ABSTRACT
The Escherichia coli membrane protein DsbD functions as an electron hub that dispatches electrons received from the cytoplasmic thioredoxin system to periplasmic oxidoreductases involved in protein disulfide isomerization, cytochrome c biogenesis, and sulfenic acid reduction. Here, we describe a new class of DsbD proteins, named ScsB, whose members are found in proteobacteria and Chlamydia. ScsB has a domain organization similar to that of DsbD, but its amino-terminal domain differs significantly. In DsbD, this domain directly interacts with substrates to reduce them, which suggests that ScsB acts on a different array of substrates. Using Caulobacter crescentus as a model organism, we searched for the substrates of ScsB. We discovered that ScsB provides electrons to the first peroxide reduction pathway identified in the bacterial cell envelope. The reduction pathway comprises a thioredoxin-like protein, TlpA, and a peroxiredoxin, PprX. We show that PprX is a thiol-dependent peroxidase that efficiently reduces both hydrogen peroxide and organic peroxides. Moreover, we identified two additional proteins that depend on ScsB for reduction, a peroxiredoxin-like protein, PrxL, and a novel protein disulfide isomerase, ScsC. Altogether, our results reveal that the array of proteins involved in reductive pathways in the oxidative cell envelope is significantly broader than was previously thought. Moreover, the identification of a new periplasmic peroxiredoxin indicates that in some bacteria, it is important to directly scavenge peroxides in the cell envelope even before they reach the cytoplasm.
IMPORTANCE
Peroxides are reactive oxygen species (ROS) that damage cellular components such as lipids, proteins, and nucleic acids. The presence of protection mechanisms against ROS is essential for cell survival. Bacteria express cytoplasmic catalases and thiol-dependent peroxidases to directly scavenge harmful peroxides. We report the identification of a peroxide reduction pathway active in the periplasm of Caulobacter crescentus, which reveals that, in some bacteria, it is important to directly scavenge peroxides in the cell envelope even before they reach the cytoplasm. The electrons required for peroxide reduction are delivered to this pathway by ScsB, a new type of membrane electron transporter. We also identified two additional likely ScsB substrates, including a novel protein disulfide isomerase. Our results reveal that the array of proteins involved in reductive pathways in the oxidative environment of the cell envelope is significantly broader than was previously thought.
Introduction
Until 20 years ago, the presence of redox reactions in the bacterial cell envelope was thought to be quite limited. However, this picture changed with the discovery of sets of enzymes that use the chemical properties of cysteines to form or reduce disulfide bonds in the bacterial periplasm (1). The oxidative enzymatic process that promotes protein disulfide bond formation derives its power from the transfer of electrons to membrane bound quinones. DsbA, a thioredoxin-like protein with a Cys-X-X-Cys active site, is the direct catalyst of the joining of two cysteines in a substrate protein in the periplasm (2). DsbA becomes reduced as a result of its oxidative reaction and is reoxidized via two pairs of redox-active cysteines of the membrane protein DsbB (3, 4). DsbB itself is reoxidized by transferring electrons to the quinones (5).
In addition, a number of enzymes were discovered in the periplasm that employ cysteines to provide electrons to various substrates. These enzymes include DsbC, a protein that is involved in disulfide bond isomerization (6–8), DsbG, which restores the reduced form of cysteines that have been oxidized to sulfenic acid (9), and CcmG, which reduces the oxidized cysteines of apo-cytochrome c so as to allow heme binding (10, 11). In Escherichia coli, these three proteins derive their reducing power from cytoplasmic thioredoxin via the cytoplasmic membrane protein DsbD. DsbD consists of three domains: an N-terminal periplasmic domain (DsbDα), a C-terminal periplasmic domain (DsbDγ), and an intervening hydrophobic membrane-embedded domain (DsbDβ) (Fig. 1). Each of the three domains contains two redox-active cysteines, essential for the protein’s function (12). DsbD homologues are found in many other bacteria (13). However, some bacteria express a protein homologue that comprises only the cytoplasmic membrane domain of DsbD. The prototype of this protein, CcdA of Rhodobacter capsulatus, may be more limited in its specificity than DsbD, although it does reduce proteins that are homologues of CcmG (13, 14).
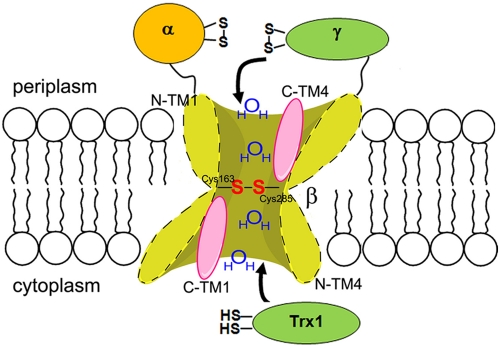
FIG 1 .
Structural model of DsbD. The model shows that DsbDβ adopts an hourglass-like structure where Cys163 and Cys285 are in the middle of the two water-exposed cavities. Halves of C-terminal TM1 and TM4 (C-TM1 and C-TM4) are water exposed (pink), while those of N-terminal ones (N-TM1 and N-TM4) are not (17, 18). Two thioredoxin modules (thioredoxin-1 and DsbDγ) may interact with Cys163 and Cys285 through these two water-exposed cavities. S, sulfur of thiol in a cysteine residue.
The transfer of electrons across the membrane by the protein DsbD occurs by a cascade of disulfide exchange reactions. This cascade begins with the reduction by cytoplasmic thioredoxin-1 of an intramembraneous disulfide bond in the hydrophobic DsbDβ domain. Electrons are then transferred from the reduced cysteines of DsbDβ to the cysteine pair of the periplasmic thioredoxin-like domain, DsbDγ (15, 16). This electron transfer occurs by an unusual mechanism whereby the two cysteines in DsbDβ are accessible to both thioredoxin-1 in the cytoplasm and the thioredoxin-like DsbDγ in the periplasm (Fig. 1) (17, 18). From DsbDγ, electrons are transferred to the immunoglobulin-like DsbDα domain, which then reduces its substrates, DsbC, DsbG, and CcmG (15, 19–21).
We have been investigating the extent to which reductive proteins related to DsbD are present in various bacterial species. Here, using bioinformatic programs to seek homologues of DsbD, we detected a third distinct class of reductive proteins (in addition to DsbD and CcdA). The prototype of this new class is Salmonella enterica serovar Typhimurium ScsB (suppression of copper sensitivity), a protein which has been shown to confer copper tolerance to copper-sensitive E. coli mutants (22).
The ScsB class has domains comparable to those of DsbD. However, the N-terminal periplasmic domain of ScsB (ScsBα) is quite different from DsbDα, suggesting that it acts on an array of substrate proteins different from those already known for DsbD. Indeed, we found that ScsB transfers electrons to a thioredoxin-like protein (TlpA), which in turn reduces a peroxidase (peroxiredoxin-Q type), PprX. PprX is the first peroxiredoxin identified in the bacterial cell envelope. ScsB also reduces a peroxiredoxin-like1 protein (PrxL) whose function remains unknown and a novel bacterial protein, disulfide isomerase (ScsC). We have established the in vivo functions of some of these proteins and their interactions with ScsB using a model organism, Caulobacter crescentus. Our results reveal that the range of such electron transfer reactions taking place in the periplasm is much broader than was previously thought. Moreover, the identification of a periplasmic peroxiredoxin indicates that in some bacteria, it is important to directly scavenge peroxides in the cell envelope before they even reach the cytoplasm.
RESULTS
A new class of DsbD family, ScsB.
We showed previously that the DsbD family is divided into two classes, the DsbD-like and CcdA-like proteins (13). The results presented here indicate that there is a third distinct class which comprises DsbD homologues with quite different α domains [Cluster of Orthologous Groups (COG) 4233] (23). The most studied member of this third class has been identified in Salmonella Typhimurium (22). Because the gene for this protein is a member of an operon that confers suppression of copper sensitivity to E. coli, the operon was annotated as scs, and scsB encodes the homologue of DsbD. Although it seemed possible that ScsB is a new type of DsbD, there has been no information on the reductive reactions it may carry out and how it may differ functionally from the other DsbD families.
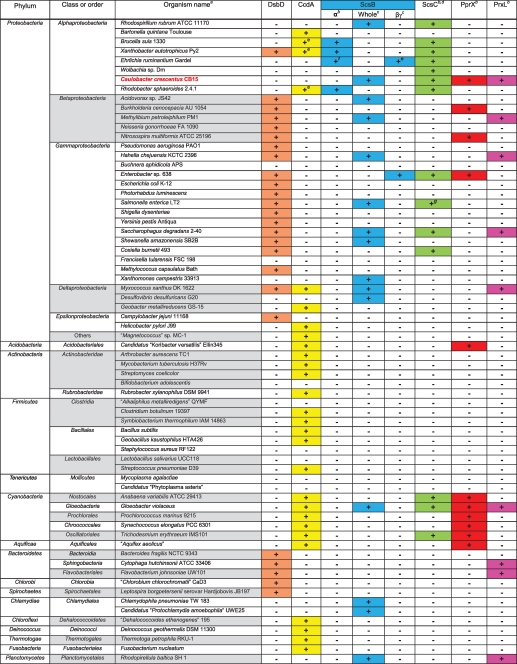
We have identified ScsBs in many other bacteria by doing a blast search for homologues of the S. Typhimurium ScsB (Table 1; also, see Table S1 in the supplemental material [Table 1 is a summary table of Table S1]) (24). To show the relationship between members of the DsbD, CcdA, and ScsB classes, we constructed a phylogenetic tree using members of each class (Fig. 2). The tree shows that ScsB proteins form a group distinct from the DsbD and CcdA classes of reductive enzymes. ScsB is positioned close to DsbD in the phylogenetic tree because both DsbDs and ScsBs have a similar domain organization (the α, β, and γ domains) and eight transmembrane segments in the β domain (see Fig. S1A and B in the supplemental material). The distribution of ScsB is quite diverse among bacteria. Most proteobacteria, excluding the epsilonproteobacteria, and all Chlamydia strains tested have scsB (Table 1; also, see Table S1). Many gammaproteobacteria, including S. Typhimurium and some E. coli strains (e.g., strain TA280), have both scsB and dsbD on their chromosomes.
TABLE 1 .
Distribution of DsbD, CcdA, ScsB, ScsC, PprX, and PrxL among diverse bacteria
|
The organisms were chosen so that they become representatives of the branches in the bacterial phylogenetic tree in Table S1 in the supplemental material.
The presence of signal sequence or N-terminal transmembrane segment was analyzed using SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/) or TMHMM 2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/), respectively.
In ScsBβ, the transmembrane segment 1, which contains the first putative redox-active cysteine, appears to be highly conserved, as shown in Fig. S1A in the supplemental material. A Lys residue in the 8th position at the C terminus of the first cysteine in ScsBβ is especially conserved and typical in ScsB (not found in DsbD and CcdA). All the tested ScsBβ proteins have this Lys residue. Therefore, the presence of this residue was used as one method to distinguish ScsBβ from DsbDβ and CcdA.
Selected are ScsC homologues which have the N-terminal helical domain (see the text). psi-blast and STRING analysis were used for the primary selection, and then the homologous sequence motif with EHPE was used for the secondary selection since this sequence motif is well conserved in the putative hinge region of the N-terminal helical domain as shown in Fig. S6A. Many ScsC variants, including prototypic ScsCs, are found in diverse bacteria. It appears that ScsC is widely used for many kinds of redox reactions in many ways in bacteria. Because of this diversity of ScsC and the established relationships between CcScsB and CcScsC, we included only the CcScsC type in Table 1 and Table S1, and its occurrence correlation with ScsB appears to be very good.
These organisms containing CcdA or ScsBβγ contain separate ScsBα but not full-length ScsB in most cases (see the text).
The ScsBα domain appears to have two subdomains (see the text). The N-terminal subdomain is relatively well conserved, while the C-terminal subdomain is not. Related to this observation, we could find ScsBα in some bacteria which do not have the C-terminal subdomain.
In Salmonella, in addition to StScsC, which is not included in this table, another ScsC homologue is found which has the N-terminal helical domain like CcScsC. The gene is annotated as bcfH, a gene in a bcf operon to encode proteins for fimbriae.
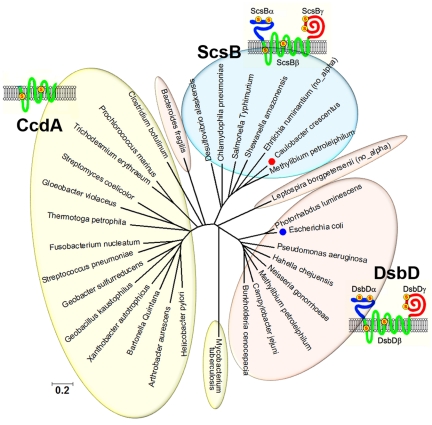
FIG 2 .
Phylogenetic analysis of the DsbD superfamily in eubacteria. The tree was constructed using the neighbor-joining method from the MEGA5 sequence analysis program (60) based on the amino acid sequences of the DsbD superfamily members listed in Table 1. A more detailed phylogenetic tree can be found in Fig. S1E in the supplemental material. The scale bar indicates the number of substitutions per site. All the family members are represented by the organism names. CcdA family members are circled in light yellow, DsbD family members in light red, and ScsB family members in light blue. We colored the DsbD homologues from Bacteroides and Leptospira and that from Mycobacterium as belonging to the DsbD and CcdA subgroup, respectively, because of their domain compositions and sequence similarities. However, these proteins appear to form distinct branches within the DsbD superfamily. Moreover, we observed that CcdA family members appear to be quite diverse compared to others. In the schematic drawings, the two essential cysteine residues in each domain of DsbD and ScsB or those in CcdA are depicted as “S” in red in yellow circles. “No_alpha” means that the corresponding DsbD or ScsB protein has no α domain. Caulobacter crescentus CB15N has ScsB and is the organism used for further study (red circle). Escherichia coli K-12 DsbD is indicated by a blue circle. Sequences are provided in Text S1 in the supplemental material.
Interestingly, we observed that many alphaproteobacteria do not have full-length ScsB but appear to express ScsBα as a separate protein (Table 1; also, see Table S1 and Fig. S1C in the supplemental material), suggesting that the full-length ScsB class of proteins may have evolved from fusion of independently expressed polypeptide chains corresponding to the three domains of the protein. We had previously postulated (13), based on subcloning of DsbD domains and bioinformatic analysis, that DsbD had evolved as a fusion protein between three different proteins (the α, β, and γ domains). Therefore, we searched for genes encoding the other two domains of ScsB, ScsBβ and ScsBγ, in organisms that expressed an independent ScsBα. We found that all but one of those organisms contain genes that encode, in addition to ScsBα, a membrane protein corresponding to either CcdA (the homologue of DsbDβ) or a ScsBβγ fusion protein (Table 1; also, see Table S1). In the former case, while we did not find any separately expressed ScsBγ homologues, we think it likely that a thioredoxin-like protein predicted to be exported would replace the function of ScsBγ in those organisms. The finding of ScsBβγ, in addition to ScsBα, is consistent with evolution of these proteins by gene fusion and suggests a possible order of gene fusion events (Table 1; also, see Table S1). Thus, ScsBα as a separate protein might represent a component of a more primitive electron transpon which CcdA (β) transfers electrons to a thioredoxin-like protein(γ) and thence to ScsBα (α), by analogy to the order of electron transfer in the three-domain protein DsbD.
While the DsbD and ScsB classes of proteins show different conservation throughout much of their polypeptide sequences (see Fig. S1A and B in the supplemental material), they differ particularly in their α domains, which cannot be aligned at all. Furthermore, the number of amino acid residues of ScsBα is greater than that of DsbDα (compare Fig. S1A and B in the supplemental material; for details, see the Discussion). Nevertheless, ScsBα is predicted to have a largely beta-sheet structure like that of DsbDα (see Fig. S1D). The position of the two cysteines is conserved in the ScsBα family, and they are presumably redox active in that domain for electron transfer (see Fig. S1A and C).
Predicting substrates of ScsB.
Since ScsBα appears to have a secondary structure similar to that of DsbDα, and ScsB has a domain organization similar to that of DsbD, we postulate that ScsBα likely plays the same role as DsbDα, which is to donate electrons to substrate proteins. However, given the difference in sequence conservation and length of these two domains, we suspected that ScsB may have a different array of substrate proteins than DsbD. Since scsB forms an scs operon with scsC, which encodes thioredoxin fold proteins in some bacteria (22), we considered ScsC a likely substrate of ScsB. No significant homologues of ScsC are found in E. coli K-12 by a BLAST search (see Fig. S2A and S2B in the supplemental material; DsbG and DsbC are the closest homologues, but the expectation values are 3.5 and 7.5, respectively). Therefore, we chose ScsC as a potential substrate of ScsB for further characterization below.
In a number of bacteria, we have also found a protein with a putative thioredoxin fold encoded by a gene near scsB on the chromosome (Table 1; also, see Table S1 in the supplemental material). The gene product is annotated as a peroxiredoxin-like1 (PrxL) by Conserved Domains (CDD) in National Center for Biotechnology Information (NCBI) and it has a signal sequence for export. PrxL has not been characterized before. Therefore, we chose PrxL as a second potential substrate of ScsB for further characterization.
As another means of identifying possible substrates of ScsB, we used STRING, an interacting-protein database, entering the amino acid sequence of Caulobacter crescentus ScsB (CC0217) (see Fig. S2C in the supplemental material; the choice of C. crescentus ScsB is explained below). The branches revealed by this analysis could represent directly interacting partners, regulators, or false positives. Among potential partners found were ScsC and PrxL, showing the likely utility of a STRING analysis for this purpose. In addition, the STRING analysis pointed to a second potential protein interacting with ScsB, a C. crescentus peroxiredoxin homologue (CC1673) which has a signal sequence for export. Peroxiredoxins (Prx) are peroxidases that utilize redox-active cysteines to detoxify peroxides (25, 26). While Prxs are widespread amongst bacteria throughout nature and protect against oxidative stress, no Prx has been identified in the bacterial cell envelope. [E. coli Tpx (27) was originally reported as a periplasmic Prx. However, Tpx does not possess a signal sequence and was subsequently demonstrated to be located in the cytoplasm (28).] For this reason, we also chose this interesting periplasmic Prx as a substrate of ScsB for further characterization. For clarity and simplicity, we designate C. crescentus periplasmic Prx as PprX.
CcmG homologues are also found in ScsB-containing organisms. However, we decided not to investigate them further because they have already been well studied in diverse bacteria as substrates of CcdA or DsbD.
ScsB is essential in C. crescentus and maintains ScsC, PprX, and PrxL reduced in the periplasm.
We proceeded to seek direct evidence that the putative substrate proteins identified in the search described above depend on ScsB for reduction. Since ScsB is likely to be involved in reductive pathways in the bacterial cell envelope, the substrate proteins would be ones that are maintained by ScsB in the reduced state in vivo. Thus, deletion of scsB should lead to oxidation of the substrate proteins. This has previously been shown for E. coli DsbD and its substrate proteins (8, 29). Among the organisms with a scsB gene, C. crescentus appeared the most amenable to genetic analysis. Furthermore, it has no ccdA and no dsbD, having only one scsB gene along with each of the candidate substrate genes mentioned above (Table 1).
Initial attempts to knock out scsB by inserting an antibiotic cassette failed, suggesting that ScsB may be essential. However, we were able to obtain the knockout mutations of scsB in the presence of a complementing plasmid (Fig. S3 in the supplemental material shows the essentiality of ScsB; also, see the Discussion). When we then sought to assess the redox state of potential substrate proteins after depletion of ScsB by removing an inducer, we found that the basal level of expression was too leaky. Therefore, we chose an alternative approach in which we grew the scsB strains carrying the complementing plasmid in the absence of antibiotic, under which conditions the plasmid is reduced in copy number, as it does not have a partitioning system (30). This approach allows the cells to reduce by attrition the copy number of the plasmid to the point where they maintain the smallest number of plasmids needed to survive with the scsB null mutation.
Thus, we depleted cells of ScsB in this way and analyzed the redox state of the potential substrates ScsC, PrxL, and PprX. Each of these proteins has only two putative redox-active cysteines. To detect the proteins and analyze the oxidation state of their cysteines by Western blotting, we tagged the three candidate substrates with Flag epitopes and alkylated cysteine residues with 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS) (12). AMS adds 0.5 kDa of molecular mass and causes a shift in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (31). When reduction of the three putative substrate proteins by dithiothreitol (DTT) is followed by AMS treatment and cell extracts are run on SDS-PAGE, all three proteins run with lower mobility (Fig. 3, lane 5) than they do in samples without DTT and AMS treatment (lane 4). In the case of PrxL and PprX, the shifts are bigger than that of ScsC because disulfide bonding of the two distantly located cysteines in the primary sequences probably leads to more significant conformational changes and a more compact protein structure. Our results show that in both the wild-type strain and the scsB-null strain expressing high levels of ScsB from the plasmid, most or significant portions of ScsC, PrxL, and PprX are in the reduced state (Fig. 3, lanes 1 and 3, respectively), suggesting that they are involved in reductive pathways. In contrast, significant portions of the three proteins are oxidized when cells are depleted of ScsB by removal of the antibiotic and inducer (Fig. 3, lane 2), indicating that they are normally reduced by ScsB. The dependence of PprX, PrxL, and ScsC on ScsB for reduction has also been confirmed by heterologous expression of these proteins in E. coli (see Fig. 5 for PprX; see Fig. S7 in the supplemental material for ScsC; data not shown for PrxL). Altogether, these results indicate that ScsB provides electrons for reduction of these periplasmic proteins.
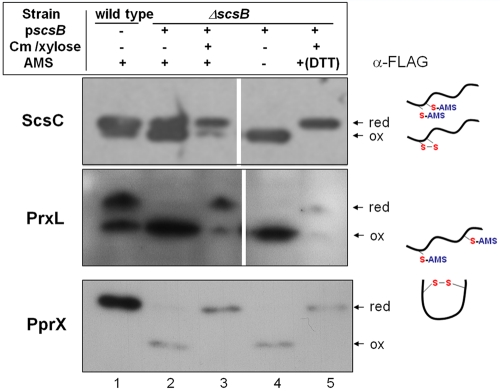
FIG 3 .
In vivo redox states of CcScsC, CcPrxL, and PprX in ScsB-depleted C. crescentus strains. Structural genes for CcScsC, CcPrxL, and PprX were fused with a Flag tag and expressed from pSC160, pSC171, and pSC159 plasmids, respectively. Each cell was cotransformed with a ScsB-expressing plasmid (pscsB; pSC161) (lanes 2 to 5) or left untransformed (lane 1). Cells were induced by 0.2% d-xylose for pscsB or 0.5 mM vanillate for the other plasmids and harvested at mid-log phase, and then proteins were precipitated with TCA and subjected to AMS alkylation. Proteins were then separated by SDS-PAGE under nonreducing conditions and visualized by Western blotting using an anti-Flag antibody. AMS was used to treat samples in lanes 1 to 3 and lane 5. In lane 5, treatment of samples with 50 mM DTT was followed by AMS alkylation. For depletion of ScsB, chloramphenicol (Cm) and d-xylose were not added (lanes 2 and 4). The strain backgrounds used were C. crescentus CB15N (wild type; lane 1) and SEN 233 (the scsB mutant with pscsB). Cartoons explain the shifts in the mobility of proteins which reflect either cysteine modification by AMS (higher bands) or a modified and more compact structure due to disulfide bond formation. CcPrxL and PprX have distantly located cysteines such that disulfide bond formation causes more compact structures and therefore a fast mobility in gels compared to the structure of AMS-modified reduced proteins, whereas CcScsC has CXXC, leading to a smaller mobility shift caused solely or primarily by AMS modification. The depletion experiment was repeated several times. Although the quantities of protein recovered after the TCA precipitation varied from sample to sample, the results were always consistent with those shown in this figure.
Discovery of a new periplasmic peroxiredoxin (PprX).
We describe above the detection of a gene encoding a protein that appears to be a periplasmic Prx, PprX. We have pursued characterization of this protein since, while Prxs have been found in the cytoplasm of bacteria, none have been confirmed to be expressed in the bacterial cell envelope. These enzymes utilize their peroxidatic cysteine residue to attack the O—O bond of the hydroperoxide substrate, thus becoming oxidized to a sulfenic acid form. A second, resolving cysteine, attacks the cysteine-sulfenic acid, generating an inter- or intrachain disulfide bond. The activity of the Prx enzyme is then regenerated by reduction of the disulfide bond, usually by thioredoxin-like proteins. In E. coli, three Prxs, alkyl hydroperoxide reductase (AhpC), bacterioferritin-comigratory protein (BCP), and thiol peroxidase (Tpx), are present in the cytoplasm and are considered major scavengers of hydrogen peroxide (28, 32–34). The closest homologue of C. crescentus PprX in E. coli is the Prx BCP, and a bioinformatic analysis based on active site profiling places this Prx clearly in the BCP/PrxQ subfamily of Prx (http://csb.wfu.edu/prex) (35, 36). Sequence alignment of some PrxQ members, including these two proteins, shows strong sequence conservation, including the absolutely conserved PXXXTXXC active site, which contains the cysteine that attacks H2O2, and a critical arginine residue which is conserved across the broad Prx family (Fig. 4A). However, the position of the resolving cysteine is different, as is the case for a subset of members of this subfamily (35, 37). The 20-amino-acid extension present at the N terminus of PprX corresponds to a signal sequence, indicating that this possible Prx is an exported protein. We have also found PprX homologues in bacteria other than C. crescentus, some among members of the alpha- and betaproteobacteria, and in most cyanobacteria (Table 1 and Table S1; see details in the Discussion).
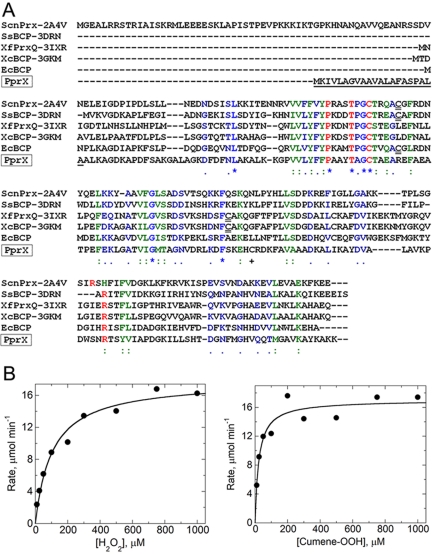
FIG 4 .
PprX is a true, thiol-dependent peroxidase. (A) Sequence alignment of PprX (CC1673) and five other Prxs from the BCP/PrxQ subfamily. Absolutely conserved Prx residues (Pro, Thr, Cys, and Arg) are shown in red. The putative signal sequence of PprX from C. crescentus is underlined. Known resolving cysteines of other BCP/PrxQ members are double-underlined. Sequences shown are from Saccharomyces cerevisiae (nuclear Prx, UniProt accession P40553), Sulfolobus solfataricus (Q97WP9), Xylella fastidiosa (Q9PER7), Xanthomonas campestris pv. campestris (Q8P9V9), and E. coli (POAE52). PDB identifiers are shown in parentheses for the first four for which structures have been determined. (B) Thiol-dependent peroxidase activity of the purified PprX. A coupled assay was used to show peroxidase activity. The assay uses thioredoxin-1 and thioredoxin reductase from E. coli as the electron donating system and H2O2 or cumene hydroperoxide (OOH) as the oxidizing substrate. The NADPH consumption rate was monitored to measure the activity. Kinetic parameters determined under these conditions for H2O2 and cumene OOH, respectively, were 17.8 ± 0.8 and 17.0 ± 0.8 µmol/min for Vmax,app and 104 ± 18 and 23 ± 6 for Km,app.
C. crescentus PprX is a thiol-dependent peroxidase.
We asked whether PprX actually had thiol peroxidase activity and what its activity was toward various peroxides. To do this, we purified a signal sequence-less engineered version of PprX and performed a coupled assay, using H2O2 or cumene hydroperoxide as the substrate in the presence of thioredoxin-1 and thioredoxin reductase from E. coli to maintain thiol peroxidase activity (Fig. 4B). PprX shows almost identical apparent Vmax values for both H2O2 and cumene hydroperoxide reactions. There is a difference in the apparent Km values for the two peroxide substrates, with the enzyme displaying a lower Km for the bulkier substrate, cumene hydroperoxide (Km,app of 23 µM versus 104 µM for H2O2). PprX shows peroxidase activity that is similar (within a factor of 2) to that measured for the E. coli BCP at the 10 µM concentration of thioredoxin-1 used in the assays (38). These results indicate that PprX is a thiol-dependent peroxidase.
Electron donor of PprX.
The in vitro results reported above suggest that the direct in vivo electron donor for PprX in the periplasm is a thioredoxin-like protein, as is generally the case for peroxiredoxins located in the cytoplasm. If the pathway for electron transfer by ScsB is the same as in DsbD, one might expect that the domain that donates electrons to substrates would be ScsBα; however, ScsBα does not have a thioredoxin fold and is predicted to be largely a beta-sheet structure (see Fig. S1D in the supplemental material) (20, 39). To determine whether ScsBα can directly reduce PprX, we coexpressed C. crescentus ScsB (CcScsB) along with PprX in an E. coli dsbD mutant. However, we observed that PprX cannot be reduced by CcScsB (Fig. 5, lane 3), suggesting that there is a missing component, presumably another thioredoxin-like protein, that mediates electron transfer from ScsB to PprX in C. crescentus.
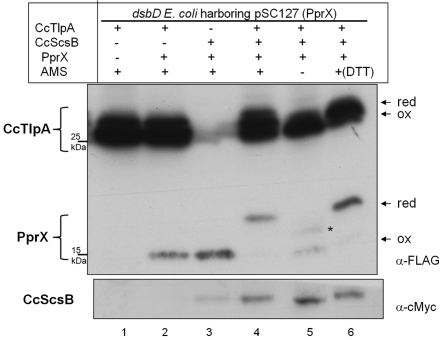
FIG 5 .
In vivo redox states of PprX expressed in dsbD E. coli strain assessed in the presence of CcTlpA or CcScsB. Cells were induced by 0.2% l-arabinose for ptlpA encoding CcTlpA with triple Flag tags and pscsB encoding CcScsB with a c-Myc tag, and 250 µM IPTG for pSC127 (PprX with a Flag tag). All strains were transformed with pSC127 and cotransformed with a second plasmid, pBAD18 (a vector control instead of pscsB), and a third plasmid, ptlpA (lanes 1 and 2), pBAD43 (a vector control instead of ptlpA) and pscsB (lane 3), or ptlpA and pscsB (lanes 4–6). To distinguish the PprX band from that of TlpA, no IPTG was added in lane 1. Cells were treated to determine redox states of proteins as described for Fig. 3. Proteins were visualized by Western blots using an anti-Flag and an anti-c-Myc antibody. The strain backgrounds used were FED126 (the dsbD mutant). The asterisk indicates portions of reduced proteins which are not alkylated by AMS.
We then set out to identify the missing link between ScsB and PprX by searching the C. crescentus genome for thioredoxin-like proteins with export signals. Several proteins that meet the criteria were identified (see Fig. S4A in the supplemental material). Among those, TlpA (thioredoxin-like protein A) drew our attention because, in certain microorganisms, this protein is fused to a periplasmic methionine sulfoxide reductase based on STRING analysis (see Fig. S4B in the supplemental material). Methionine sulfoxide reductase is an antioxidant enzyme that reduces oxidized methionine residues and, like Prx, also requires a thioredoxin-like protein as an electron donor (40). Therefore, we decided to test whether CcTlpA can provide electrons to PprX by introducing a CcTlpA expression plasmid in the E. coli dsbD strain expressing CcScsB and PprX. Interestingly, we observed that the expression of CcTlpA leads to the reduction of PprX (Fig. 5, compare lane 4 with lane 3). The reduction is dependent on the presence of CcScsB (Fig. 5, compare lane 4 with lane 2). Consistently, a larger fraction of CcTlpA is in the reduced state in the presence of CcScsB (Fig. 5, compare lane 4 with lanes 1, 2, 5, and 6). Thus, we can conclude from these experiments that ScsB transfers electrons to PprX via TlpA in C. crescentus.
CcPrxL does not have a detectable peroxidase activity.
While the second potential peroxiredoxin-like (PrxL) protein (cd02969) that is a likely substrate of ScsB does not belong to the Prx family (cd02971) according to the classification of CDD and the PREX database (this protein has two Cys residues but lacks any of the other stringently conserved residues mentioned above), the name originated from its sequence similarity to Prx family members according to the description in CDD. Therefore, we also attempted to measure thiol-dependent peroxidase activity for PrxL in vitro. Using the FOX assay to detect loss of peroxide and DTT as a nonspecific reductant of the potential peroxiredoxin, we could not detect significant thiol-dependent peroxidase activity. Thus, it appears that PrxL, which is probably not a peroxidase, has a yet-to-be defined redox function.
ScsC is a bacterial protein disulfide isomerase in C. crescentus.
The third potential ScsB substrate we studied is ScsC. Results presented above suggested that ScsC, which has a thioredoxin fold, is involved in a reductive pathway in the envelope of C. crescentus. C. crescentus has two putative dsbAs (CC0374 and CC0375) and one putative dsbB (CC2196) (41, 42) but no dsbC or dsbG in its genome (see the next section for details; also, see Fig. S5A in the supplemental material) (13). However, the closest homologue of CcScsC in E. coli K-12 is DsbC (see Fig. S5A). Therefore, we hypothesized that the in vivo role of ScsC in C. crescentus is protein disulfide isomerization.
In E. coli, several proteins have nonconsecutive disulfide bonds whose formation depends on DsbC (43–46). To detect in vivo protein disulfide isomerase activity of CcScsC, we sought to identify C. crescentus periplasmic proteins with nonconsecutive disulfide bonds. We found a periplasmic ribonuclease (RNase) (CC0030; RNase I) which is homologous to a tobacco RNase that has nonconsecutive disulfide bonds (47). Two of the cysteines that are involved in the formation of a nonconsecutive disulfide bond are conserved in C. crescentus RNase I (Fig. 6A), strongly suggesting that C. crescentus RNase I also contains a nonconsecutive disulfide in its native conformation (Fig. 6A).
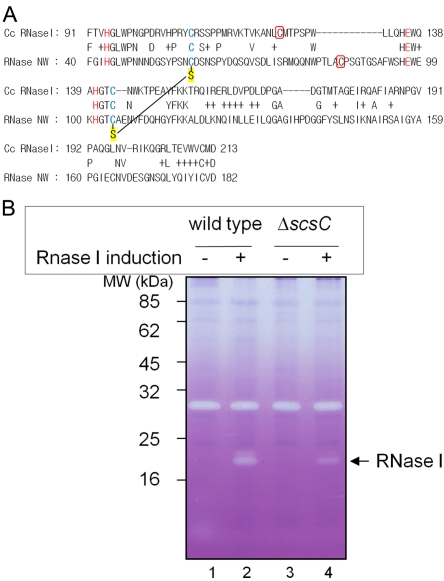
FIG 6 .
In vivo RNase I activity depends on scsC in C. crescentus. (A) Sequence alignment of C. crescentus RNase I (CC0030) and a tobacco RNase (RNase NW) after Jpred3 analysis. The high-resolution structure of the latter has been solved, and disulfide bonds have been assigned from the structure (PDB: 1iyb). The letters in red in the alignment are predicted to be catalytic residues in the active sites of both RNases. Two cysteine residues in blue are conserved in both RNases, and those from RNase NW form a disulfide bond which is represented with an S (sulfur in thiol) in a yellow background. One additional cysteine residue between these conserved cysteines in each of the C. crescentus RNase I and RNase NW proteins is indicated in a red box. (B) Zymogram to show RNase activity in the wild-type and the scsC C. crescentus strains. Cells were induced by 0.5 mM vanillate for expression of C. crescentus RNase I from pSC164. Cells were harvested at mid-log phase, broken by sonication, and subjected to SDS-PAGE. Poly(C) as an RNA substrate and toluidine blue as an intercalating dye were added to visualize RNase activity on a gel. The strain backgrounds used were C. crescentus CB15N (wild type; lanes 1 and 2) and SEN 224 (the scsC mutant; lanes 3 and 4).
To ask whether CcScsC is involved in RNase I folding, we generated an scsC mutant of C. crescentus, cloned rnaI for expression in C. crescentus, and tested for RNase activity in wild-type and scsC strains. Using a zymogram to assess RNase activity, we found that the activity was significantly reduced in the scsC mutant compared to the wild type (Fig. 6B, compare lane 4 and lane 2). This result suggests that CcScsC is involved in RNase I folding, likely via disulfide bond rearrangement. The protein disulfide isomerization activity of CcScsC was further confirmed by expressing it in a dsbC E. coli mutant and finding that it restores the folding of RcsF, an outer membrane lipoprotein with two nonconsecutive disulfide bonds whose assembly normally requires DsbC (for details, see Fig. S5B in the supplemental material) (44).
C. crescentus ScsC forms a homodimer in a different way from DsbC.
The dimeric nature of DsbC is important for its protein disulfide isomerization activity (48–50). To determine whether CcScsC also forms dimers, we performed gel filtration chromatography to estimate the molecular weight of CcScsC. CcScsC with a His tag (25 kDa in monomer size) runs similarly to DsbC with a His tag (25 kDa in monomer size) and elutes with an apparent molecular mass of 47 ±10 kDa, indicating that it forms a dimer in solution (see Fig. S6B in the supplemental material). We then predicted the secondary structure of CcScsC to see whether this protein contains a dimerization domain similar to that of DsbC (see Fig. S6A). The C-terminal domain of CcScsC is predicted to have a thioredoxin-like fold like that of DsbC. The dimerization domain of DsbC is located at the N terminus of the protein and consists of beta-strands. However, the N terminus of CcScsC is predicted to form a long alpha helix (see Fig. S6A). We hypothesize that this region mediates dimerization of CcScsC. S. Typhimurium ScsC (StScsC) does not have the long N-terminal alpha helices found in CcScsC (see Fig. S6A). Interestingly, it behaves in gel filtration chromatography like a monomer (see Fig. S6C). This result suggests that the N-terminal helical domain of CcScsC mediates homodimerization of the molecule.
DISCUSSION
We present evidence that a family of prevalent electron transporters in bacterial membranes can be divided into the DsbD, CcdA, and ScsB classes. Each of these protein families uses two similarly positioned redox-active cysteines in its membrane-embedded domain to transfer electrons from the cytoplasmic thioredoxin system to a periplasmic thioredoxin-like protein or domain. The three families differ first in that the CcdA protein comprises only the membrane-embedded domain, while, usually, DsbD and ScsB both contain an amino-terminal periplasmic domain (the α domain) and a carboxy-terminal periplasmic domain (the γ domain). DsbD and ScsB themselves differ in that their α domains show no evidence of homology to each other.
Our studies of ScsB expand the range of protein classes that are substrates of these reductive enzymes. Specifically, we have shown (i) that periplasmic peroxiredoxin homologues exist in bacteria, (ii) that they are maintained in the active state by a reducing system that involves ScsB and a thioredoxin-like protein (TlpA), and (iii) that a novel type of protein disulfide isomerase depends on ScsB for reduction. In an example similar to the periplasmic peroxiredoxins, it has been shown that Neisseria gonorrhoeae expresses a periplasmic methionine sulfoxide reductase, an enzymatic activity previously considered to be restricted to the cytoplasm (51, 52). Like cytoplasmic peroxiredoxins, cytoplasmic methionine sulfoxide reductases are reduced by thioredoxins. And by analogy to periplasmic methionine sulfoxide reductase, periplasmic peroxiredoxins can be reduced via electrons transferred from DsbD or ScsB. In many ways, the functioning of the membrane domain of members of the DsbD family fills the same role in the periplasm as thioredoxin reductase does in the cytoplasm.
Subdomain structure of ScsBα.
The number of amino acid residues of ScsBα is greater than that of DsbDα (compare Fig. S1A and B in the supplemental material). Among ScsBα domains, especially within the N-terminal portion, which has the two conserved cysteines, the sequences are relatively well conserved compared to those in the C-terminal part (see Fig. S1A and C). This finding, along with the observation that Ehrlichia ruminantium has ScsBα lacking the less-conserved C terminus, suggests that ScsBα may have two subdomains (the N subdomain and the C subdomain) (Table 1; see Table S1 in the supplemental material for more examples; see also Fig. S1C). One possibility is that the C subdomain of CcScsBα modulates the substrate specificity of the N subdomain, which on its own would function as a general electron donor. Some of our data are consistent with this idea. E. coli DSbDα (EcDsbDα) has only one domain and is missing the hypothetical modulator domain. EcDsbDα appears to have broader substrate specificity than CcScsB, since EcDsbDα can reduce either EcDsbC or CcScsC, while CcScsBα can reduce only CcScsC (see Fig. S7A, lanes 2 and 3). It would be interesting to see whether ScsBα without the C subdomain has broader substrate specificity than the ScsBα with the two subdomains.
Periplasmic peroxiredoxin in bacteria.
Detoxification of peroxides in the cytoplasm has been extensively studied in bacteria. Prxs are very efficient scavengers for peroxides generated at physiological concentration (micromolar range), while catalase can deal with peroxides only at much higher concentrations (millimolar range) (25). In E. coli, two of the three Prxs, all cytoplasmic, are particularly specialized in efficiently reducing either H2O2 (AhpC) (53) or bulkier hydroperoxide substrates (Tpx) (32, 33). The third, BCP, exhibits broader specificity but also lowered catalytic efficiencies (from about 107 M−1 s−1 for AhpC and Tpx with their best substrates to around 105 M−1 s−1 for BCP with the same substrates, hydrogen peroxide and cumene hydroperoxide) (38). Another member of this BCP/PrxQ subfamily, PrxQ from Xylella fastidiosa, also exhibited a catalytic efficiency (kcat/Km for peroxides) of ~105 M−1 s−1 but was shown in rapid-reaction studies to degrade hydroperoxides and peroxynitrite with second-order rate constants of ~106 to 107 M−1 s−1 (54).
Several reports highlight the importance of oxidative stress defense mechanisms to protect envelope proteins from oxidative damage, including the Neisseria gonorrhoeae methionine sulfoxide reductase described above (52). Moreover, DsbG, a protein homologous to DsbC, regenerates cysteines in oxidatively damaged (sulfenylated) proteins in the E. coli periplasm (9). Tpx, a peroxiredoxin family member in E. coli was originally reported to be a periplasmic protein (27) but has since been shown conclusively to be cytoplasmically localized (28). Here, we report the identification of a new periplasmic peroxiredoxin, which acts as a direct scavenger of peroxides in the bacterial cell envelope. Deletion of pprX does not seem to increase the sensitivity of C. crescentus to peroxides (data not shown), and the in vivo significance of PrxP needs further investigation.
Electron donor of PprX.
We have presented evidence that CC2210 directly donates electrons to PprX. CC2210 is a TlpA (thioredoxin-like protein) homologue which is known to be involved in the maturation of aa3-type cytochrome oxidase in Bradyrhizobium japonicum (55) (Fig. S4C), but the molecular mechanism involved has not been explored. B. japonicum is an alphaproteobacterium, like C. crescentus, so it would be interesting to see whether deletion of CcTlpA affects the maturation of aa3-type cytochrome oxidase in C. crescentus.
In the experiments to show the interactions of PprX, CcScsB, and CcTlpA, we used the wild-type E. coli strain which expresses DsbD as a control (see Fig. S7B in the supplemental material). Surprisingly, portions of PprX are in the reduced state in the absence of CcTlpA. Overexpressed DsbD significantly increased the reduction of PprX. Since DsbD appears to reduce PprX, we asked whether it might do so through its DsbDγ, thioredoxin-like domain. This turned out to be the case: when we expressed a DsbD with a Cys mutation in DsbDα and another with Cys mutations in DsbDγ, we found that the former was still able to reduce PprX, while the latter was not. These results show that the periplasmic thioredoxin-like domain of DsbD can reduce PprX but the amino-terminal periplasmic domain cannot. Although this is an electron transfer by DsbD that is not known to take place in E. coli, we propose that such a mechanism of electron transfer by DsbD may exist in other bacteria, since certain bacteria containing PprX, for example Bordetella and Burkholderia, do not have ScsB but have DsbD as an electron donor (Table 1; also, see Table S1 in the supplemental material).
Diversity of ScsC.
Our search for ScsC homologues using the StScsC sequence has revealed relatives of the protein in many bacteria, including the CcScsC (Table 1; also, see Table S1 in the supplemental material). These ScsC homologues showed significant diversity, the molecular difference in oligomeric state between CcScsC and StScsC described above being one example.
Essentiality of ScsB.
While DsbD in E. coli is dispensable for growth under normal laboratory growth conditions, ScsB in C. crescentus is essential for growth, which was also recently reported in another study (56). We asked whether any of the three identified substrate proteins of ScsB are essential in C. crescentus. However, null mutants of each of the three genes still grow (data not shown). Interestingly, DsbD in Neisseria meningitidis has also been reported to be essential (57). Neisseria and Caulobacter share common membrane electron transport components which are not present in E. coli. These components involve the cytochrome bc1 complex and the cytochrome c oxidase, which use cytochrome c as an electron acceptor and donor, respectively, for aerobic respiration. Interestingly, the synthesis of cytochrome c is known to require electrons donated by a periplasmic thioredoxin-like protein (CcmG). Therefore, it is likely that the nonviability of the N. meningitidis dsbD and CcscsB mutants is caused by disruption of the cytochrome c synthesis pathway in these bacteria. Consistent with this hypothesis, E. coli uses the cytochrome bd or bo oxidase for aerobic respiration. These enzymes do not depend on cytochrome c, explaining why an E. coli dsbD null mutant is viable. To test this hypothesis, we asked whether R. capsulatus CcdA (RcCcdA) can complement the growth defect of a scsB mutant. The complementation was successful, which suggests that the essentiality of CcScsB is due to a defect in aerobic respiration (see Fig. S3 in the supplemental material). However, we cannot exclude the possibility of the existence of another, yet-unknown essential reductive pathway(s) which can receive electrons from RcCcdA.
MATERIALS AND METHODS
Strains and media.
Strains and plasmids used in this work are listed in Table S2 in the supplemental material. E. coli cells were grown in NZ medium (7) at 37°C. When needed, 200 µg/ml of ampicillin, 30 µg/ml of chloramphenicol, and 100 µg/ml of spectinomycin were added. Expression of the genes was induced for 1 h by adding 0.2% l-arabinose or appropriate concentrations of isopropyl-β-d-thiogalactopyranoside (IPTG). C. crescentus cells were grown in PYE medium (58) at 30°C. When needed, media were supplemented with the following antibiotics (concentrations in liquid and solid media, in µg/ml); tetracycline (1 and 2), spectinomycin (25 and 100), chloramphenicol (2 and 1), and kanamycin (5 and 25). Expression of the genes was induced for 4 h by adding 0.2% d-xylose or 0.5 mM vanillate (Sigma)–NaOH (pH 7.5) (30).
Gene deletion in C. crescentus.
To delete a gene in C. crescentus, we used a method described previously (59). A brief description can be found in the supplemental material.
Thiol-redox state analyses.
To analyze the in vivo redox states of the proteins, free thiols were acid-trapped by trichloroacetic acid (TCA) and alkylated with AMS (Invitrogen) as previously described (12). When indicated, 50 mM DTT was added to reduce samples or AMS and DTT were not added to samples to obtain oxidized control proteins. Anti-Flag mouse monoclonal antibody (M2; Sigma) was used for Western blot analysis.
Thiol peroxidase assay using purified PprX and PrxL.
To assess the in vitro thiol peroxidase activity of PprX and PrxL, we overexpressed the two proteins containing His tag but no signal sequences in E. coli and purified them using nickel affinity chromatography following the standard manufacturer’s protocol (Novagen). Construction of the expression plasmids is described in the supplemental material.
To measure thiol peroxidase activity of PprX, a coupled assay using thioredoxin-1 (8 µM) and thioredoxin reductase (0.5 µM) from E. coli and 100 µM NADPH was used essentially as previously described (33). Briefly, various concentrations (5 to 500 µM) of H2O2 or cumene hydroperoxide were added to the 600-µl reaction mixtures. The reaction was initiated by the addition of 0.5 µM PprX. The NADPH consumption rate was measured at 340 nm. For each reaction, the rate of 340 nm absorbance change was corrected for the rate measured before addition of the PprX (typically 10% of the PprX-dependent rate). All the rates are the mean of at least two determinations. A more direct FOX assay to monitor the disappearance of peroxide (see Text S1 in the supplemental material) was used to detect any peroxidase activity of PrxL.
Bioinformatic analyses.
To generate a phylogenetic tree of DsbD family proteins, DsbD, ScsB, and CcdA family members were randomly chosen from bacteria which are distantly related according to the phylogenetic classification of bacteria. The tree was constructed by the neighbor-joining method based on amino acid sequences using MEGA5 (60). For the sequence alignment, we used the internal MUSCLE alignment program from MEGA5.
STRING (http://string-db.org/) analysis was used to predict interacting proteins of a query protein. To determine the distribution of DsbD, CcdA, ScsB, PprX, ScsC, and PrxL in eubacteria, the “occurrence view” option in STRING analysis was used. To complement the STRING analysis for the distribution of homologues, protein-protein searches were performed using the algorithm PSI-BLAST (position-specific iterated blast) at the NCBI (61). The PSI-BLAST analysis was also used to obtain the amino acid sequence alignment of two specific proteins.
Secondary structures of proteins were predicted using PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/) (62). High-resolution structures of homologues of interest were found using Jpred3 analysis (http://www.compbio.dundee.ac.uk/www-jpred/) by an amino acid sequence query of a protein of interest. The presence of signal sequence or N-terminal transmembrane segment was analyzed using SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/) or TMHMM 2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/), respectively (63).
Zymogram for RNase activity.
A zymogram to show RNase activity was performed according to a previous report (64). Briefly, cells expressing C. crescentus RNase I were harvested at mid-log phase, broken by sonication, and subjected to SDS-PAGE. Poly(C) (Sigma) as an RNA substrate was added to the solution for SDS-PAGE, and toluidine blue as an intercalating dye was added after electrophoresis to the gel-staining solution.
SUPPLEMENTAL MATERIAL
Supplemental materials and methods. Download Text S1, DOC file, 0.1 MB.
Amino acid sequence analyses of ScsB and DsbD. Sequence alignments of ScsB (A), DsbD (B), and ScsBα (C). The alignments were performed using ClustalW (http://www.ebi.ac.uk/Tools/msa/clustalw2/). In panels A and B, two putative reactive cysteines in each domain are indicated in red and bold on top of the alignments. Putative linker regions separate the three domains (from the N-terminal α to the C-terminal γ through β in the middle), which are shown in transparent gray in the alignments to discriminate each domain. The transparent yellow boxes in panels A and B show relatively well conserved regions in ScsBα and DsbDα, respectively. The transparent green box in panel A shows the presence of the additional subdomain in ScsBα compared to DsbDα. Yellow bars in panel A indicate the predicted transmembrane segments of ScsBβ using HMMTOP (http://www.enzim.hu/hmmtop/) (G. E. Tusnady and I. Simon, Bioinformatics 17:849-850, 2001). Esc_col represents Escherichia coli NP_418559 (DDBJ/EMBL/GenBank accession number); Cam_jej, Campylobacter jejuni NP_281786; Nei_men, Neisseria meningitidis NP_274527; Bur_xen, Burkholderia xenovorans YP_560642; Bor_par, Bordetella parapertussis NP_882425; Myx_xan, Myxococcus xanthus YP_628569; She_one, Shewanella oneidensis NP_716329; Chl_tep, Chlorobium tepidum NP_661966; Cox_bur, Coxiella burnetii NP_820704; Chl_tra, Chlamydia trachomatis YP_328419; Cau_cre, Caulobacter crescentus NP_419036; Sal_typ, Salmonella Typhimurium NP_460087; Sac_deg, Saccharophagus degradans YP_526043; Rho_rub, Rhodospirillum rubrum YP_428140; Glo_vio, Gloeobacter violaceus NP_926909; Hyp_nep, Hyphomonas neptunium YP_761830. In panel C, the sequences of separately existing ScsBαs are selected from Brucella suis 1330, Rhodobacter sphaeroides 2.4.1, Agrobacterium tumefaciens strain C58, Ehrlichia ruminant strain Welgevonden. The two putative reactive cysteines are indicated in red on top of the alignment. (D) Secondary structure prediction of E. coli DsbDα (EcDsbDα) and C. crescentus ScsBα (CcScsBα) using PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/). The upper symbols for the α-helices and the beta-sheets in EcDsbDα are adopted from the structure of EcDsbDα (C. W. Goulding et al., Biochemistry 41:6920-6927, 2002; P. W. Haebel et al. EMBO J. 21:4774-4784, 2002). In both sequences, the signal sequences are not included. (E) Phylogenetic analysis of the DsbD superfamily in eubacteria. See the legend to Fig. 2 for further details. Note that the amino acid sequences of all the DsbD superfamily members listed in Table 1 and in Text S1 were used to build this tree. Download Figure S1, PDF file, 2.3 MB.
Prediction of substrate proteins of CcScsB. Sequence alignment of Salmonella Typhimurium ScsC (StScsC) and EcDsbG (A), and that of EcDsbC and StScsC (B) using BLAST analyses. (C) STRING analysis of CcScsB (http://string-db.org/). CC0220, CC1879, and CC1673 are indicated as PrxL, ScsC, and PprX in red, respectively. Download Figure S2, PDF file, 0.5 MB.
Essentiality of ScsB in C. crescentus and complementation by Rhodobacter capsulatus CcdA (RcCcdA). The C. crescentus scsB deletion strain could be obtained only in the presence of a complementing plasmid, suggesting that scsB is essential in that bacterium. Note that the plasmid used to generate the mutant strain has a chloramphenicol (Cm) resistance gene [pscsB (Cm): pSC161]. To further test the essentiality of ScsB in C. crescentus, we transformed the scsB mutant harboring pSC161 with pRVMCS-2 [vector (km)]. pRVMCS-2, which carries a kanamycin (Km) resistance gene, has the same replication origin as pSC161. We selected the transformants on a kanamycin-containing plate. Because pSC161 and pRVMCS-2 have the same replication origin and because the scsB mutant colonies were selected on kanamycin, only pRVMCS-2 should remain in the bacteria if ScsB does not have an essential function in C. crescentus. In contrast, if scsB is essential, we expect the bacteria to keep the minimal number of copies of pSC161 required for growth together with pRVMCS-2. As a positive control, pRVMCS-2 carrying a copy of scsB [pscsB (Km): pSC166] was also transformed. As shown in the upper panel (the upper two panels are the same data but have a different contrast for visualization), we observed that the strain transformed with pRVMCS-2 [vector (Km)] grew more poorly on a Km plate than the strain transformed with pscsB (Km) (pSC166) [it took more days for it to form colonies than for the strain transformed with pscsB (Km)]. Then, to test whether the transformed bacteria kept the original pSC161 plasmid [pscsB (Cm)], seven colonies from the differently transformed strains were streaked on a Cm plate as shown in the lower panel. We found that all the colonies originating from the bacteria transformed with the empty pRVMCS-2 plasmid were able to grow on chloramphenicol, which indicates that they still contain the original pSC161 plasmid. In contrast, six of the seven colonies corresponding to the bacteria transformed with pRVMCS-2 carrying the scsB gene were not able to grow on chloramphenicol. This validates our plasmid-replacing experiment and further suggests that ScsB is essential in C. crescentus. We also tested whether R. capsulatus ccdA can complement the growth defect of a scsB deletion strain using the same approach. As shown in this figure, the scsB strain complemented with pRVMCS-2 carrying the ccdA gene [pccdA (Km)] grew less efficiently than the strain complemented with pscsB (Km) but better than the strain transformed with the empty pRVMCS-2 plasmid (upper panel). Moreover, none of the colonies tested was able to grow on chloramphenicol (lower panel), indicating that the bacteria lost the original pSC161 plasmid (Cm) and suggesting that RcccdA can complement scsB in C. crescentus. The essentiality of ScsB was recently confirmed by Christen et al., who identified 480 essential open reading frames, including scsB, in the genome of C. crescentus using hypersaturated transposon mutagenesis coupled with high-throughput sequencing (B. Christen et al., Mol. Syst. Biol. 7:528, 2011). Download Figure S3, PDF file, 1.1 MB.
Prediction of the electron donor of PprX. (A) Thioredoxin-like proteins predicted to be exported in C. crescentus. CC0374, CC0375, CC1879, and CC0220 are already described in the main text. For the others, we used the sequences of known thioredoxin-like proteins as queries for BLAST analysis. Their potential export to the envelope was predicted by the presence of signal sequences or transmembrane segments using SignalP 3.0 or TMHMM 2.0, as described in Materials and Methods. The sequences in bold with underlines represent signal sequences or transmembrane segments. The two cysteines in Cys–Xaa–Xaa–Cys or Cys–Xaa–Xaa–Xaa–Cys are indicated in red. (B) STRING analysis of CcTlpA (CC2210). The reason why STRING analysis shows MsrB as an interacting protein is that a homologue of TlpA in a certain organism forms a fusion protein with a methionine sulfoxide reductase. MsrB is indicated by an arrow in blue. (C) Sequence alignment of Bradyrhizobium japonicum TlpA (BrTlpA) and CcTlpA using BLAST. Download Figure S4, PDF file, 0.4 MB.
CcscsC can complement the mucoid phenotype in dsbC mdoG E. coli. (A) Sequence alignment of CcScsC and EcDsbC using BLAST. The letters in yellow boxes in the alignment indicate Cys–Xaa–Xaa–Cys and cis-Pro regions in the active sites of the Trx family. The box lined in red shows the big mismatches of the two proteins in their N termini. EcDsbC and CcScsC are homologous in their thioredoxin domains, but there are many gaps in the alignment, and the N-terminal domain is not aligned at all, which gives a high expectation value of 0.57, suggesting they have very low homology. (B) Protein disulfide isomerase activity of CcScsC. To further support the idea that CcScsC is a protein disulfide isomerase, we decided to test the activity in E. coli using the mucoid phenotype, which depends on DsbC (P. Leverrier et al. J. Biol. Chem. 286:16734-16742, 2011). Briefly, when mdoG E. coli cells are grown in a minimal medium, colonies become mucoid and the mucoidy depends on RcsF (M. P. Castanie-Cornet et al., J. Bacteriol. 188:4264-4270, 2006). We recently showed that RcsF has nonconsecutive disulfide bonds and its folding depends on protein disulfide isomerase activity of DsbC, so the dsbC mdoG strain is not mucoid (Leverrier et al., J. Biol. Chem. 286:16734-16742). Therefore, if CcScsC has a protein disulfide isomerization activity, it would restore mucoidy phenotype in dsbC mdoG strain. Since CcScsC appears to be a cytoplasmic-membrane lipoprotein, its functionality in E. coli might be affected by tethering it in the cytoplasmic membrane. Therefore, we engineered CcScsC so that the signal sequence is replaced by that of E. coli DsbA (DsbAss) to make it a free soluble protein in the periplasm (C. F. Schierle et al., J. Bacteriol. 185:5706-5713, 2003). A plasmid encoding DsbAss-CcScsC (left panel; pSC108) or a vector (right panel) was transformed into a dsbC mdoG E. coli strain. Cells were grown on M63 minimal medium with 0.2% glucose as a carbon source and 10 µM IPTG as an inducer of the protein. While the mdoG dsbC strain transformed with a vector does not exhibit mucoidy, the strain transformed with a CcscsC plasmid is mucoid, showing the activity of protein disulfide isomerization of CcScsC. Download Figure S5, PDF file, 0.7 MB.
CcScsC forms a homodimer. (A) Secondary structure prediction of EcDsbC, CcScsC, and StScsC using PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/). The boxes lined in red indicate the dimerization domain of DsbC and the putative one of CcScsC. The boxes lined in light green shows Cys–Xaa–Xaa–Cys and cis-Pro regions in the active sites of thioredoxin domains. The box lined in blue indicates a conserved putative hinge motif (EHPE) found in the N-terminal helical domain of CcScsC. In all the sequences, the signal sequences are not included. Gel filtration analyses of CcScsC (B) and StScsC (C) with DsbC. We expressed CcScsC and StScsC with a His tag but without signal sequence in E. coli (the calculated molecular masses of the monomers are 25 kDa and 22 kDa, respectively), purified them, and performed gel filtration chromatography in which the two ScsCs were coinjected with the purified DsbC. In the other gel filtration experiments to obtain molecular masses using standard proteins, the calculated molecular masses of CcScsC and StScsC with a His tag were 37 to 57 kDa, depending on the standards and conditions, and around 20 kDa, respectively. Download Figure S6, PDF file, 0.8 MB.
In vivo redox states of CcScsC and EcDsbC (A) and PprX (B) expressed in the dsbD E. coli strain. As indicated, additional wild-type or cysteine-mutant EcDsbD or CcScsB was expressed. Cells were induced by 0.2% l-arabinose for pdsbD (pEJS92) and pscsB (pSC27) and by 10 µM IPTG for expression of CcScsC (pSC129) and pSC127 (PprX). (A) Strains transformed with pSC129 were cotransformed with pBAD43 as a vector control (lane 1), pdsbD (lane 2), or pscsB (lanes 3 and 4). (B) All strains were transformed with pSC127 and cotransformed with a second vector (pBAD43; lanes 1 and 2), pdsbD encoding wild-type DsbD (pEJS92; lanes 3, 6, and 7), pdsbD encoding DsbD C103A (pEJS94; lane 4), or pdsbD encoding DsbD C461A/C464A (pEJS96; lane 5). Cells were treated to determine redox states of proteins as described for Fig. 3. Proteins were visualized by Western blotting using an antibody against the Flag epitope or an antibody against EcDsbC. The strain background used was FED126 (the dsbD mutant), except that E. coli MC1000 (wild type) was used for lane 1 in panel B. *, indicates small portions of reduced proteins. Download Figure S7, PDF file, 0.1 MB.
Distribution of DsbD, CcdA, ScsB, ScsC, PprX, and PrxL from 529 bacteria.
Strains and plasmids (A) and primers (B) used in this study.
ACKNOWLEDGMENTS
We gratefully acknowledge Michael Laub and Celeste Peterson at MIT. They kindly donated all plasmids used for C. crescentus and shared many molecular biology techniques for C. crescentus with us. We also acknowledge Pauline Leverrier for helping us to perform the physiological assay for protein disulfide isomerase.
This work was supported by a grant to J.B. from the National Institutes of Health GMO41883, and by the European Research Council (FP7/2007 –2013) ERC independent researcher starting grant 282335—sulfenic to J.F.C. J.F.C. is a Chercheur Qualifié of the Belgian FNRS. J.B. is an American Cancer Society Professor. Financial support from NIH GM050389 to L.B.P. is also acknowledged.
Footnotes
Citation Cho S-H, et al. 2012. A new family of membrane electron transporters and its substrates, including a new cell envelope peroxiredoxin, reveal a broadened reductive capacity of the oxidative bacterial cell envelope. mBio 3(2):e00291-11. doi:10.1128/mBio.00291-11.
REFERENCES
- 1. Kadokura H, Katzen F, Beckwith J. 2003. Protein disulfide bond formation in prokaryotes. Annu. Rev. Biochem. 72:111–135 [DOI] [PubMed] [Google Scholar]
- 2. Bardwell JC, McGovern K, Beckwith J. 1991. Identification of a protein required for disulfide bond formation in vivo. Cell 67:581–589 [DOI] [PubMed] [Google Scholar]
- 3. Bardwell JC, et al. 1993. A pathway for disulfide bond formation in vivo. Proc. Natl. Acad. Sci. U. S. A. 90:1038–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guilhot C, Jander G, Martin NL, Beckwith J. 1995. Evidence that the pathway of disulfide bond formation in Escherichia coli involves interactions between the cysteines of DsbB and DsbA. Proc. Natl. Acad. Sci. U. S. A. 92:9895–9899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bader M, Muse W, Ballou DP, Gassner C, Bardwell JC. 1999. Oxidative protein folding is driven by the electron transport system. Cell 98:217–227 [DOI] [PubMed] [Google Scholar]
- 6. Zapun A, Missiakas D, Raina S, Creighton TE. 1995. Structural and functional characterization of DsbC, a protein involved in disulfide bond formation in Escherichia coli. Biochemistry 34:5075–5089 [DOI] [PubMed] [Google Scholar]
- 7. Rietsch A, Belin D, Martin N, Beckwith J. 1996. An in vivo pathway for disulfide bond isomerization in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 93:13048–13053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Joly JC, Swartz JR. 1997. In vitro and in vivo redox states of the Escherichia coli periplasmic oxidoreductases DsbA and DsbC. Biochemistry 36:10067–10072 [DOI] [PubMed] [Google Scholar]
- 9. Depuydt M, et al. 2009. A periplasmic reducing system protects single cysteine residues from oxidation. Science 326:1109–1111 [DOI] [PubMed] [Google Scholar]
- 10. Metheringham R, Griffiths L, Crooke H, Forsythe S, Cole J. 1995. An essential role for DsbA in cytochrome c synthesis and formate-dependent nitrite reduction by Escherichia coli K-12. Arch. Microbiol. 164:301–307 [DOI] [PubMed] [Google Scholar]
- 11. Fabianek RA, Hennecke H, Thöny-Meyer L. 1998. The active-site cysteines of the periplasmic thioredoxin-like protein CcmG of Escherichia coli are important but not essential for cytochrome c maturation in vivo. J. Bacteriol. 180:1947–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stewart EJ, Katzen F, Beckwith J. 1999. Six conserved cysteines of the membrane protein DsbD are required for the transfer of electrons from the cytoplasm to the periplasm of Escherichia coli. EMBO J. 18:5963–5971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Katzen F, Deshmukh M, Daldal F, Beckwith J. 2002. Evolutionary domain fusion expanded the substrate specificity of the transmembrane electron transporter DsbD. EMBO J. 21:3960–3969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Deshmukh M, Brasseur G, Daldal F. 2000. Novel Rhodobacter capsulatus genes required for the biogenesis of various c-type cytochromes. Mol. Microbiol. 35:123–138 [DOI] [PubMed] [Google Scholar]
- 15. Katzen F, Beckwith J. 2000. Transmembrane electron transfer by the membrane protein DsbD occurs via a disulfide bond cascade. Cell 103:769–779 [DOI] [PubMed] [Google Scholar]
- 16. Kim JH, Kim SJ, Jeong DG, Son JH, Ryu SE. 2003. Crystal structure of DsbDgamma reveals the mechanism of redox potential shift and substrate specificity. FEBS Lett. 543:164–169 [DOI] [PubMed] [Google Scholar]
- 17. Cho SH, Porat A, Ye J, Beckwith J. 2007. Redox-active cysteines of a membrane electron transporter DsbD show dual compartment accessibility. EMBO J. 26:3509–3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cho SH, Beckwith J. 2009. Two snapshots of electron transport across the membrane: insights into the structure and function of DsbD. J. Biol. Chem. 284:11416–11424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Collet JF, Riemer J, Bader MW, Bardwell JC. 2002. Reconstitution of a disulfide isomerization system. J. Biol. Chem. 277:26886–26892 [DOI] [PubMed] [Google Scholar]
- 20. Haebel PW, Goldstone D, Katzen F, Beckwith J, Metcalf P. 2002. The disulfide bond isomerase DsbC is activated by an immunoglobulin-fold thiol oxidoreductase: crystal structure of the DsbC-DsbDalpha complex. EMBO J. 21:4774–4784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stirnimann CU, et al. 2005. Structural basis and kinetics of DsbD-dependent cytochrome c maturation. Structure 13:985–993 [DOI] [PubMed] [Google Scholar]
- 22. Gupta SD, Wu HC, Rick PD. 1997. A Salmonella typhimurium genetic locus which confers copper tolerance on copper-sensitive mutants of Escherichia coli. J. Bacteriol. 179:4977–4984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Porat A, Cho SH, Beckwith J. 2004. The unusual transmembrane electron transporter DsbD and its homologues: a bacterial family of disulfide reductases. Res. Microbiol. 155:617–622 [DOI] [PubMed] [Google Scholar]
- 24. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 25. Seaver LC, Imlay JA. 2001. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J. Bacteriol. 183:7173–7181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wood ZA, Schröder E, Robin Harris J, Poole LB. 2003. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem. Sci. 28:32–40 [DOI] [PubMed] [Google Scholar]
- 27. Cha MK, Kim HK, Kim IH. 1995. Thioredoxin-linked “thiol peroxidase” from periplasmic space of Escherichia coli. J. Biol. Chem. 270:28635–28641 [DOI] [PubMed] [Google Scholar]
- 28. Tao K. 2008. Subcellular localization and in vivo oxidation-reduction kinetics of thiol peroxidase in Escherichia coli. FEMS Microbiol. Lett. 289:41–45 [DOI] [PubMed] [Google Scholar]
- 29. Rietsch A, Bessette P, Georgiou G, Beckwith J. 1997. Reduction of the periplasmic disulfide bond isomerase, DsbC, occurs by passage of electrons from cytoplasmic thioredoxin. J. Bacteriol. 179:6602–6608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thanbichler M, Iniesta AA, Shapiro L. 2007. A comprehensive set of plasmids for vanillate- and xylose-inducible gene expression in Caulobacter crescentus. Nucleic Acids Res. 35:e137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vestweber D, Schatz G. 1988. Mitochondria can import artificial precursor proteins containing a branched polypeptide chain or a carboxy-terminal stilbene disulfonate. J. Cell Biol. 107:2045–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cha MK, Kim WC, Lim CJ, Kim K, Kim IH. 2004. Escherichia coli periplasmic thiol peroxidase acts as lipid hydroperoxide peroxidase and the principal antioxidative function during anaerobic growth. J. Biol. Chem. 279:8769–8778 [DOI] [PubMed] [Google Scholar]
- 33. Baker LM, Poole LB. 2003. Catalytic mechanism of thiol peroxidase from Escherichia coli. Sulfenic acid formation and overoxidation of essential CYS61. J. Biol. Chem. 278:9203–9211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jeong W, Cha MK, Kim IH. 2000. Thioredoxin-dependent hydroperoxide peroxidase activity of bacterioferritin comigratory protein (BCP) as a new member of the thiol-specific antioxidant protein (TSA)/alkyl hydroperoxide peroxidase C (AhpC) family. J. Biol. Chem. 275:2924–2930 [DOI] [PubMed] [Google Scholar]
- 35. Nelson KJ, et al. 2011. Analysis of the peroxiredoxin family: using active-site structure and sequence information for global classification and residue analysis. Proteins 79:947–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Soito L, et al. 2011. PREX: peroxiredoxin classification indEX, a database of subfamily assignments across the diverse peroxiredoxin family. Nucleic Acids Res. 39:D332–D337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liao SJ, Yang CY, Chin KH, Wang AH, Chou SH. 2009. Insights into the alkyl peroxide reduction pathway of Xanthomonas campestris bacterioferritin comigratory protein from the trapped intermediate-ligand complex structures. J. Mol. Biol. 390:951–966 [DOI] [PubMed] [Google Scholar]
- 38. Reeves SA, Parsonage D, Nelson KJ, Poole LB. 2011. Kinetic and thermodynamic features reveal that E. coli BCP is an unusually versatile peroxiredoxin. Biochemistry 50:8970–8981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Goulding CW, et al. 2002. Thiol-disulfide exchange in an immunoglobulin-like fold: structure of the N-terminal domain of DsbD. Biochemistry 41:6920–6927 [DOI] [PubMed] [Google Scholar]
- 40. Stadtman ER, Moskovitz J, Levine RL. 2003. Oxidation of methionine residues of proteins: biological consequences. Antioxid. Redox Signal. 5:577–582 [DOI] [PubMed] [Google Scholar]
- 41. Kurz M, et al. 2008. Cloning, expression, purification and characterization of a DsbA-like protein from Wolbachia pipientis. Protein Expr. Purif. 59:266–273 [DOI] [PubMed] [Google Scholar]
- 42. Sevier CS, et al. 2005. The prokaryotic enzyme DsbB may share key structural features with eukaryotic disulfide bond forming oxidoreductases. Protein Sci. 14:1630–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Denoncin K, Vertommen D, Paek E, Collet JF. 2010. The protein-disulfide isomerase DsbC cooperates with SurA and DsbA in the assembly of the essential beta-barrel protein LptD. J. Biol. Chem. 285:29425–29433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Leverrier P, et al. 2011. Crystal structure of the outer membrane protein RCSF, a new substrate for the periplasmic protein disulfide isomerase DSBC. J. Biol. Chem. 286:16734–16742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hiniker A, Bardwell JC. 2004. In vivo substrate specificity of periplasmic disulfide oxidoreductases. J. Biol. Chem. 279:12967–12973 [DOI] [PubMed] [Google Scholar]
- 46. Berkmen M, Boyd D, Beckwith J. 2005. The nonconsecutive disulfide bond of Escherichia coli phytase (AppA) renders it dependent on the protein-disulfide isomerase, DsbC. J. Biol. Chem. 280:11387–11394 [DOI] [PubMed] [Google Scholar]
- 47. Kawano S, Kakuta Y, Kimura M. 2002. Guanine binding site of the Nicotiana glutinosa ribonuclease NW revealed by X-ray crystallography. Biochemistry 41:15195–15202 [DOI] [PubMed] [Google Scholar]
- 48. Sun XX, Wang CC. 2000. The N-terminal sequence (residues 1–65) is essential for dimerization, activities, and peptide binding of Escherichia coli DsbC. J. Biol. Chem. 275:22743–22749 [DOI] [PubMed] [Google Scholar]
- 49. Zhao Z, Peng Y, Hao SF, Zeng ZH, Wang CC. 2003. Dimerization by domain hybridization bestows chaperone and isomerase activities. J. Biol. Chem. 278:43292–43298 [DOI] [PubMed] [Google Scholar]
- 50. Segatori L, Paukstelis PJ, Gilbert HF, Georgiou G. 2004. Engineered DsbC chimeras catalyze both protein oxidation and disulfide-bond isomerization in Escherichia coli: reconciling two competing pathways. Proc. Natl. Acad. Sci. U. S. A. 101:10018–10023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Brot N, et al. 2006. The thioredoxin domain of Neisseria gonorrhoeae PilB can use electrons from DsbD to reduce downstream methionine sulfoxide reductases. J. Biol. Chem. 281:32668–32675 [DOI] [PubMed] [Google Scholar]
- 52. Skaar EP, et al. 2002. The outer membrane localization of the Neisseria gonorrhoeae MsrA/B is involved in survival against reactive oxygen species. Proc. Natl. Acad. Sci. U. S. A. 99:10108–10113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Parsonage D, et al. 2005. Analysis of the link between enzymatic activity and oligomeric state in AhpC, a bacterial peroxiredoxin. Biochemistry 44:10583–10592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Horta BB, de Oliveira MA, Discola KF, Cussiol JR, Netto LE. 2010. Structural and biochemical characterization of peroxiredoxin Qbeta from Xylella fastidiosa: catalytic mechanism and high reactivity. J. Biol. Chem. 285:16051–16065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Loferer H, Bott M, Hennecke H. 1993. Bradyrhizobium japonicum TlpA, a novel membrane-anchored thioredoxin-like protein involved in the biogenesis of cytochrome aa3 and development of symbiosis. EMBO J. 12:3373–3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Christen B, et al. 2011. The essential genome of a bacterium. Mol. Syst. Biol. 7:528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kumar P, Sannigrahi S, Scoullar J, Kahler CM, Tzeng YL. 2011. Characterization of DsbD in Neisseria meningitidis. Mol. Microbiol. 79:1557–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ely B. 1991. Genetics of Caulobacter crescentus. Methods Enzymol. 204:372–384 [DOI] [PubMed] [Google Scholar]
- 59. Gora KG, et al. 2010. A cell-type-specific protein-protein interaction modulates transcriptional activity of a master regulator in Caulobacter crescentus. Mol. Cell 39:455–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Altschul SF, et al. 1997. Gapped BLAST and psi-blast: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bryson K, et al. 2005. Protein structure prediction servers at university college London. Nucleic Acids Res. 33:W36–W38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bendtsen JD, Nielsen H, von Heijne G, Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783–795 [DOI] [PubMed] [Google Scholar]
- 64. Bravo J, Fernández E, Ribó M, de Llorens R, Cuchillo CM. 1994. A versatile negative-staining ribonuclease zymogram. Anal. Biochem. 219:82–86 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental materials and methods. Download Text S1, DOC file, 0.1 MB.
Amino acid sequence analyses of ScsB and DsbD. Sequence alignments of ScsB (A), DsbD (B), and ScsBα (C). The alignments were performed using ClustalW (http://www.ebi.ac.uk/Tools/msa/clustalw2/). In panels A and B, two putative reactive cysteines in each domain are indicated in red and bold on top of the alignments. Putative linker regions separate the three domains (from the N-terminal α to the C-terminal γ through β in the middle), which are shown in transparent gray in the alignments to discriminate each domain. The transparent yellow boxes in panels A and B show relatively well conserved regions in ScsBα and DsbDα, respectively. The transparent green box in panel A shows the presence of the additional subdomain in ScsBα compared to DsbDα. Yellow bars in panel A indicate the predicted transmembrane segments of ScsBβ using HMMTOP (http://www.enzim.hu/hmmtop/) (G. E. Tusnady and I. Simon, Bioinformatics 17:849-850, 2001). Esc_col represents Escherichia coli NP_418559 (DDBJ/EMBL/GenBank accession number); Cam_jej, Campylobacter jejuni NP_281786; Nei_men, Neisseria meningitidis NP_274527; Bur_xen, Burkholderia xenovorans YP_560642; Bor_par, Bordetella parapertussis NP_882425; Myx_xan, Myxococcus xanthus YP_628569; She_one, Shewanella oneidensis NP_716329; Chl_tep, Chlorobium tepidum NP_661966; Cox_bur, Coxiella burnetii NP_820704; Chl_tra, Chlamydia trachomatis YP_328419; Cau_cre, Caulobacter crescentus NP_419036; Sal_typ, Salmonella Typhimurium NP_460087; Sac_deg, Saccharophagus degradans YP_526043; Rho_rub, Rhodospirillum rubrum YP_428140; Glo_vio, Gloeobacter violaceus NP_926909; Hyp_nep, Hyphomonas neptunium YP_761830. In panel C, the sequences of separately existing ScsBαs are selected from Brucella suis 1330, Rhodobacter sphaeroides 2.4.1, Agrobacterium tumefaciens strain C58, Ehrlichia ruminant strain Welgevonden. The two putative reactive cysteines are indicated in red on top of the alignment. (D) Secondary structure prediction of E. coli DsbDα (EcDsbDα) and C. crescentus ScsBα (CcScsBα) using PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/). The upper symbols for the α-helices and the beta-sheets in EcDsbDα are adopted from the structure of EcDsbDα (C. W. Goulding et al., Biochemistry 41:6920-6927, 2002; P. W. Haebel et al. EMBO J. 21:4774-4784, 2002). In both sequences, the signal sequences are not included. (E) Phylogenetic analysis of the DsbD superfamily in eubacteria. See the legend to Fig. 2 for further details. Note that the amino acid sequences of all the DsbD superfamily members listed in Table 1 and in Text S1 were used to build this tree. Download Figure S1, PDF file, 2.3 MB.
Prediction of substrate proteins of CcScsB. Sequence alignment of Salmonella Typhimurium ScsC (StScsC) and EcDsbG (A), and that of EcDsbC and StScsC (B) using BLAST analyses. (C) STRING analysis of CcScsB (http://string-db.org/). CC0220, CC1879, and CC1673 are indicated as PrxL, ScsC, and PprX in red, respectively. Download Figure S2, PDF file, 0.5 MB.
Essentiality of ScsB in C. crescentus and complementation by Rhodobacter capsulatus CcdA (RcCcdA). The C. crescentus scsB deletion strain could be obtained only in the presence of a complementing plasmid, suggesting that scsB is essential in that bacterium. Note that the plasmid used to generate the mutant strain has a chloramphenicol (Cm) resistance gene [pscsB (Cm): pSC161]. To further test the essentiality of ScsB in C. crescentus, we transformed the scsB mutant harboring pSC161 with pRVMCS-2 [vector (km)]. pRVMCS-2, which carries a kanamycin (Km) resistance gene, has the same replication origin as pSC161. We selected the transformants on a kanamycin-containing plate. Because pSC161 and pRVMCS-2 have the same replication origin and because the scsB mutant colonies were selected on kanamycin, only pRVMCS-2 should remain in the bacteria if ScsB does not have an essential function in C. crescentus. In contrast, if scsB is essential, we expect the bacteria to keep the minimal number of copies of pSC161 required for growth together with pRVMCS-2. As a positive control, pRVMCS-2 carrying a copy of scsB [pscsB (Km): pSC166] was also transformed. As shown in the upper panel (the upper two panels are the same data but have a different contrast for visualization), we observed that the strain transformed with pRVMCS-2 [vector (Km)] grew more poorly on a Km plate than the strain transformed with pscsB (Km) (pSC166) [it took more days for it to form colonies than for the strain transformed with pscsB (Km)]. Then, to test whether the transformed bacteria kept the original pSC161 plasmid [pscsB (Cm)], seven colonies from the differently transformed strains were streaked on a Cm plate as shown in the lower panel. We found that all the colonies originating from the bacteria transformed with the empty pRVMCS-2 plasmid were able to grow on chloramphenicol, which indicates that they still contain the original pSC161 plasmid. In contrast, six of the seven colonies corresponding to the bacteria transformed with pRVMCS-2 carrying the scsB gene were not able to grow on chloramphenicol. This validates our plasmid-replacing experiment and further suggests that ScsB is essential in C. crescentus. We also tested whether R. capsulatus ccdA can complement the growth defect of a scsB deletion strain using the same approach. As shown in this figure, the scsB strain complemented with pRVMCS-2 carrying the ccdA gene [pccdA (Km)] grew less efficiently than the strain complemented with pscsB (Km) but better than the strain transformed with the empty pRVMCS-2 plasmid (upper panel). Moreover, none of the colonies tested was able to grow on chloramphenicol (lower panel), indicating that the bacteria lost the original pSC161 plasmid (Cm) and suggesting that RcccdA can complement scsB in C. crescentus. The essentiality of ScsB was recently confirmed by Christen et al., who identified 480 essential open reading frames, including scsB, in the genome of C. crescentus using hypersaturated transposon mutagenesis coupled with high-throughput sequencing (B. Christen et al., Mol. Syst. Biol. 7:528, 2011). Download Figure S3, PDF file, 1.1 MB.
Prediction of the electron donor of PprX. (A) Thioredoxin-like proteins predicted to be exported in C. crescentus. CC0374, CC0375, CC1879, and CC0220 are already described in the main text. For the others, we used the sequences of known thioredoxin-like proteins as queries for BLAST analysis. Their potential export to the envelope was predicted by the presence of signal sequences or transmembrane segments using SignalP 3.0 or TMHMM 2.0, as described in Materials and Methods. The sequences in bold with underlines represent signal sequences or transmembrane segments. The two cysteines in Cys–Xaa–Xaa–Cys or Cys–Xaa–Xaa–Xaa–Cys are indicated in red. (B) STRING analysis of CcTlpA (CC2210). The reason why STRING analysis shows MsrB as an interacting protein is that a homologue of TlpA in a certain organism forms a fusion protein with a methionine sulfoxide reductase. MsrB is indicated by an arrow in blue. (C) Sequence alignment of Bradyrhizobium japonicum TlpA (BrTlpA) and CcTlpA using BLAST. Download Figure S4, PDF file, 0.4 MB.
CcscsC can complement the mucoid phenotype in dsbC mdoG E. coli. (A) Sequence alignment of CcScsC and EcDsbC using BLAST. The letters in yellow boxes in the alignment indicate Cys–Xaa–Xaa–Cys and cis-Pro regions in the active sites of the Trx family. The box lined in red shows the big mismatches of the two proteins in their N termini. EcDsbC and CcScsC are homologous in their thioredoxin domains, but there are many gaps in the alignment, and the N-terminal domain is not aligned at all, which gives a high expectation value of 0.57, suggesting they have very low homology. (B) Protein disulfide isomerase activity of CcScsC. To further support the idea that CcScsC is a protein disulfide isomerase, we decided to test the activity in E. coli using the mucoid phenotype, which depends on DsbC (P. Leverrier et al. J. Biol. Chem. 286:16734-16742, 2011). Briefly, when mdoG E. coli cells are grown in a minimal medium, colonies become mucoid and the mucoidy depends on RcsF (M. P. Castanie-Cornet et al., J. Bacteriol. 188:4264-4270, 2006). We recently showed that RcsF has nonconsecutive disulfide bonds and its folding depends on protein disulfide isomerase activity of DsbC, so the dsbC mdoG strain is not mucoid (Leverrier et al., J. Biol. Chem. 286:16734-16742). Therefore, if CcScsC has a protein disulfide isomerization activity, it would restore mucoidy phenotype in dsbC mdoG strain. Since CcScsC appears to be a cytoplasmic-membrane lipoprotein, its functionality in E. coli might be affected by tethering it in the cytoplasmic membrane. Therefore, we engineered CcScsC so that the signal sequence is replaced by that of E. coli DsbA (DsbAss) to make it a free soluble protein in the periplasm (C. F. Schierle et al., J. Bacteriol. 185:5706-5713, 2003). A plasmid encoding DsbAss-CcScsC (left panel; pSC108) or a vector (right panel) was transformed into a dsbC mdoG E. coli strain. Cells were grown on M63 minimal medium with 0.2% glucose as a carbon source and 10 µM IPTG as an inducer of the protein. While the mdoG dsbC strain transformed with a vector does not exhibit mucoidy, the strain transformed with a CcscsC plasmid is mucoid, showing the activity of protein disulfide isomerization of CcScsC. Download Figure S5, PDF file, 0.7 MB.
CcScsC forms a homodimer. (A) Secondary structure prediction of EcDsbC, CcScsC, and StScsC using PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/). The boxes lined in red indicate the dimerization domain of DsbC and the putative one of CcScsC. The boxes lined in light green shows Cys–Xaa–Xaa–Cys and cis-Pro regions in the active sites of thioredoxin domains. The box lined in blue indicates a conserved putative hinge motif (EHPE) found in the N-terminal helical domain of CcScsC. In all the sequences, the signal sequences are not included. Gel filtration analyses of CcScsC (B) and StScsC (C) with DsbC. We expressed CcScsC and StScsC with a His tag but without signal sequence in E. coli (the calculated molecular masses of the monomers are 25 kDa and 22 kDa, respectively), purified them, and performed gel filtration chromatography in which the two ScsCs were coinjected with the purified DsbC. In the other gel filtration experiments to obtain molecular masses using standard proteins, the calculated molecular masses of CcScsC and StScsC with a His tag were 37 to 57 kDa, depending on the standards and conditions, and around 20 kDa, respectively. Download Figure S6, PDF file, 0.8 MB.
In vivo redox states of CcScsC and EcDsbC (A) and PprX (B) expressed in the dsbD E. coli strain. As indicated, additional wild-type or cysteine-mutant EcDsbD or CcScsB was expressed. Cells were induced by 0.2% l-arabinose for pdsbD (pEJS92) and pscsB (pSC27) and by 10 µM IPTG for expression of CcScsC (pSC129) and pSC127 (PprX). (A) Strains transformed with pSC129 were cotransformed with pBAD43 as a vector control (lane 1), pdsbD (lane 2), or pscsB (lanes 3 and 4). (B) All strains were transformed with pSC127 and cotransformed with a second vector (pBAD43; lanes 1 and 2), pdsbD encoding wild-type DsbD (pEJS92; lanes 3, 6, and 7), pdsbD encoding DsbD C103A (pEJS94; lane 4), or pdsbD encoding DsbD C461A/C464A (pEJS96; lane 5). Cells were treated to determine redox states of proteins as described for Fig. 3. Proteins were visualized by Western blotting using an antibody against the Flag epitope or an antibody against EcDsbC. The strain background used was FED126 (the dsbD mutant), except that E. coli MC1000 (wild type) was used for lane 1 in panel B. *, indicates small portions of reduced proteins. Download Figure S7, PDF file, 0.1 MB.
Distribution of DsbD, CcdA, ScsB, ScsC, PprX, and PrxL from 529 bacteria.
Strains and plasmids (A) and primers (B) used in this study.