ABSTRACT
Mycoplasma hyopneumoniae causes enormous economic losses to swine production worldwide by colonizing the ciliated epithelium in the porcine respiratory tract, resulting in widespread damage to the mucociliary escalator, prolonged inflammation, reduced weight gain, and secondary infections. Protein Mhp684 (P146) comprises 1,317 amino acids, and while the N-terminal 400 residues display significant sequence identity to the archetype cilium adhesin P97, the remainder of the molecule is novel and displays unusual motifs. Proteome analysis shows that P146 preprotein is endogenously cleaved into three major fragments identified here as P50P146, P40P146, and P85P146 that reside on the cell surface. Liquid chromatography with tandem mass spectrometry (LC-MS/MS) identified a semitryptic peptide that delineated a major cleavage site in Mhp684. Cleavage occurred at the phenylalanine residue within sequence 672ATEF↓QQ677, consistent with a cleavage motif resembling S/T-X-F↓X-D/E recently identified in Mhp683 and other P97/P102 family members. Biotinylated surface proteins recovered by avidin chromatography and separated by two-dimensional gel electrophoresis (2-D GE) showed that more-extensive endoproteolytic cleavage of P146 occurs. Recombinant fragments F1P146-F3P146 that mimic P50P146, P40P146, and P85P146 were constructed and shown to bind porcine epithelial cilia and biotinylated heparin with physiologically relevant affinity. Recombinant versions of F3P146 generated from M. hyopneumoniae strain J and 232 sequences strongly bind porcine plasminogen, and the removal of their respective C-terminal lysine and arginine residues significantly reduces this interaction. These data reveal that P146 is an extensively processed, multifunctional adhesin of M. hyopneumoniae. Extensive cleavage coupled with variable cleavage efficiency provides a mechanism by which M. hyopneumoniae regulates protein topography.
IMPORTANCE
Vaccines used to control Mycoplasma hyopneumoniae infection provide only partial protection. Proteins of the P97/P102 families are highly expressed, functionally redundant molecules that are substrates of endoproteases that generate multifunctional adhesin fragments on the cell surface. We show that P146 displays a chimeric structure consisting of an N terminus, which shares sequence identity with P97, and novel central and C-terminal regions. P146 is endoproteolytically processed at multiple sites, generating at least nine fragments on the surface of M. hyopneumoniae. Dominant cleavage events occurred at S/T-X-F↓X-D/E-like sites generating P50P146, P40P146, and P85P146. Recombinant proteins designed to mimic the major cleavage fragments bind porcine cilia, heparin, and plasminogen. P146 undergoes endoproteolytic processing events at multiple sites and with differential processing efficiency, generating combinatorial diversity on the surface of M. hyopneumoniae.
INTRODUCTION
Genome-reduced pathogens typically display host-restricted behavior but retain the capacity to deploy complex mechanisms to effectively colonize mucosal sites critical for their dissemination to naive hosts. Mycoplasma species have evolved strategies to vary their surface topography to avoid detection and eradication by the host immune response (1). Mycoplasmas are the smallest self-replicating life forms with most genomes ranging in size from 500 to 1,300 kb. Gene families encoding proteins displayed on the surface with reiterated sequences in their promoter or coding regions are highly mutable due to slip strand mispairing and have been reported in a number of Mycoplasma species (2–4). Mutations in these reiterated regions generate phase (on/off) and size variants that contribute to surface diversity and immune evasion. Mycoplasma hyopneumoniae is unusual in this regard with analysis of four complete genome sequences failing to find evidence of reiterated sequences capable of inducing variable expression of most dominant surface proteins (5–7). Despite this, M. hyopneumoniae causes porcine enzootic pneumonia, a chronic geographically widespread respiratory disease that inflicts severe economic losses to pig production (8, 9).
Within the confines of commercial swine-rearing facilities, M. hyopneumoniae enters naive animals via the inhalation of mucorespiratory droplets expelled during bouts of coughing from M. hyopneumoniae-infected swine (10). Upon inhalation, M. hyopneumoniae must overcome the mucociliary escalator and traverse layers of heavily glycosylated mucins produced as decoys for bacterial adhesins that exploit surface glycoconjugates and extracellular matrix components during colonization. M. hyopneumoniae is highly adept at colonizing ciliated respiratory epithelia (11, 12) where it initially causes ciliostasis but eventually destroys cilia and induces epithelial cell death. Heparin effectively blocks the binding of M. hyopneumoniae to porcine cilial preparations (13), indicating that M. hyopneumoniae is reliant on the presentation of heparin-binding proteins on its cell surface. Proteoglycans with highly sulfated glycosaminoglycan (GAG) side chains are prominently displayed on the surface of ciliated epithelium lining the porcine respiratory tract (14). Enzootic pneumonia is a chronic disease, and adaptive immune responses must be evaded for M. hyopneumoniae to survive and proliferate within host tissues. Consequently, the ability to adhere tightly to respiratory cilia is a key strategy employed by pathogens to overcome the mucociliary escalator (11, 13, 15, 16).
Functional redundancy within key surface protein families is identified in multiple pathogens and may be a strategy to avoid immune responses and regulate both adhesion to host tissues and cellular invasion (17, 18). In M. hyopneumoniae, multifunctional adhesins belonging to the P97/P102 families include Mhp182 (P102), Mhp183 (P97), Mhp493 (P159), Mhp494 (P216), Mhp683 (P135), Mhp271, Mhp107, and Mhp108 (P116). These adhesins are prominently expressed during broth culture (19–29), and their expression in vivo is indicated by reverse transcription-PCR (RT-PCR) of M. hyopneumoniae mRNA recovered from the bronchoalveolar lavage (BAL) fluid of experimentally infected swine (30). P97/P102 paralog family members on the cell surface of M. hyopneumoniae are targets of endoproteolytic processing events that generate a substantial combinatorial repertoire of cleavage fragments. Fragments of P97, P216, P135, Mhp271, Mhp107, and Mhp108 have been shown to bind porcine cilia (21–26), underscoring their important role in colonizing the respiratory tracts of swine. Additionally, fragments derived from P97, P159, P216, P135, Mhp271, and Mhp107 are potent adhesins of heparin (19, 21, 22, 24, 26, 31). Interactions between microbial virulence factors and host GAGs underpin a plethora of pathogenic mechanisms including adhesion, invasion, and tissue tropism (18). These findings suggest that endoproteolytic fragments derived from P97/P102 paralog family members profoundly influence how M. hyopneumoniae interacts with its host.
Binding of plasminogen and fibronectin to the surface of M. hyopneumoniae is dose dependent and saturable (24–26), indicating that these two host proteins are also important targets during the infection process. Surface proteins P102 (27), P102 paralog Mhp108 (P116) (25), and P97 paralog Mhp107 (26) are strong plasminogen- and fibronectin-binding proteins, and the P97 paralog Mhp271 also binds fibronectin (24). Plasminogen is freely available at ciliated epithelial sites and in BAL fluid samples recovered from healthy and M. hyopneumoniae-infected swine. However, plasmin activity in BAL fluid samples increase significantly after challenge with M. hyopneumoniae (27). These observations indicate that M. hyopneumoniae engages the plasminogen activation system during colonization of the respiratory tract. Moreover, plasminogen exposed to recombinant P102 is more readily activated to plasmin in the presence of tissue-specific plasminogen activator (tPA). These data suggest that plasminogen-binding members of the P97/P102 paralog families can influence the tertiary structure of plasminogen, making it more susceptible to activation by plasminogen activators (27). Collectively, our studies show that P97/P102 paralog family members display discrete functional domains with multifunctional attributes and are key molecules in the pathogenic arsenal of M. hyopneumoniae.
Endoproteolytic processing is a well-known mechanism used by bacterial pathogens to convert immature preproteins into mature functioning virulence molecules; however, in the M. hyopneumoniae P97/P102 adhesin families, posttranslational cleavage is extensive and unorthodox. Most of the P97/P102 family members previously examined undergo at least one cleavage event and in some cases two or more cleavage events (19–22, 28, 29). The archetype cilium adhesin P97 itself undergoes extensive processing, creating many novel endoproteolytic fragments, but many of these fragments have not been experimentally characterized (20). Cleavage fragments reside on the surfaces of M. hyopneumoniae cells despite the lack of putative transmembrane domains (19–27, 30). Recently, the identification of an endoproteolytic cleavage motif with sequence S/T-X-F↓X-D/E has facilitated the prediction of cleavage sites among members of the P97/P102 families (22); however, the biological rationale for cleavage and the protease(s) responsible are yet to be determined.
The P146 adhesin-like-protein (Mhp684) is a paralog of the P97 cilium adhesin, and its corresponding gene is situated within a two-gene operon with the cilium- and heparin-binding protein P135 (Mhp683) (6). The gene itself is transcribed in vivo (30) and has garnered interest as a target for genotyping M. hyopneumoniae, due to its extensive variability in strains (32). A polyserine repeat (P146R3) is observed to vary considerably among the sequenced strains of M. hyopneumoniae (5–7). In this study, we have characterized binding functions of P146 and determined how it is endoproteolytically processed.
RESULTS
P146 is extensively processed on the surface of M. hyopneumoniae.
In pathogenic strain 232 of M. hyopneumoniae, the p97 (mhp183) paralog p146 (mhp684) encodes a 148.2-kDa protein with a theoretical pI of 8.79. Homologs of p146 have been identified in the genome sequences of M. hyopneumoniae strains J (mhj_0663), 7448 (mhp7448_0663), and 168 (mhp168_676) (5–7). Additionally, p146 homologs have been identified and sequenced in strains F7.2C and BQ14 and partially sequenced in strains PMS and 7422 (32, 33). The six full-length p146 gene sequences share >85% identity. The TMHMM algorithm identified a transmembrane domain (P = 0.999) spanning residues 7 to 29; however, SignalP 4.0 did not conclusively identify the presence of a putative signal peptide (D score of 0.44; signal peptide cutoff of D score of >0.45).
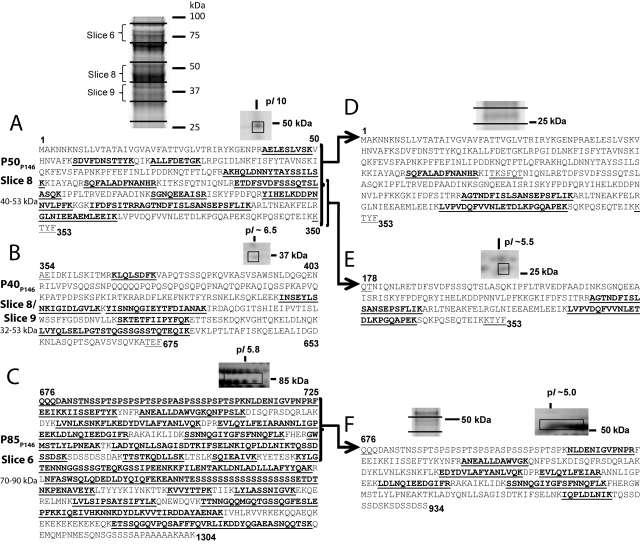
Several members of the P97 and P102 families undergo extensive posttranslational cleavage (19–22, 24, 25, 28, 29). Expression of P146 by M. hyopneumoniae strain J in broth culture was established by using liquid chromatography with tandem mass spectrometry (LC-MS/MS) to identify tryptic peptides derived from proteins separated by SDS-PAGE (Fig. 1). Consistent with previously characterized P97/P102 family members and proteomic studies of M. hyopneumoniae (28, 29), we were unable to identify any peptides unique to P146 near its predicted mass of 148.2 kDa. Tryptic peptides spanning P146 mapped to three endoproteolytic fragments, indicating that P146 is cleaved twice. A protein that migrated with mass between 40 and 53 kDa (slice 8) mapped to the N-terminal region of P146 (Fig. 1A) (P50P146). Peptides matching the central region of the P146 sequence were identified in slices 8 (40 to 53 kDa) and 9 (39 to 42 kDa) (Fig. 1B) (P40P146), and peptides matching the C-terminal region were identified in slice 6 (70 to 90 kDa) (Fig. 1C) (P85P146). Recently, we proposed a cleavage motif with sequence S/T-X-F↓X-D/E that can be used to predict cleavage sites in the P97 and P102 paralog families (22). Two sequences that closely resemble this motif were identified within the cleavage regions defined by mass spectrometry. The first of these sequences, TYFAE, resides between amino acids 315 and 366 that separate matched peptides defining P50P146 and P40P146. However, we have been unable to experimentally confirm cleavage at the phenylalanine residue in TYFAE. The second motif, with sequence TEFQQ, resides between residues 630 and 675 that separate matched peptides that define P40P146 and P85P146. Analysis of spectra derived from P85P146 revealed a semitryptic peptide 676QQQDANSTNSSPTSPSPSPTSPSPASPSSSPSPTSPK712 that defines the N terminus of P85P146 (see Fig. S1 in the supplemental material) and confirms the TEF↓QQ cleavage site.
FIG 1 .
Mass spectrometry analysis of P146. M. hyopneumoniae strain J proteins were examined using the following two methods. In method 1, lysates were subjected to 1-D SDS-PAGE, the gels were sectioned into pieces representing 14 molecular mass ranges, and the proteins in each gel slice were examined using LC-MS/MS. In method 2, surface proteins were labeled by surface biotinylation and enriched by affinity chromatography of the cell lysate. Purified biotinylated proteins were separated by either 1-D SDS-PAGE or 2-D GE and identified by LC-MS/MS. Peptides identified by LC-MS/MS are shown in bold type and underlined. The numbers above or below the sequence refer to amino acid positions. (A to C) Peptides unique to three regions of the P146 sequence were identified in both experiments. Terminal sequences matching an M. hyopneumoniae cleavage motif defined previously are underlined but not bold. (A) Fragment P50P146 and peptides unique to the N-terminal region of P146. (B) Fragment P40P146 and peptides unique to the central region of P146. (C) Fragment P85P146 and peptides unique to the C-terminal region of P146. The N terminus of P85P146 was identified from a semitryptic peptide. (D to F) Additional proteins were identified by method 2 at masses significantly lower than those identified by method 1. Terminal sequences matching or almost matching an M. hyopneumoniae cleavage motif defined previously (22) are underlined but not bold. (D) Peptides unique to P50P146 were identified in a 1-D SDS-polyacrylamide gel slice containing proteins migrating at ~25 kDa. (E) Peptides unique to one-half of P50P146 identified in a 2-D GE protein spot migrating at 25 kDa and pI of ~5.5. (F) Peptides unique to P85P146 identified in 2-D GE protein spots migrating at 50 kDa and pI of ~5.0.
To determine whether fragments of P146 reside on the surface of the bacterium, we biotinylated intact M. hyopneumoniae cells and enriched biotin-conjugated proteins from whole-cell extracts using a monomeric avidin column. Biotinylated surface proteins were then separated using both 1- and 2-D GE. Peptides unique to P50P146, P40P146, and P85P146 were identified from 2-D GE at molecular masses similar to those from previous 1-D SDS-PAGE with LC-MS/MS experiments (Fig. 1A to C). However, peptides unique to P146 fragments were also identified at masses significantly lower than those identified previously. Tryptic peptides mapping to P50P146 were identified from proteins with masses of ~25 kDa (Fig. 1D), indicating that a second, less efficient cleavage event may occur to separate the P50P146 fragment into two halves. Confirming this, peptides unique to the second half of P50P146 were identified from 2-D GE of biotinylated surface proteins at ~25 kDa (Fig. 1E). A peptide sequence, TKSFQT, similar to but not matching the S/T-X-F↓X-D/E cleavage motif, is found within the P50P146 fragment, indicating a possible cleavage site. Peptides unique to the P85P146 fragment were also identified from 2-D GE of biotinylated surface proteins in spots at ~50 kDa (Fig. 1F), which indicates further cleavage of these fragments.
While surface exposure of P146 endoproteolytic fragments was observed in biotinylation experiments, we also performed whole-cell digestion experiments with trypsin. Antisera generated against recombinant polyhistidine-tagged proteins F1P146, F2bP146, and F3P146, which recognize P50P146, P40P146, and P85P146, respectively (Fig. 2), were used to probe blots containing lysates of M. hyopneumoniae strain J cells exposed to different concentrations of trypsin for 15 min. The blots show that all P146 fragments are degraded by trypsin, confirming their surface location (see Fig. S2A in the supplemental material). In all immunoblots, trypsin digestion products are observed in concentrations as low as 0.5 µg ml−1, and major cleavage products are almost completely digested at a concentration of 50 µg ml−1. Immunoblots containing identical lysates probed with antisera raised against the ribosomal protein L7/L12 showed that this protein was still detected at a trypsin concentration of 300 µg ml−1. Consistent with previous experiments (19, 21, 22, 25–27), these data indicate that the cell membrane remained intact. We also performed mass spectrometry on peptides released from the surface of M. hyopneumoniae after trypsin digestion. Multiple peptides unique to all major cleavage fragments of P146 were identified by LC-MS/MS (see Fig. S2B in the supplemental material). Live/dead staining and flow cytometry of M. hyopneumoniae cells confirmed the absence of excessive cell lysis during trypsin and surface biotinylation experiments (data not shown).
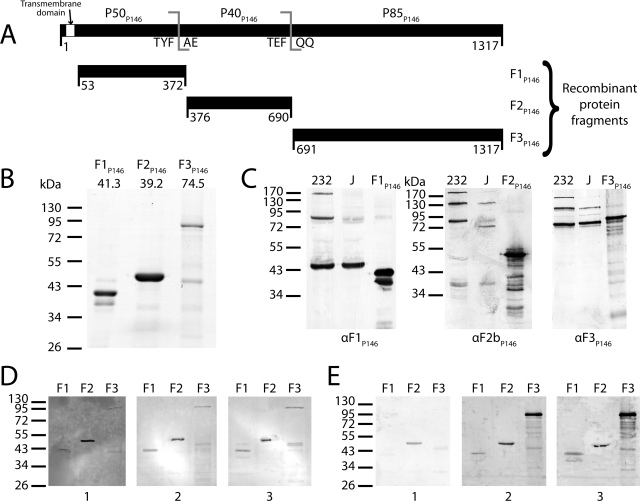
FIG 2 .
Cloning of p146 and immunoblot analysis of its cleavage fragments. (A) p146 (3954 bp encoding 1317 residues) was cloned from M. hyopneumoniae strain 232 homolog mhp684 in three fragments (F1P146/232 to F3P146/232) closely matching native cleavage as observed by mass spectrometry. A hydrophobic transmembrane domain predicted by TMHMM (53 residues) was removed from the N terminus of F1P146/232. All in-frame TGA codons were mutated to TGG using overlap extension PCR. (B) Coomassie blue-stained SDS-PAGE of the three P146 recombinant proteins. The predicted mass (in kilodaltons) is shown above each gel lane. Recombinant protein F2P146/232 runs at a higher molecular mass than predicted. (C) Immunoblots of recombinant proteins (F1P146/232 to F3P146/232) and whole-cell lysates from M. hyopneumoniae strains 232 and J, probed with anti-F1P146 (αF1P146) and anti-F3P146 (αF3P146) sera. Sera labeled αF1P146 and αF3P146 were extracted from New Zealand White rabbits challenged with recombinants F1P146/232 and F3P146/232; anti-F2bP146 (αF2bP146) serum was generated in the same manner from a recombinant protein representing residues 586 to 690 of P146. Minor bands present in recombinant protein lanes (F1P146/232 to F3P146/232) may be either products of recombinant protein degradation or truncated proteins resulting from mRNA instability. (D) Immunoblots of recombinant proteins (F1P146/232 to F3P146/232) probed with porcine serum sourced from a high-health-status herd (blot 1) and sera from convalescent pigs (blots 2 and 3). (E) Immunoblots of recombinant proteins (F1P146/232 to F3P146/232) probed with sera obtained from a single pig prior to treatment (blot 1), after vaccination with the commercial bacterin vaccine Suvaxyn (blot 2), or 6 weeks after challenge with M. hyopneumoniae strain Hillcrest (blot 3).
Antibodies from vaccinated, challenged, and convalescent pigs recognize P146.
To determine whether P146 sequences were recognized by the porcine humoral immune response, sera from pigs recovering from porcine enzootic pneumonia (convalescent-phase sera) were used to probe immunoblots of P146 recombinant proteins F1P146, F2P146, and F3P146. The results were compared with results for serum from a pig from a healthy herd (high-health-status sera) (Fig. 2D). Immunoblots probed with high-health-status sera (negative control) showed a strong reaction to F2P146 and weak reactions to F1P146 and F3P146. Immunoblots probed with sera from two separate convalescent pigs both showed increased reactions to all three recombinant proteins compared to the high-health-status sera. In addition to convalescent porcine sera, we also examined serum from a single pig vaccinated with the commercial bacterin vaccine Suvaxyn and serum from the same pig after subsequent challenge with a pathogenic strain of M. hyopneumoniae (Fig. 2E). Immunoblots probed with sera collected prior to vaccination again showed a strong reaction to F2P146 but no significant reactions to F1P146 and F3P146. All three recombinant proteins showed strong reactions when probed with sera collected after treatment with Suvaxyn and 6 weeks after challenge with virulent M. hyopneumoniae strain Hillcrest.
P146 is processed variably in M. hyopneumoniae field isolates.
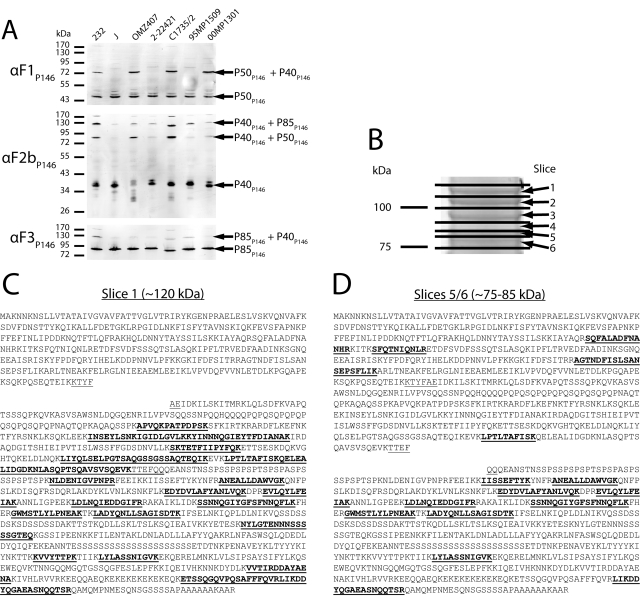
To determine whether the cleavage observed in P146 was consistent across multiple M. hyopneumoniae strains, cell lysates of laboratory strains and field isolates from varied geographical locations were analyzed by immunoblotting with anti-F1P146, anti-F2bP146, and anti-F3P146 sera. Processing of the P85P146, P50P146, and P40P146 fragments varied among strains (Fig. 3A). In strain J and field isolates 2-22421 and 95MP1509, strong bands at 40 kDa and 50 kDa were dominant when the blots were probed with anti-F1P146 and anti-F2bP146 sera, respectively. However, in strain 232 and field isolates OMZ407, C1735/2, and 00MP1301, dominant bands were also observed at ~80 and ~125 kDa with these antisera, indicating that cleavage of P146 may be less efficient in these strains. An ~80-kDa band that appears in immunoblots with anti-F1P146 and anti-F2bP146 sera suggests inefficient cleavage between P50P146 and P40P146, whereas a ~125-kDa band that appears in immunoblots with anti-F2bP146 and anti-F3P146 sera suggests inefficient cleavage between P40P146 and P85P146. To confirm this, we performed SDS-PAGE on a cell lysate of M. hyopneumoniae strain 232, sectioned the gel with molecular mass range from 75 to 125 kDa into 6 slices, digested them with trypsin, and examined the tryptic peptides eluted from each slice using LC-MS/MS (Fig. 3B). In gel slice 1 (~110 to 125 kDa), peptides unique to both P40P146 and P85P146 were identified, indicating that an uncleaved fragment made of P40P146 and P85P146 was present (Fig. 3C). In gel slices 5 and 6 (~75 to 85 kDa), peptides unique to P50P146, P40P146, and P85P146 were identified, indicating that an uncleaved fragment made of P50P146 and P40P146 was present migrating at a molecular mass similar to that of fragment P85P146 (Fig. 3D). In gel slice 4 (~85 to 90 kDa), only peptides unique to fragment P85P146 were detected. Analysis of M. hyopneumoniae genome sequences reveals that both the TEFQQ and putative TYFAE motifs are present and unchanged in both strains J and 232 (data not shown).
FIG 3 .
P146 is processed variably in M. hyopneumoniae laboratory strains and field isolates. (A) Immunoblots of whole-cell lysates of different M. hyopneumoniae strains and field isolates. The blots were separately probed with anti-F1P146 (αF1P146), anti-F2bP146 (αF2bP146), and anti-F3P146 (αF3P146) sera in order to assess the consistency of protein expression and processing. Cleavage fragments equivalent to P50P146, P40P146, and P85P146 are conserved across all isolates in this study, but some also show distinct bands at higher masses that correspond to fragments that would result from inefficient cleavage. (B) 1-D SDS-polyacrylamide gel (molecular mass region ~75 to 125 kDa) of an M. hyopneumoniae strain 232 cell lysate was sectioned into 6 slices, and each slice was examined using LC-MS/MS. (C) Slice 1 (~110 to 125 kDa) was found to contain peptides unique to regions of P146 corresponding to P40P146 and P85P146 fragments and matched a band found at a similar mass in strain 232 immunoblots. (D) Slices 5 and 6 (~75 to 85 kDa) were found to contain peptides unique to all major fragments of P146, suggesting that a cleavage event separated the protein into two separate ~80-kDa fragments.
Molecular analysis of P146.
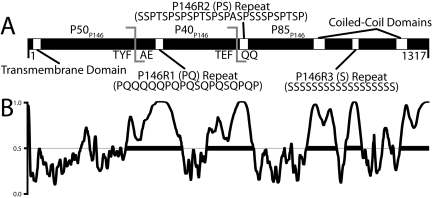
P146 contains many striking sequence features, including three variable repeat regions, a KEKE repeat motif, and in strain J, a C terminus with the sequence SSSSAPAAAAAKAAK (Fig. 4A). Three repeat regions identified previously include a proline-glutamine repeat (P146R1) spanning residues 415 to 444 of the strain 232 sequence, a proline-serine repeat (P146R2) spanning residues 696 to 722, and a serine repeat (P146R3) spanning residues 1065 to 1085 (33). All three regions vary in length in strains (33). The serine repeat varies from 44 consecutive residues in strain 7448, 19 in strain J, and 21 residues in strain 232 (6, 7).
FIG 4 .
In silico analysis of P146. (A) Sequence features of P146. The TMHMM algorithm predicts that P146 contains a transmembrane domain (P = 0.999). P146 contains three defined repeat regions: P146R1 (PQ), P146R2 (PS), and P146R3 (S). Multiple coiled-coil prediction algorithms identified two putative coiled-coils flanking the P146R3 repeat region in P85P146. The experimentally determined cleavage site (ATEFQQ) and predicted site (KTYFAE) are also shown. (B) PONDR VSL1 analysis of P146 232 homolog (Mhp684). The regions above the line at 0.5 denote regions predicted to be structurally disordered within P146. The thick black bars denote disordered regions spanning 40 or more amino acids. P146 is predicted to contain five regions of significant disorder, two corresponding with the experimental and predicted cleavage sites. The remaining disordered regions correspond to putative coiled-coil domains, the C-terminal region, and the P146R3 repeat region.
P146 shares significant sequence identity with P97 family members of the M. hyopneumoniae genome, but this is essentially restricted to a region spanning ~400 amino acids from the N terminus. The remainder of the P146 protein is largely unique (6). Outside of the M. hyopneumoniae genome, BLASTP analysis determines that P146 shares the most similarity with hypothetical proteins of Mycoplasma conjunctivae: MCJ_004230, MCJ_004150, MCJ_005100, MCJ_003150, and the LppS adhesin; this is unsurprising considering previously reported similarity between adhesins of these Mycoplasma species (34). Analysis of P146 with coiled-coil prediction algorithms reveals two putative coiled-coil regions spanning residues 939 to 975 (coil 1) and 1210 to 1245 (coil 2) of the strain 232 sequence. The Paircoil2 (35) (coil 1 P score = 0.0078, coil 2 P score = 0.0155; coiled-coil predicted at a P score of <0.025) and COILS algorithms (36) (coil 1 P score = 0.802, coil 2 P score = 1.000) both indicated that coil 1 and coil 2 regions form coiled-coil structures. The coil 2 region corresponds to a KEKE motif, and interestingly, a putative coiled-coil linked to a KEKE motif has been observed previously in the P146 operon partner Mhp683 (22).
Previously we have shown that dominant cleavage sites in P97 and P102 paralogs were located within regions of predicted protein disorder (22). Five regions of significant structural disorder were predicted in P146 using the VSL1 algorithm (37) spanning residues 324 to 508, 589 to 742, 917 to 1016, 1045 to 1098, and 1224 to 1317 (Fig. 4B). Cleavage motifs TYFAE (P50P146/P40P146) and TEFQQ (P40P146/P85P146) reside within two predicted disordered regions spanning amino acids 324 to 508 and 589 to 742. Interestingly, most of P40P146 is predicted to be disordered, and this fragment is clearly the most unstable compared to P50P146 and P85P146. This prediction suggests that P40P146 displays greater accessibility to endoproteolytic cleavage.
P146 fragments bind heparin.
Members of the P97/P102 families have a propensity to bind sulfated GAGs (19, 21, 22, 24, 26, 31). Recombinant fragments F1P146/232 and F3P146/232 bound heparin in a dose-dependent and saturable manner with physiologically relevant affinity constants, as assessed in microtiter plate-based enzyme-linked immunosorbent assays (ELISAs) (see Fig. S3A in the supplemental material). F2P146/232 did not significantly bind heparin in microtiter plate assays (data not shown). F1P146/232 and F3P146/232 binding to heparin was largely specific, as the interactions were almost completely (F1P146/232) or mostly (F3P146/232) inhibited by the addition of a 50× excess unlabeled heparin. Affinity constants (Kd) of 53 ± 10 nM and 520 ± 80 nM were calculated for F1P146/232 and F3P146/232, respectively. Preincubation of biotinylated heparin with unlabeled heparin or fucoidan inhibited binding of F1P146/232 and F3P146/232, while preincubation with porcine mucin or chondroitin sulfate B had limited or no significant effect (see Fig. S3B in the supplemental material).
F3P146 binds plasminogen.
In M. hyopneumoniae and other bacterial pathogens, surface proteins possessing a C-terminal lysine residue have been correlated with the ability to bind the serine protease precursor plasminogen (25–27, 38). P146 homologs in strains J, F7.2C, and BQ14 possess a C-terminal lysine residue (-SSSSAPAAAAAKAAK); however, in strains 232, 7448, and 168, the terminating residue is an arginine (-SSSSAPAAAAAAKAAR). To determine whether this sequence variability influences interactions with plasminogen, we cloned F3P146 from both strains 232 and J and examined their binding to plasminogen using dot blotting and surface plasmon resonance (SPR). In dot blotting experiments, the J and 232 versions (F3P146/J and F3P146/232) bound biotinylated plasminogen similarly (see Fig. S4A and S4B in the supplemental material). SPR experiments indicated that both also bound plasminogen in a dose-dependent manner with Kd = 39 ± 3 nM and association rate (ka) = (4.6 ± 0.5) × 104 M−1 s−1 for F3P146/J and Kd = 46 ± 6 nM and ka = (1.7 ± 0.5) × 105 M−1 s−1 for F3P146/232 (see Fig. S4C and S4D in the supplemental material). This suggests that whether the C-terminal basic residue is arginine or lysine has little effect on binding affinity; it does modestly affect the kinetics of interaction, with recognition of arginine being ~4-fold more efficient than lysine.
To examine this further, we cloned strain J and 232 versions of F3P146 without their respective C-terminal residues (F3P146/JΔK and F3P146/232ΔK); these two proteins have near identical C-terminal sequences. Both displayed 5-fold-reduced affinity for plasminogen and as expected showed identical binding affinity and kinetics; Kd = 210 ± 33 nM and ka = (1.3 ± 0.2) × 104 M−1 s−1 for F3P146/JΔK and Kd of 220 ± 33 nM and ka = (1.3 ± 0.2) × 104 M−1 s−1 for F3P146/232ΔR (see Fig. S4E and S4F in the supplemental material). These data clearly quantify the significance of having a C-terminal basic residue (a 5-fold contribution to affinity) and whether it is a lysine or an arginine (an effect on binding kinetics).
P146 fragments bind porcine cilia.
A microtiter plate-based assay used previously to identify cilium-binding proteins showed that P146 recombinant proteins F1P146/232-F3P146/232 reproducibly bound cilia (see Fig. S5 in the supplemental material). The recombinant protein F4Mhp385, derived from the C terminus of Mhp385 and previously observed to display low cilium-binding properties (39) did not bind porcine cilia and was included as a negative binding control in these studies. F2P97, a recombinant protein that carries the R1 cilium-binding domain of cilium adhesin P97, was used as a positive control and bound to porcine cilia as expected.
DISCUSSION
Members of the P97/P102 paralog families are secreted to the surface of M. hyopneumoniae where they play key roles in interactions with host cells, extracellular matrix components, and circulating host proteins (19, 21–27, 30). There are three overarching features that define members of these two protein families. The first feature is the presence of a single putative transmembrane domain at the N terminus that is not removed by signal peptidase 1 (19, 20, 22). Second, these molecules undergo endoproteolytic processing whereby N-terminal cleavage fragments presumably remain tethered to the cell membrane via the N-terminal transmembrane domain while central and C-terminal endoproteolytic fragments are released. Despite the absence of transmembrane domains distant from the N terminus, central and C-terminal cleavage fragments remain attached to the cell surface of M. hyopneumoniae, presumably by protein-protein interactions that are poorly understood. Third, P97 and P102 paralogs are multifunctional proteins that bind host cell-associated and circulatory molecules and present a variety of structural motifs with largely unknown functions (19–22, 24–27).
Here we show that P146, a paralog of P97, contains several structurally distinctive motifs including a polyserine repeat of unknown function and a serine/alanine-rich C-terminal motif [SSSSAAAAAKAA(K/R)] whose terminal K or R (strain-dependent) residue plays an important role in binding plasminogen (see Fig. S4 in the supplemental material). We also identified two putative coiled-coil regions, one of which carries a KEKE motif, and a reiterated PQ motif, all of unknown function. Interestingly, these features are localized to the C-terminal 85 kDa of P146 (P85P146), while sequence similarity with P97 is restricted to the N-terminal 400 amino acids. Recombinant fragments spanning different regions of P146 identified at least three cilium-binding domains located in F1P146/232, F2P146/232, and F3P146/232, while GAG binding localized to F1P146/232 and F3P146/232. Residual high-affinity plasminogen binding retained after removal of C-terminal K/R residues in homologs from strains J and 232 indicate that additional plasminogen-binding sites are present. These data show that P146 is a multifunctional protein that binds porcine respiratory cilia, various GAGs, and plasminogen.
We have previously established that plasminogen is freely available in the bronchoalveolar regions of the porcine lung (27). M. hyopneumoniae displays surface receptors that bind plasminogen in a process that facilitates conversion to plasmin by mammalian plasminogen activators (e.g., tPA). Importantly, plasmin activity is significantly increased in the BAL fluid of pigs following infection with M. hyopneumoniae (27). Consequently, M. hyopneumoniae colonizing the ciliated epithelia is likely to sequester plasminogen on its cell surface and facilitate its conversion to plasmin. This process is likely to have ramifications for tissue invasion and systemic infection. To date, including P146, we have characterized four members of the P97 and P102 families as plasminogen-binding proteins, three of which contain C-terminal lysine residues (25–27). P146 is particularly interesting due to the variation of the C-terminal residue; the C-terminal residue was arginine in strains 232, 7448, and 168 and lysine in strains J, F7.2C, and BQ14. Lysine and arginine are both positively charged at physiological pH. We have shown that both strain J and 232 versions of F3P146 bind plasminogen using dot blotting and SPR. While the binding of plasminogen by proteins with C-terminal lysines is well established, binding of plasminogen by proteins with a C-terminal arginine residue is unusual. However, the Streptococcus pyogenes Prp protein has been shown to bind plasminogen via an internal arginine residue (40), and certain arginyl residues have been shown to bind human plasminogen kringle 4 (41). Thrombin-cleaved prourokinase has also been shown to bind human plasminogen via a C-terminal arginine present at the thrombin cleavage site (42).
F3P146/232 and F3P146/J affinity for plasminogen is reduced 5-fold upon removal of the C-terminal Lys/Arg but is not eliminated, indicating the presence of other binding motifs within the F3P146 sequences. Both F3P146/232 and F3P146/J are rich in internal lysine residues (66 and 68 lysines, respectively). Additionally, all known homologs of P146 possess a second lysine, 4 or 5 residues from the C terminus within a string of 7 or 8 alanine residues depending upon the strain. This near-C-terminal lysine or other internal lysines may be responsible for residual plasminogen binding observed in the F3P146/JΔK and F3P146/232ΔR mutants. Plasminogen binding by enolase from group A streptococci has previously been probed by mutation of C-terminal and internal lysines (43). Removal or replacement of C-terminal lysines with retention of internal lysines resulted in a significant reduction in the extent of binding (43). However, removal of some internal lysines also resulted in a significant reduction in the extent of binding, indicating that internal lysines also play an import role in plasminogen binding by enolase (43). Removal of internal lysines by site-directed mutagenesis and subsequent binding assays may also determine whether these residues have a role in F3P146 adherence to plasminogen.
LC-MS/MS of proteins resolved by 1- and 2-D gel electrophoresis identified endoproteolytic cleavage fragments representing P50P146, P40P146, and P85P146. Two sequences that conform to the S/T-X-F↓X-D/E cleavage motif described recently (22) were identified in regions spanning tryptic peptides that define these cleavage fragments. We confirmed the identity of one cleavage site by LC-MS/MS analysis of the semitryptic peptide 676QQQDANSTNSSPTSPSPSPTSPSPASPSSSPSPTSPK712 which defines the N terminus of the C-terminal cleavage fragment P85P146. The cleavage motif 673TEFQQ677 displays a S/T residue in the −3 position, and cleavage occurs at the Phe residue residing at position −1. Although no D/E is present in the +2 position, these features are in accord with cleavage sites in other members of the P97 and P102 paralogs (22). We were unable to experimentally confirm the putative cleavage site 351TYFAE355 predicted to generate P50P146 and P40P146.
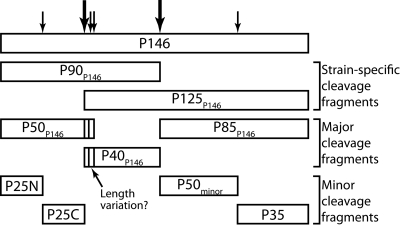
Immunoblots of cell lysates of M. hyopneumoniae strains from different geographic locations probed separately with anti-F1P146 and anti-F3P146 sera showed that cleavage in strain 232 and field isolates OMZ407, C1735/2, and 00MP1301 is less efficient than in strain J and field isolates 2-22421 and 95MP1509. We have previously indicated that P97 (Mhp183) (20) and P159 (Mhp494) (19) undergo strain-specific processing events. However, the molecular identities of the strain-specific products were not identified. For the first time, we show here that strains of M. hyopneumoniae vary in their ability to efficiently cleave P146. We identified large endoproteolytic products comprising a C-terminal 120-kDa fragment (P85P146 plus P40P146), derived from an inefficient cleavage event at the 673TEFQQ677 site and an N-terminal P90P146 fragment (P50P146 plus P40P146), derived from inefficient processing at the putative 351TYFAE355 cleavage site (Fig. 5). P120P146 and P90P146 sequences were identified from high-molecular-mass regions of cell lysates of strain 232 by LC-MS/MS (Fig. 1B to D) and by probing cell lysates with anti-F1P146 and anti-F3P146 sera (Fig. 3A). We also identified minor endoproteolytic cleavage fragments with masses smaller than the dominant cleavage species P50P146, P40P146, and P85P146 by enriching biotinylated proteins with avidin chromatography (Fig. 5). LC-MS/MS of biotinylated proteins resolved by 2-D gel electrophoresis confirmed that P50P146 (predicted pI of 9.13) is cleaved to generate two fragments ~25 kDa in size. The C-terminal half of P50P146 is enriched in acidic amino acids (D or E residues). LC-MS/MS of a protein spot that migrated with a mass of ~25 kDa and pI of 5.5 confirmed that the protein represented the C terminus of P50P146. We have not determined the precise site of cleavage in P50P146. Minor fragments of P50P146 and P40P146 with different molecular masses were detected with anti-F1P146 and anti-F2bP146 sera, respectively (Fig. 3). This may indicate variation at the P50P146 C terminus and P40P146 N terminus caused by minor cleavage events (Fig. 5). LC-MS/MS identified a 50-kDa N-terminal fragment of P85P146 with a pI of ~5.0. It is interesting that the minor cleavage fragments identified by LC-MS/MS display predicted pI values that are considerably more acidic than those of P50P146 (pI = 9.13), P40P146 (9.33), and P85P146 (7.07), and of intact P146 (8.90). These data suggest that products of these minor cleavage events are not random breakdown products but represent domains enriched in acidic amino acids embedded within larger preproteins with strongly basic pIs. Furthermore, as noted for P159 (Mhp494) and P216 (Mhp493), regions enriched in acidic amino acid residues are a feature of members of the P97 and P102 paralog families (19, 21) and underscore their modular structure.
FIG 5 .
Summary of P146 cleavage. Cleavage fragments can be split into three groups as shown. Strain-specific cleavage fragments are prevalent fragments in selected strains (e.g., strain 232). Major cleavage fragments P50P146, P40P146, and P85P146 are abundant in all strains. Minor cleavage fragments are present in small amounts and are detected only by enrichment of surface proteins.
In total, we identified nine proteolytic cleavage fragments representing different regions of P146. These fragments are sensitive to digestion by trypsin, and many were recovered by avidin chromatography after the surfaces of M. hyopneumoniae cells were labeled with biotin. These data show that complex endoproteolytic processing events occur either on the surface of M. hyopneumoniae or prior to transport across the cell membrane. Processing is modulated by the efficiency of cleavage at dominant cleavage sites because different strains present different cleavage fragments on their cell surface. The mechanism by which M. hyopneumoniae regulates endoproteolytic cleavage remains unknown. Nonetheless, we have provided further evidence that cleavage often occurs at sites that conform to the S/T-X-F↓X-D/E cleavage motif described recently (22). P97 is also subject to extensive endoproteolysis, but so far the events that precipitate cleavage and the precise composition of many of the endoproteolytic fragments have not been experimentally determined (20). Endoproteolytic processing occurs with all members of the P97 and P102 paralog families, and in at least two cases (P97 and P146), processing is more extensive than originally envisaged. Endoproteolytic processing significantly increases surface protein diversity by altering the presentation of functional domains on the cell surface and may be a mechanism used to regulate adhesion to host tissues, potentiate host cell invasion, and avoid immune clearance.
MATERIALS AND METHODS
Porcine sera and Mycoplasma hyopneumoniae strains and culture.
Convalescent porcine sera were obtained from finisher pigs from a herd infected with M. hyopneumoniae in an area where M. hyopneumoniae is endemic (Joadja, Australia). High-health-status serum was obtained from a high-health-status herd and tested negative for M. hyopneumoniae infection using a commercial ELISA kit. Pretreatment (negative-control) serum was obtained from a 9-week-old pig that tested negative to M. hyopneumoniae by quantitative PCR (qPCR) (44). The pig was subsequently vaccinated at 9, 12, and 15 weeks with Suvaxyn. Postvaccination serum was collected 15 weeks after the third vaccination. The pig was challenged with M. hyopneumoniae strain Hillcrest at week 17 and postchallenge serum was collected at week 23. The sources and conditions used to culture and harvest M. hyopneumoniae strains J, 232, and Hillcrest and field isolates OMZ407, 00MP1301, 2-2241, C1735-2, and 95MP1509 are as previously described (22, 24, 45).
Proteomics and surface localization.
The materials and methods used for 1- and 2-D GE, immunoblotting, trypsin digestion, Zip-Tip cleanup, reduction, and alkylation have been previously described (20, 24, 46–48) with the following adjustments. For 2-D GE, first-dimension immobilized pH gradient (IPG) strips (ReadyStrip IPG strips, 110 mm long, pH 3 to 10; Bio-Rad) were used. The strips were rehydrated overnight with 300 µg of biotinylated surface protein in 210 µl of solubilization buffer [7 M urea, 2 M thiourea, 1% (wt/vol) 3-(4-heptyl)phenyl-3-hydroxypropyl)dimethylammoniopropanesulfonate] and overlaid with paraffin oil. M. hyopneumoniae proteins were reduced prior to 1-D SDS-PAGE for immunoblotting and reduced and alkylated with iodoacetamide prior to 1-D SDS-PAGE for LC-MS/MS. Gel slices prepared from 1-D SDS-PAGE for LC-MS/MS analysis were processed as previously described (24). LC-MS/MS analysis of 1- and 2-D GE excisions were done as previously described (22, 49), as were graded trypsin digestions of intact M. hyopneumoniae cells (22) and trypsin digestion experiments for mass spectrometry and biotinylation of the M. hyopneumoniae surface (22). Cell integrity for trypsin and biotinylation experiments was evaluated by flow cytometry with a BDTM LSR II (BD Biosciences, NJ) and YO-PRO-1 iodide dead cell dye (Invitrogen, CA) with data analyzed using BD FACSDiva software (BD Biosciences). Bioinformatic analysis of P146 used online resources: BLASTP (50), ProtParam (51), COILS (36), Paircoil2 (35), TMHMM (52), SignalP (53), and PONDR VSL1 (37).
Expression of recombinant proteins and creation of polyclonal antisera.
Cloning of p146 was performed by using three fragments of different lengths (Fig. 2A) and based on the M. hyopneumoniae strain 232 genome sequence (6); fragments were constructed to mimic the P146 cleavage pattern as determined by preliminary mass spectrometry experiments. The fragments were denoted F1P146/232, F2P146/232, and F3P146/232 and ranged from amino acids 53 to 372, 376 to 690, and 691 to 1317, respectively. F3P146/J and F3P146/JΔK were generated from M. hyopneumoniae strain J genome sequence (7). In-frame TGA codons were substituted by TGG and C-terminal residues (F3P146/JΔK and F3P146/232ΔR) were removed by use of overlap extension PCR or site-directed mutagenesis using mutagenic primers (see Table S1 in the supplemental material). The fragments were amplified by PCR from M. hyopneumoniae strain 232 chromosomal DNA using Pfu polymerase (Aglient Technologies, CA) and cloned into the pET100/GW/D-TOPO vector (Invitrogen) as described previously (24). Protein expression was performed using Escherichia coli BL21Star (DE3) as previously described (27) with the following alterations: protein expression was performed using either LB broth, Terrific broth, or 2× Terrific broth depending on the amount of proteolytic cleavage observed in preliminary experiments. Expression of recombinant proteins was initiated by incubating cells at 18°C for 30 min prior to the addition of isopropyl-β-d-thiogalactopyranoside (IPTG), followed by incubation at 18°C overnight. Recombinant proteins were purified natively by nickel affinity chromatography and dialyzed in phosphate-buffered saline (PBS) (Fig. 2B); their concentrations were estimated as described previously (31). Polyclonal antisera (anti-F1P146/232 and anti-F3P146/232 sera) to recombinant proteins F1P146/232 and F3P146/232 were prepared by immunization of New Zealand White rabbits as previously described (54). Polyclonal antiserum anti-F2bP146 was prepared similarly with a recombinant protein representing residues 586 to 690 of P146. For some immunoblots, purified anti-F1P146/232 antibodies were used to remove cross-reactions and were prepared as described previously (55). All antisera were tested for activity by immunoblotting with recombinant protein (Fig. 2C).
Binding assays.
Heparin-binding, inhibition, and competitive immunoassays were performed in 96-well, flat-bottomed microtiter plates (Linbro/Titertek; ICN Biomedicals Inc., Aurora, OH) as described elsewhere (31). The binding of F1P146/232-F3P146/232 to porcine cilia was examined using a microtiter plate adherence assay developed for the identification of the cilium-binding protein P97 (21). SPR analyses were performed using a BIAcore T100 instrument (Biacore AB, Sweden), operated at 20°C. The preparation of analyte proteins, ligand immobilization, kinetics assays, porcine plasminogen purification, and biotinylation of plasminogen have been described previously (25). Plasminogen was immobilized as a ligand for SPR assays; binding of recombinant P146 proteins was measured with a concentration range of 0 to 200 nM. A blank immobilized flow cell was used as a reference cell. F3P146 association kinetics with plasminogen were analyzed as the sum of two exponentials for a heterogeneous surface as described previously (24). Plasminogen dot blots were performed (24) with 5 µg ml−1 biotinylated porcine plasminogen as described previously.
SUPPLEMENTAL MATERIAL
MS/MS spectrum of 676QQQDANSTNSSPTSPSPSPTSPSPASPSSSPSPTSPK712. Ions corresponding to fragmentation are denoted according to the nomenclature system of P. Roepstorff and J. Fohlman (Biomed. Mass Spectrom. 11:601, 1984). Download Figure S1, TIF file, 0.1 MB.
P146 fragments are present on the surface of M. hyopneumoniae. (A) Intact and washed M. hyopneumoniae (strain J) cells were exposed to different concentrations of trypsin (listed above the blots in micrograms per milliliter) for 15 min. Immunoblots were probed with anti-F1P146 (αF1P146), anti-F2P146 (αF2bP146), and anti-F3P146 (αF3P146) sera detecting the P50P146, P40P146, and P85P146 fragments, respectively. Identical lysates were subsequently probed with an intracellular control protein (ribosomal protein L7/L12) to confirm cell integrity. (B) Peptides unique to P146 (MHJ_0663) obtained by shaving washed, intact M. hyopneumoniae (strain J) with 50 µg ml−1 trypsin for 5 min. Underlined sequences indicate peptides identified by LC-MS/MS. Download Figure S2, TIF file, 1.8 MB.
Recombinant fragments F1P146/232 and F3P146/232 bind heparin. (A) Binding of recombinant protein to biotinylated heparin. Total binding was determined by coating recombinant proteins F1P146/232 and F3P146/232 (10 µg ml−1) to the wells on a 96-well microtiter plate followed by incubation with increasing concentrations of biotinylated heparin (x axis). Nonspecific binding was determined by binding with unlabeled heparin in a 50-fold excess to biotinylated heparin. Specific binding was determined by subtracting the nonspecific binding from the total binding. Kd values for recombinant proteins F1P146/232 and F3P146/232 were determined from the specific binding data. (B) Inhibition of heparin binding by GAGs. Recombinant protein (10 µg ml−1) was coated onto the wells of a 96-well microtiter plate and incubated with biotinylated heparin in the presence of an increasing excess of inhibitor (x axis). Inhibitors included unlabeled heparin, fucoidan, chondroitin sulfate B, and porcine type II mucin. Download Figure S3, TIF file, 0.2 MB.
Recombinant fragments F3P146/232 and F3P146/J bind plasminogen. (A and B) Dot blots of recombinant proteins F3P146/232 and F3P146/J and controls (bovine serum albumin [BSA] and PBS). Each dot contains 1 µg of protein. Blots were stained with amido black (A) or incubated with 10 µg ml−1 biotinylated porcine plasminogen and developed with streptavidin-peroxidase and diaminobenzidine (DAB)-hydrogen peroxide (B). (C and D) SPR sensorgrams of F3P146 derived from M. hyopneumoniae strain J (C) and strain 232 (D) injected over immobilized plasminogen at concentrations of 5 (bottom line), 10, 20, 50, 100, and 200 nM (top line). (E and F) SPR sensorgrams of F3P146 derived from M. hyopneumoniae strain J (E) and strain 232 (F) without native C-terminal residues (lysine and arginine, respectively) injected over immobilized plasminogen at concentrations of 5 (bottom line), 10, 20, 50, 100 and 200 nM (top line). In all SPR experiments, F3P146 homologs were injected at a flow rate of 10 µl min−1 for 720 s. The arrows indicate the end of the injection period and initiation of F3P146 dissociation from the ligand. Download Figure S4, TIF file, 0.5 MB.
Recombinant fragments F1P146/232 to F3P146/232 bind porcine epithelial cilia. The results of microtiter plate binding assay showing P146 recombinant fragments binding to porcine cilia are depicted. Recombinant fragment derived from P97 encoding the cilium-binding domain R1 and R2 (F2P97) acted as a positive control for cilium binding in this experiment (Deutscher AT, et al., J. Proteome Res. 11:1924–1936, 2012). The recombinant fragment that has previously been reported to have low cilium binding (F4Mhp385) acted as a negative control for cilium binding in this experiment (A. T. Deutscher et al., submitted for publication). Values are means plus least significant differences at P < 0.05 (error bars). Means with any superscript (a to e) in common are not statistically different; means with no superscript in common are different at P < 0.05. Download Figure S5, TIF file, 0.1 MB.
Sequences of primers used in this study
ACKNOWLEDGMENTS
Daniel R. Bogema is the recipient of an Australian Postgraduate Award (Industry). This work was funded by ARC-Linkage grant LP776711, the ithree Institute, University of Technology at Sydney (UTS), and a grant from the McGarvie Smith Trust to Steven. P. Djordjevic. J. L. Tacchi and B. B. A. Raymond are recipients of UTS postgraduate scholarships.
Footnotes
Citation Bogema DR, et al. 2012. Characterization of cleavage events in the multifunctional cilium adhesin Mhp684 (P146) reveals a mechanism by which Mycoplasma hyopneumoniae regulates surface topography. mBio 3(2):e00282-11. doi:10.1128/mBio.00282-11.
REFERENCES
- 1. Browning GF, Marenda MS, Noormohammadi AH, Markham PF. 2011. The central role of lipoproteins in the pathogenesis of mycoplasmoses. Vet. Microbiol. 153:44–50 [DOI] [PubMed] [Google Scholar]
- 2. Rosengarten R, Wise KS. 1991. The Vlp system of Mycoplasma hyorhinis: combinatorial expression of distinct size variant lipoproteins generating high-frequency surface antigenic variation. J. Bacteriol. 173:4782–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Citti C, Watson-McKown R, Droesse M, Wise KS. 2000. Gene families encoding phase- and size-variable surface lipoproteins of Mycoplasma hyorhinis. J. Bacteriol. 182:1356–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Noormohammadi AH, et al. 1997. Mycoplasma synoviae has two distinct phase-variable major membrane antigens, one of which is a putative hemagglutinin. Infect. Immun. 65:2542–2547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu W, et al. 2011. Complete genome sequence of Mycoplasma hyopneumoniae strain 168. J. Bacteriol. 193:1016–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Minion FC, et al. 2004. The genome sequence of Mycoplasma hyopneumoniae strain 232, the agent of swine mycoplasmosis. J. Bacteriol. 186:7123–7133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vasconcelos AT, et al. 2005. Swine and poultry pathogens: the complete genome sequences of two strains of Mycoplasma hyopneumoniae and a strain of Mycoplasma synoviae. J. Bacteriol. 187:5568–5577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clark LK, et al. 1991. Investigating the transmission of Mycoplasma hyopneumoniae in a swine herd with enzootic pneumonia. Vet. Med. 86:543–550 [Google Scholar]
- 9. Haesebrouck F, et al. 2004. Efficacy of vaccines against bacterial diseases in swine: what can we expect? Vet. Microbiol. 100:255–268 [DOI] [PubMed] [Google Scholar]
- 10. Desrosiers R. 2011. Transmission of swine pathogens: different means, different needs. Anim. Health Res. Rev. 12:1–13 [DOI] [PubMed] [Google Scholar]
- 11. Tajima M, Yagihashi T. 1982. Interaction of Mycoplasma hyopneumoniae with the porcine respiratory epithelium as observed by electron microscopy. Infect. Immun. 37:1162–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. DeBey MC, Ross RF. 1994. Ciliostasis and loss of cilia induced by Mycoplasma hyopneumoniae in porcine tracheal organ cultures. Infect. Immun. 62:5312–5318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang Q, Young TF, Ross RF. 1994. Microtiter plate adherence assay and receptor analogs for Mycoplasma hyopneumoniae. Infect. Immun. 62:1616–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Erlinger R. 1995. Glycosaminoglycans in porcine lung: an ultrastructural study using cupromeronic blue. Cell Tissue Res. 281:473–483 [DOI] [PubMed] [Google Scholar]
- 15. Soane MC, et al. 2000. Interaction of Bordetella pertussis with human respiratory mucosa in vitro. Respir. Med. 94:791–799 [DOI] [PubMed] [Google Scholar]
- 16. Edwards JA, Groathouse NA, Boitano S. 2005. Bordetella bronchiseptica adherence to cilia is mediated by multiple adhesin factors and blocked by surfactant protein A. Infect. Immun. 73:3618–3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kuespert K, Roth A, Hauck CR. 2011. Neisseria meningitidis has two independent modes of recognizing its human receptor CEACAM1. PLoS One 6:e14609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bartlett AH, Park PW. 2010. Proteoglycans in host-pathogen interactions: molecular mechanisms and therapeutic implications. Expert Rev. Mol. Med. 12:e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Burnett TA, et al. 2006. P159 is a proteolytically processed, surface adhesin of Mycoplasma hyopneumoniae: defined domains of P159 bind heparin and promote adherence to eukaryote cells. Mol. Microbiol. 60:669–686 [DOI] [PubMed] [Google Scholar]
- 20. Djordjevic SP, Cordwell SJ, Djordjevic MA, Wilton J, Minion FC. 2004. Proteolytic processing of the Mycoplasma hyopneumoniae cilium adhesin. Infect. Immun. 72:2791–2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wilton J, et al. 2009. Mhp493 (P216) is a proteolytically processed, cilium and heparin binding protein of Mycoplasma hyopneumoniae. Mol. Microbiol. 71:566–582 [DOI] [PubMed] [Google Scholar]
- 22. Bogema DR, et al. 2011. Sequence TTKF ↓ QE defines the site of proteolytic cleavage in Mhp683 protein, a novel glycosaminoglycan and cilium adhesin of Mycoplasma hyopneumoniae. J. Biol. Chem. 286:41217–41229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang Q, Young TF, Ross RF. 1995. Identification and characterization of a Mycoplasma hyopneumoniae adhesin. Infect. Immun. 63:1013–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Deutscher AT, et al. 2010. Repeat regions R1 and R2 in the P97 paralogue Mhp271 of Mycoplasma hyopneumoniae bind heparin, fibronectin and porcine cilia. Mol. Microbiol. 78:444–458 [DOI] [PubMed] [Google Scholar]
- 25. Seymour LM, et al. 2010. A processed multidomain Mycoplasma hyopneumoniae adhesin binds fibronectin, plasminogen, and swine respiratory cilia. J. Biol. Chem. 285:33971–33978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Seymour LM, et al. 2011. Mhp107 is a member of the multifunctional adhesin family of Mycoplasma hyopneumoniae. J. Biol. Chem. 286:10097–10104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Seymour LM, et al. 2012. Mhp182 (P102) binds fibronectin and contributes to the recruitment of plasmin(ogen) to the Mycoplasma hyopneumoniae cell surface. Cell. Microbiol. 14:81–94 [DOI] [PubMed] [Google Scholar]
- 28. Pinto PM, et al. 2007. Proteomic survey of the pathogenic Mycoplasma hyopneumoniae strain 7448 and identification of novel post-translationally modified and antigenic proteins. Vet. Microbiol. 121:83–93 [DOI] [PubMed] [Google Scholar]
- 29. Pinto PM, Klein CS, Zaha A, Ferreira HB. 2009. Comparative proteomic analysis of pathogenic and non-pathogenic strains from the swine pathogen Mycoplasma hyopneumoniae. Proteome Sci. 7:45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Adams C, Pitzer J, Minion FC. 2005. In vivo expression analysis of the P97 and P102 paralog families of Mycoplasma hyopneumoniae. Infect. Immun. 73:7784–7787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jenkins C, et al. 2006. Two domains within the Mycoplasma hyopneumoniae cilium adhesin bind heparin. Infect. Immun. 74:481–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stakenborg T, et al. 2006. Comparison of molecular techniques for the typing of Mycoplasma hyopneumoniae isolates. J. Microbiol. Methods 66:263–275 [DOI] [PubMed] [Google Scholar]
- 33. de Castro LA, et al. 2006. Variable number of tandem amino acid repeats in adhesion-related CDS products in Mycoplasma hyopneumoniae strains. Vet. Microbiol. 116:258–269 [DOI] [PubMed] [Google Scholar]
- 34. Belloy L, Vilei EM, Giacometti M, Frey J. 2003. Characterization of LppS, an adhesin of Mycoplasma conjunctivae. Microbiology 149:185–193 [DOI] [PubMed] [Google Scholar]
- 35. McDonnell AV, Jiang T, Keating AE, Berger B. 2006. Paircoil2: improved prediction of coiled coils from sequence. Bioinformatics 22:356–358 [DOI] [PubMed] [Google Scholar]
- 36. Lupas A, Van Dyke M, Stock J. 1991. Predicting coiled coils from protein sequences. Science 252:1162–1164 [DOI] [PubMed] [Google Scholar]
- 37. Obradovic Z, et al. 2005. Exploiting heterogeneous sequence properties improves prediction of protein disorder. Proteins 61(Suppl. 7):176–182 [DOI] [PubMed] [Google Scholar]
- 38. Lähteenmäki K, Kuusela P, Korhonen TK. 2001. Bacterial plasminogen activators and receptors. FEMS Microbiol. Rev. 25:531–552 [DOI] [PubMed] [Google Scholar]
- 39. Deutscher AT, et al. 2012. Mycoplasma hyopneumoniae surface proteins Mhp385 and Mhp384 bind host cilia and glycosaminoglycans and are endoproteolytically processed by proteases that recognize different cleavage motifs. J. Proteome Res. 11:1924–1936 [DOI] [PubMed] [Google Scholar]
- 40. Sanderson-Smith ML, Dowton M, Ranson M, Walker MJ. 2007. The plasminogen-binding group A streptococcal M protein-related protein Prp binds plasminogen via arginine and histidine residues. J. Bacteriol. 189:1435–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu JN, Gurewich V. 1993. The kinetics of plasminogen activation by thrombin-cleaved pro-urokinase and promotion of its activity by fibrin fragment E-2 and by tissue plasminogen activator. Blood 81:980–987 [PubMed] [Google Scholar]
- 42. Rejante MR, Byeon IJ, Llinás M. 1991. Ligand specificity of human plasminogen kringle 4. Biochemistry 30:11081–11092 [DOI] [PubMed] [Google Scholar]
- 43. Cork AJ, et al. 2009. Defining the structural basis of human plasminogen binding by streptococcal surface enolase. J. Biol. Chem. 284:17129–17137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Strait EL, et al. 2008. Real-time PCR assays to address genetic diversity among strains of Mycoplasma hyopneumoniae. J. Clin. Microbiol. 46:2491–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Scarman AL, et al. 1997. Identification of novel species-specific antigens of Mycoplasma hyopneumoniae by preparative SDS-PAGE ELISA profiling. Microbiology 143(Part 2):663–673 [DOI] [PubMed] [Google Scholar]
- 46. Scott NE, et al. 2010. Mass spectrometric characterization of the Campylobacter jejuni adherence factor CadF reveals post-translational processing that removes immunogenicity while retaining fibronectin binding. Proteomics 10:277–288 [DOI] [PubMed] [Google Scholar]
- 47. Cordwell SJ, et al. 2008. Identification of membrane-associated proteins from Campylobacter jejuni strains using complementary proteomics technologies. Proteomics 8:122–139 [DOI] [PubMed] [Google Scholar]
- 48. Zhang K, McKinlay C, Hocart CH, Djordjevic MA. 2006. The Medicago truncatula small protein proteome and peptidome. J. Proteome Res. 5:3355–3367 [DOI] [PubMed] [Google Scholar]
- 49. Szczepanek SM, et al. 2010. Identification of lipoprotein MslA as a neoteric virulence factor of Mycoplasma gallisepticum. Infect. Immun. 78:3475–3483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Altschul SF, et al. 2005. Protein database searches using compositionally adjusted substitution matrices. FEBS J. 272:5101–5109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gasteiger E, et al. 2005. Protein identification and analysis tools on the ExPASy server, p 571–607 In Walker JM, The proteomics protocols handbook. Humana Press, New York, NY. [Google Scholar]
- 52. Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580 [DOI] [PubMed] [Google Scholar]
- 53. Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8:785–786 [DOI] [PubMed] [Google Scholar]
- 54. Scott NE, et al. 2009. Mass spectrometric characterization of the surface-associated 42 kDa lipoprotein JlpA as a glycosylated antigen in strains of Campylobacter jejuni. J. Proteome Res. 8:4654–4664 [DOI] [PubMed] [Google Scholar]
- 55. Nature Publishing Group 2005. Immunoblot affinity purification. Nat. Methods 2:797–798 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MS/MS spectrum of 676QQQDANSTNSSPTSPSPSPTSPSPASPSSSPSPTSPK712. Ions corresponding to fragmentation are denoted according to the nomenclature system of P. Roepstorff and J. Fohlman (Biomed. Mass Spectrom. 11:601, 1984). Download Figure S1, TIF file, 0.1 MB.
P146 fragments are present on the surface of M. hyopneumoniae. (A) Intact and washed M. hyopneumoniae (strain J) cells were exposed to different concentrations of trypsin (listed above the blots in micrograms per milliliter) for 15 min. Immunoblots were probed with anti-F1P146 (αF1P146), anti-F2P146 (αF2bP146), and anti-F3P146 (αF3P146) sera detecting the P50P146, P40P146, and P85P146 fragments, respectively. Identical lysates were subsequently probed with an intracellular control protein (ribosomal protein L7/L12) to confirm cell integrity. (B) Peptides unique to P146 (MHJ_0663) obtained by shaving washed, intact M. hyopneumoniae (strain J) with 50 µg ml−1 trypsin for 5 min. Underlined sequences indicate peptides identified by LC-MS/MS. Download Figure S2, TIF file, 1.8 MB.
Recombinant fragments F1P146/232 and F3P146/232 bind heparin. (A) Binding of recombinant protein to biotinylated heparin. Total binding was determined by coating recombinant proteins F1P146/232 and F3P146/232 (10 µg ml−1) to the wells on a 96-well microtiter plate followed by incubation with increasing concentrations of biotinylated heparin (x axis). Nonspecific binding was determined by binding with unlabeled heparin in a 50-fold excess to biotinylated heparin. Specific binding was determined by subtracting the nonspecific binding from the total binding. Kd values for recombinant proteins F1P146/232 and F3P146/232 were determined from the specific binding data. (B) Inhibition of heparin binding by GAGs. Recombinant protein (10 µg ml−1) was coated onto the wells of a 96-well microtiter plate and incubated with biotinylated heparin in the presence of an increasing excess of inhibitor (x axis). Inhibitors included unlabeled heparin, fucoidan, chondroitin sulfate B, and porcine type II mucin. Download Figure S3, TIF file, 0.2 MB.
Recombinant fragments F3P146/232 and F3P146/J bind plasminogen. (A and B) Dot blots of recombinant proteins F3P146/232 and F3P146/J and controls (bovine serum albumin [BSA] and PBS). Each dot contains 1 µg of protein. Blots were stained with amido black (A) or incubated with 10 µg ml−1 biotinylated porcine plasminogen and developed with streptavidin-peroxidase and diaminobenzidine (DAB)-hydrogen peroxide (B). (C and D) SPR sensorgrams of F3P146 derived from M. hyopneumoniae strain J (C) and strain 232 (D) injected over immobilized plasminogen at concentrations of 5 (bottom line), 10, 20, 50, 100, and 200 nM (top line). (E and F) SPR sensorgrams of F3P146 derived from M. hyopneumoniae strain J (E) and strain 232 (F) without native C-terminal residues (lysine and arginine, respectively) injected over immobilized plasminogen at concentrations of 5 (bottom line), 10, 20, 50, 100 and 200 nM (top line). In all SPR experiments, F3P146 homologs were injected at a flow rate of 10 µl min−1 for 720 s. The arrows indicate the end of the injection period and initiation of F3P146 dissociation from the ligand. Download Figure S4, TIF file, 0.5 MB.
Recombinant fragments F1P146/232 to F3P146/232 bind porcine epithelial cilia. The results of microtiter plate binding assay showing P146 recombinant fragments binding to porcine cilia are depicted. Recombinant fragment derived from P97 encoding the cilium-binding domain R1 and R2 (F2P97) acted as a positive control for cilium binding in this experiment (Deutscher AT, et al., J. Proteome Res. 11:1924–1936, 2012). The recombinant fragment that has previously been reported to have low cilium binding (F4Mhp385) acted as a negative control for cilium binding in this experiment (A. T. Deutscher et al., submitted for publication). Values are means plus least significant differences at P < 0.05 (error bars). Means with any superscript (a to e) in common are not statistically different; means with no superscript in common are different at P < 0.05. Download Figure S5, TIF file, 0.1 MB.
Sequences of primers used in this study