocation. J Biol Chem. 2007;282:3337–3346. doi: 10.1074/jbc.M607710200. [DOI] [PubMed] [Google Scholar]
- 40.Yin YW, Steitz TA. The structural mechanism of translocation and helicase activity in T7 RNA polymerase. Cell. 2004;116:393–404. doi: 10.1016/s0092-8674(04)00120-5. [DOI] [PubMed] [Google Scholar]
- 41.Vassylyev D, Vassylyeva M, Perederina A, Tahirov T, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 42.Westover KD, Bushnell DA, Kornberg RD. Structural basis of transcription: separation of RNA from DNA by RNA polymerase II. Science. 2004;303:1014–1016. doi: 10.1126/science.1090839. [DOI] [PubMed] [Google Scholar]
- 43.Kettenberger H, Armache KJ, Cramer P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol Cell. 2004;16:955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 44.Gnatt A. Elongation by RNA polymerase II: structure-function relationship. Biochim Biophys Acta. 2002;1577:175–190. doi: 10.1016/s0167-4781(02)00451-7. [DOI] [PubMed] [Google Scholar]
- 45.Marchand B, Gotte M. Site-specific footprinting reveals differences in the translocation status of HIV-1 reverse transcriptase. Implications for polymerase translocation and drug resistance. J Biol Chem. 2003;278:35362–35372. doi: 10.1074/jbc.M304262200. [DOI] [PubMed] [Google Scholar]
- 46.Liu C, Martin CT. Fluorescence characterization of the transcription bubble in elongation complexes of T7 RNA polymerase. J Mol Biol. 2001;308:465–475. doi: 10.1006/jmbi.2001.4601. [DOI] [PubMed] [Google Scholar]
- 47.Kashkina E, Anikin M, Brueckner F, Lehmann E, Kochetkov SN, McAllister WT, Cramer P, Temiakov D. Multisubunit RNA polymerases melt only a single DNA base pair downstream of the active site. J Biol Chem. 2007;282:21578–21582. doi: 10.1074/jbc.C700098200. [DOI] [PubMed] [Google Scholar]
- 48.Chan CL, Landick R. Dissection of the his leader pause site by base substitution reveals a multipartite signal that includes a pause RNA hairpin. J Mol Biol. 1993;233:25–42. doi: 10.1006/jmbi.1993.1482. [DOI] [PubMed] [Google Scholar]
- 49.Chan C, Wang D, Landick R. Spacing from the transcript 3′ end determines whether a nascent RNA hairpin interacts with RNA polymerase to prolong pausing or triggers termination. J Mol Biol. 1997;268:54–68. [Google Scholar]
- 50.Kireeva ML, Kashlev M. Mechanism of sequence-specific pausing of bacterial RNA polymerase. Proc Natl Acad Sci U S A. 2009;106:8900–8905. doi: 10.1073/pnas.0900407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aivazashvili VA, Bibilashvili R, Vartikian RM, Kutateladze TA. [Effect of the primary structure of RNA on the pulse character of RNA elongation in vitro by Escherichia coli RNA polymerase: a model] Mol Biol (Mosk) 1981;15:915–929. [PubMed] [Google Scholar]
- 52.Herbert KM, Greenleaf WJ, Block SM. Single-molecule studies of RNA polymerase: motoring along. Annu Rev Biochem. 2008;77:149–176. doi: 10.1146/annurev.biochem.77.073106.100741. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Oligonucleotides used in this study
Figure S1. RNA 3′ dinucleotide effects on pyrophosphorolysis are evolutionarily conserved
Figure S2. Effect of apyrase on pyrophosphorolysis of EC9UG
Figure S3. Incomplete pyrophosphorolysis caused by NTP accumulation using a complete scaffold
Figure S4. PPi-concentration dependence of pyrophosphorolysis of EcoEC9GU reconstituted on a complete nucleic acid scaffold
Figure S5. Extension of the RNA:DNA hybrid past 9bp shift translocation bias