Abstract
Aged-related macular degeneration (AMD) is one of the major causes of blindness in aging population and progresses with death of retinal pigment epithelium (RPE) and photoreceptor degeneration inducing impairment of central vision.
Discovery of human induced pluripotent stem (hiPS) cells has opened new avenues for the treatment of degenerative diseases using patient specific stem cells to generate tissues and cells for autologous cell-based therapy. Recently, RPE cells were generated from hiPS cells. However, there is no evidence that those hiPS-derived RPE possess specific RPE functions that fully distinguish them from other type of cells.
Here we show for the first time that RPE generated from hiPS under defined conditions exhibit ion transport, membrane potential, polarized VEGF secretion and gene expression profile similar to those of native RPE. The hiPS-RPE could therefore be a very good candidate for RPE replacement therapy in AMD. However, these cells show rapid telomere shortening, DNA chromosomal damage and increased p21 expression that cause cell growth arrest. This rapid senescence might affect the survival of the transplanted cells in vivo and therefore, only the very early passages should be used for regeneration therapies. Future research needs to focus on the generation of “safe” as well as viable hiPS-derived somatic cells.
Keywords: Human iPS cells, retinal pigment epithelium, hiPS-RPE, gene expression, ion transport, membrane potential, telomere shortening, growth arrest
INTRODUCTION
Recent discovery of induced pluripotent stem (iPS) cells from skin fibroblasts [1–4] has opened new avenues for the treatment of many degenerative diseases for which there is no adequate therapy and has offered the possibility to generate patient-specific stem cells that bypass the immunological and ethical problems related to human embryonic stem cells (hES) cells. One of the major limitations for the use of iPS cells in therapeutic application is the presence of the retroviruses that might induce tumor formation. Nonetheless this aspect is under rigorous investigation by different research groups to generate virus-free iPS cells, which do not have integrated retro- or lentiviruses in their genome [5–7]. More importantly, as iPS cell research is still at its initial stages, it is not well established whether once differentiated, the iPS-derived cells could behave as fully functional somatic cells and rescue all impaired functions of the degenerated native cells in vivo.
Aged-related macular degeneration (AMD) is the leading cause of blindness in people over 55 years old in developed countries with a tremendous social and financial burden on society [8]. AMD starts with the deposit of drusen between RPE and Bruch’s membrane and progresses with the death of RPE and photoreceptors degeneration and eventually loss of central vision [8]. RPE comprises of a monolayer of pigmented cells with the apical membrane facing the light-sensitive outer segments of photoreceptors and the basolateral membrane facing the fenestrated capillaries of the choroid [9, 10]. It plays many crucial roles in the retina including, formation of blood/retina barrier by tight junctions, transportation of nutrients such as glucose or vitamin A from blood to the photoreceptors, conveyance of water from subretinal space to the blood, establishment of immune privilege of the eye and a constant ion composition in the subretinal space, light absorption and isomerization of the retinal in the visual cycle, secretion of growth factors and phagocytosis of the outer segments of the photoreceptors [11, 12]. Recent clinical investigations using autologous transplantation of RPE-choroid sheet to the macula of AMD patients has shown improvement in light perception [13–18], and point out the prospective for RPE autologous cell-based therapy for AMD.
Recently, generation of RPE from hES [19–21] and hiPS [22, 23] cells has been reported. The hiPS-derived RPE have been shown to be polarized, able to phagocytose and exhibit a gene expression pattern comparable to that of native RPE [23, 24]. However, both of these studies only analyzed one function in RPE cells, i.e., phagocytosis of outer segment of photoreceptors and fall short in demonstrating other RPE specific functions such as polarized VEGF secretion, ion channel activity and membrane potential. In addition, there is insufficient evidence that these cells can maintain their long-term functionality and integrity that is crucial for their implementation in cell-based therapy in clinical application. In most studies the transplanted cells have died within two weeks and long-term survival is not achieved [21, 25, 26]. Moreover, the monolayer structure characteristic of functional RPE was not formed after transplantation.
Here we show that human iPS cells generated with OCT4, SOX2, NANOG and LIN28 can be differentiated to RPE exhibiting the phenotype, physiology and molecular signature of native RPE. Using whole-cell electrophysiology, we show that the capacity for ion transport and the membrane potential of hiPS-RPE resemble those of the native RPE. However, the hiPS-RPE undergo fast telomere shortening, DNA chromosomal damage, increased p21 expression, and present growth arrest. This may affect the survival of transplanted cells in vivo and should be taken into consideration for future clinical applications.
These observations indicate that hiPS cell technology, still at its early stages, should not only be improved in terms of viral vectors used for reprogramming that might induce malignant transformation in vivo, but also for its accelerated senescence of hiPS-derived cells.
MATERIALS AND METHODS
1. Cell Culture
The IMR90-4 hiPS cell line (WiCell, Madison, WI) was cultured in feeder free conditions (WiCell procedure # SOP-CC-005), on hESC-qualified Matrigel (BD Biosciences, San Diego, CA), in mTeSR1 medium (Stem Cell Technologies, Vancouver, Canada). Human fetal native RPE cells at passage 2 (HRPEpiC, ScienCell) were cultured on poly-D-lysine (2μg/cm2, BD Biosciences, Franklin Lakes, NJ) and laminin (4μg/ml, SIGMA) coated surfaces in EpiCM epithelial cell medium (ScienCell).
2. Differentiation, Enrichment, and Expansion of hiPS-RPE Cells
The IMR90-4 hiPS cells were transferred to Aggrewells (Stem Cell Technologies) (1,000–2,000 cells/well). EBs were formed and cultured in suspension in DMEM with 10% serum replacement, 1x NEAA, 2mM glutamine and 0.1mM β-mercaptoethanol) for one day, with 10 μM nicotinamide (NIC, SIGMA, St. Louis, MO) added the next day and supplemented with 140ng/ml recombinant human activin-A (R&D Systems, Minneapolis, MN) in the presence of NIC and SB431542 (SIGMA) for the 3rd and 4th week of differentiation. Pigmented foci were visible after the 4th week. Differentiating clusters were transferred to poly-D-lysine (2μg/cm2, BD Biosciences,) and 4μg/ml laminin (SIGMA), for attachment on the 6th week and cultured in the presence of NIC for 3–5 weeks. For enrichment and expansion, pigmented areas were manually dissected, triturated, and cultured in monolayers. After 10–12 weeks of differentiation, the hiPS-RPE cells were expanded in EpiCM medium (ScienCell, Carlsbad, CA).
3. Quantitative Real-Time Polymerase Chain Reaction
Total RNA was extracted with the RNeasy kit (Qiagen, Germantown, MD), treated with RNase-free DNase I, and reverse-transcribed with oligo-dT using the SuperScript III cDNA synthesis kit (Invitrogen). Quantitative PCR was performed with the QuantiTect SYBR Green PCR Kit (Qiagen). Specific primers were designed with PrimerQuest software (Integrated DNA Technologies, Skokie IL) (Supplementary Table 1).
4. Immunostaining
Cells grown on poly-D-lysine and laminin coated plastic 8-well chambers were stained based on established protocols for the primary and secondary antibodies (Supplementary Table 2). Immunostaining conditions for anti—phospho H2A.X were as described by Marion et al [27]. Stained cells were mounted with anti-fading mounting medium (Invitrogen) and images were captured by confocal microscopy (FV1000 Confocal Microscope, Olympus, Center Valley, PA).
5. Immunoblot Analysis
Protein samples were extracted and analyzed using the NuPAGE electrophoresis and XCell western blot systems (Invitrogen). Primary and secondary antibodies were used based on their manufacturer’s protocols (Supplementary Table 2). Immunoreactive protein bands were revealed by the SuperSignal® West Dura Chemiluminescent Substrate (PIERCE, Rockford, IL) followed by X-ray imaging. The relative intensity of the protein bands was calculated using Photoshop CS4 as explained in Supplementary data.
6. Phagocytosis assay
To test the ability of hiPS-RPE cells to phagocytose in vitro, the cells were grown to confluency as monolayers on poly-D-lysine and laminin plastic dishes, incubated for 16hrs with 1 μm green fluorescent latex beads, dissociated with trypsin, re-plated on poly-D-lysine and laminin plastic chambers for a few hours to attach, and stained with an antibody to ZO-1 (Invitrogen).
7. Polarity assays
To confirm the apical localization of Na+/K+ ATPase, polarized hiPS-RPE cells grown on Transwells, were stained with the antibody to Na+/K+ ATPase and optical cross section images were taken using the XZ-scan mode of the FV1000 Confocal Microscope (Olympus). To confirm the polarized secretion of VEGF, polarized hiPS-RPE were grown on 6-well Transwell dishes and the medium from both the upper and lower reservoirs was collected after the 4th week of culture. Media samples were shipped in dry ice to Antigen Targeting and Consulting Service (ATCS Inc, Worcester, MA) for the VEGF ELISA assays.
8. Electrophysiology
Native RPE and hiPS-RPE cells plated on a poly-D-lysine and laminin coated glass coverslip were transferred to a recording chamber (volume ~700μl) attached to the stage of a Nikon E600-FN microscope. They were continuously perfused with recording solution at room temperature (23–26°C) having the following composition (in mM): NaCl 130; KCl 5; CaCl2 2; MgCl2 2; HEPES 10 and glucose 5 (pH 7.3). Cells were identified visually by infrared-differential interference contrast (IR-DIC) and fluorescence optics via a CCD camera (Dage S-75). A 60x-water immersion objective (Nikon) was used for identifying and approaching cells. Whole-cell voltage clamp of the cells were made according to the detailed methodology (Supplementary data).
9. Telomere length measurements
Relative telomere lengths between DNA samples were determined by measuring the factor by which our experimental samples differ from the control sample (native RPE passage 3) in their ratio of telomere repeat copy number to the single-copy gene’s 36B4 copy number. This ratio is proportional to the average telomere length. Therefore, telomere repeats and the single-copy gene were detected by specific primer sets as previously described [28] in standard qPCR reactions using SYBR green. The quantities of the DNA samples were normalized to 36B4 using the ΔΔCt method.
RESULTS
1. Induced pluripotent stem (iPS) cells generated from human fibroblasts silence pluripotency genes after differentiation
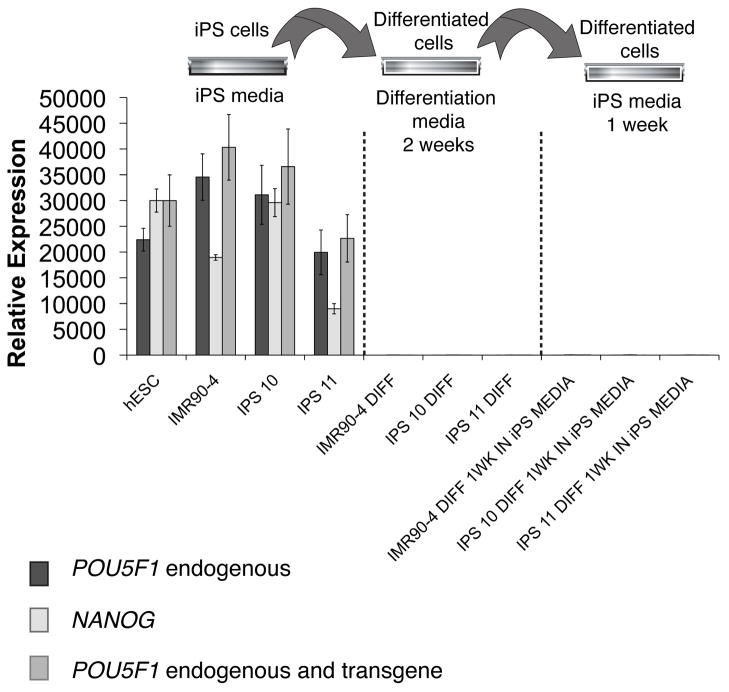
It is crucial to generate safe hiPS-RPE, with potential therapeutic applications. To check the safety of our hiPS-RPE, we first tested the ability of our hiPSCs to switch off the genes encoding pluripotency factors after culture in differentiation media. We tested two of our own hiPS cell lines (iPS10 and iPS11) that were both generated from skin fibroblasts of the same male donor using the four Yamanaka retroviral vectors for reprogramming [2], and also the iPS cell line IMR90-4 (WiCell)[29]. The IMR90-4 is a commonly used iPS cell line, with established pluripotency potential and normal karyotype (Wi-Cell). We also verified the pluripotency and chromosome stability of the cell lines iPS 10 and 11 by staining with anti-NANOG and by confirming a normal chromosome number in their metaphase spreads (Supplementary Figure 1). For differentiation, we first let the hiPSCs form EBs in suspension in DMEM containing 10%FBS for 1 week and further differentiated them in adhesion on plastic dishes in the same medium for another week. No iPS-like colonies were detected at the end of the differentiation period. Total RNA was isolated after differentiation. In parallel, to confirm that the pluripotency genes would not be re-activated in the absence of the differentiation media, iPSCs were differentiated and then cultured in iPS-media (mTeSR1) for one additional week before the RNA isolation. We then compared the levels of endogenous and transgene POU class 5 homeobox 1 (POU5F1), and endogenous Nanog homeobox (NANOG) mRNAs by qRT-PCR between undifferentiated control hiPSCs, the differentiated cells for 2 weeks, and the differentiated cells that were returned back to iPS media. To distinguish between POU5F1 endogenous and POU5F1 transgene expression, we designed primers at the 3′UTR of POU5F1, specific for the endogenous POU5F1. We detected POU5F1 endogenous expression in all three hiPS cell lines before differentiation, but POU5F1 mRNA could not be detected in the differentiated cells. As NANOG is not included in the Yamanaka reprogramming factors, it is therefore considered as a reliable endogenous marker for reprogramming. NANOG expression was also detected in the three hiPS cell lines before differentiation but was completely switched off after differentiation. In addition, returning to iPS-media after differentiation did not reactivate POU5F1 or NANOG genes (Figure 1). Therefore, our qRT-PCR data suggest that the three hiPS cell lines completely lost their pluripotency potential after differentiation and that this loss is irreversible in our culture conditions.
Figure 1. Pluripotency marker gene expression and safety test of our iPS cell lines.
qRT-PCR analysis of the expression of POU5F1 and NANOG. A standard curve was calculated for each target gene and for GAPDH with different dilutions of IMR90-4 cDNA. All samples were normalized to GAPDH. IMR90-4, iPS10 and iPS11 were cultured in mTesR1 medium for 2–3 passages before being used for this analysis. ‘IMR90-4 diff’, ‘iPS10 diff’ and ‘iPS11 diff’ are cDNAs from IMR90-4, iPS 10 and iPS 11, cultured in differentiation media for 2 weeks. Samples labeled ‘1wk in iPS media’ are from the same differentiated cells that were cultured again for an additional week in iPS media (mTeSR1) before this analysis to verify the ‘safety’ of the differentiated cells.
2. hiPS cells differentiating in RPE media gradually express RPE markers
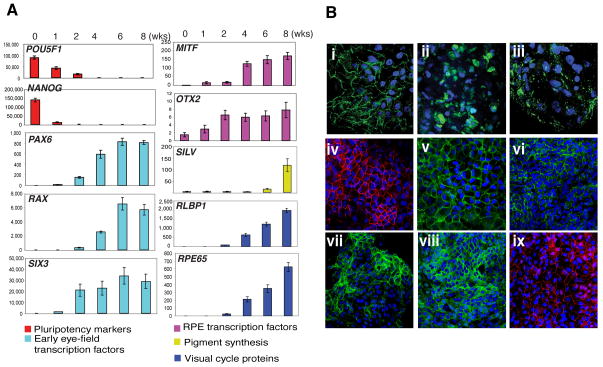
It has been reported that functional RPE can be derived from hiPS cells using IMR90-4 cell line [23]. Here we have differentiated the IMR90-4 hiPSCs to RPE using a modified protocol. The expression of several markers was monitored during the first 8 weeks of differentiation process by qRT-PCR (Figure 2A). Switching off the genes of pluripotency (POU5F1 and NANOG) seemed to be the first step in the process, occurring in the first 2 weeks. Following was the activation of the eye-field transcription factor genes, i.e., paired box 6 (PAX6), retina and anterior neural fold homeobox (RAX a.k.a RX) and SIX homeobox 3 (SIX3). Their expression levels increased significantly between the 2nd and 6th weeks and remained constant until the 8th week. We also checked the expression of genes encoding RPE transcription factors, i.e., microphthalmia-associated transcription factor (MITF) and orthodenticle homeobox 2 (OTX2). Low levels of MITF mRNA could be detected in the first two weeks after induction of differentiation, but an approximately 5-fold increase was observed at the fourth week. OTX2 gene was expressed at low levels in iPS cells, increased 2-fold in the 1st week and 4-fold in the 2nd week, and remained stable thereafter until the 8th week. As expected, genes encoding proteins responsible for the visual cycle (retinaldehyde binding protein 1, RLBP1 a.k.a. CRALBP; retinal pigment epithelium-specific protein 65kDa, RPE65) or for pigment synthesis (Silver homolog, SILV) were activated later. CRALBP and RPE65 transcripts were detected at low levels on the 2nd week, increased 20- to 30-fold at 4th week and continued to increase until the 8th week. SILV was the last gene to be activated; its mRNA was detected for the first time at low levels on the 6th week and increased ~10-fold in the 8th week. This pattern correlates with our observation of pigmented iPS-RPE after the 6th week of differentiation (Supplementary Figure 2-(E & F).
Figure 2. Monitoring the expression of pluripotency and RPE markers during differentiation of iPS to RPE.
(A), qRT-PCR analysis of the genes was performed on undifferentiated iPS cells (time 0) and during differentiation at 1wk, 2wks, 4wks, 6wks and 8wks time points. 500ng of DNAse-treated RNA was used for each sample. A standard curve was calculated for each target gene and for GAPDH with different dilutions of IMR90-4 cDNA (for POU5F1 and NANOG) or human native RPE cDNA (for all the other genes). All samples were normalized to GAPDH. Pluripotency markers were undetectable after the 2nd week of differentiation, when the early eye-field and RPE transcription factor genes started expressing. Transcripts of genes coding for visual cycle proteins (RLBP1 and RPE65) could be detected at low levels on 2nd week and their levels increased thereafter. Pigment synthesis gene SILV was activated last, starting at 6th week of differentiation, consistent with the appearance of pigmented cells in 6–8 wks old clusters. (B), RPE marker proteins detected in hiPS-RPE and in native human RPE; (i-iii), 10 weeks old clusters express ZO-1 (i), bestrophin (ii) and RPE65 (iii); (iv–vi), hiPS-RPE monolayers express ZO-1 (iv), Na+/K+ ATPase (v), Occludin (vi); (vii–ix), native human RPE expressing Na+/K+ ATPase (vii), Occludin (viii) and ZO-1 (ix). Nuclei were counter-stained with DAPI (blue). Magnification used is 60x.
In addition to gene expression at the mRNA level, we also detected several RPE protein markers on the differentiating clusters and the differentiated hiPS-RPE cells. Tight junction protein ZO-1, its binding partner Occludin, the Na+/K+ ATPase membrane channel, the visual cycle protein RPE65 and the chloride channel bestrophin (BEST1) were detected by immunostaining with specific antibodies (Figure 2B). Similar pattern of expression was observed in control experiments with native human RPE growing in monolayers (Figure 2B, vii-ix). Negative control experiments in the absence of primary antibodies were also performed (Supplementary Figure 3).
Pigmentation of differentiating hiPS-RPE cells was also monitored during differentiation in suspension culture of cell clusters and also during the expansion of differentiated hiPS-RPE in adhesion culture (Supplementary Figure 2). The hiPS cells grown in suspension in NIC-containing media form cell clusters, increasing in size and showing partial pigmentation (10–20% of cells in a cluster) after 4–6 weeks of differentiation [Supplementary Figure 2D). The average percentage of pigmented clusters in three independent differentiation experiments, for each using 16,000 hiPS cells, was 37%, with a standard deviation of 6.5%. Pigment synthesis spreads to >90% of the cells in clusters by the end of the 8th week, when the cells have also acquired a typical hexagonal shape [Supplementary Figure 2(E & F)]. For the enrichment of hiPS-RPE, pigmented clusters were trypsinized, triturated, transferred onto poly-D-lysine and laminin and cultured in expansion media. Pigmentation was initially lost in the adhesion culture (Supplementary Figure 2G), but became evident again after 4 weeks of culture in high cell density (50,000 cells / cm2), starting as pigmentation foci (Supplementary Figure 2H) and spreading to 70–80% of the cells in the monolayer of hiPS-RPE in the following 2–3 weeks (Supplementary Figure 2I)
4. hiPS-RPE are polarized and able to phagocytose
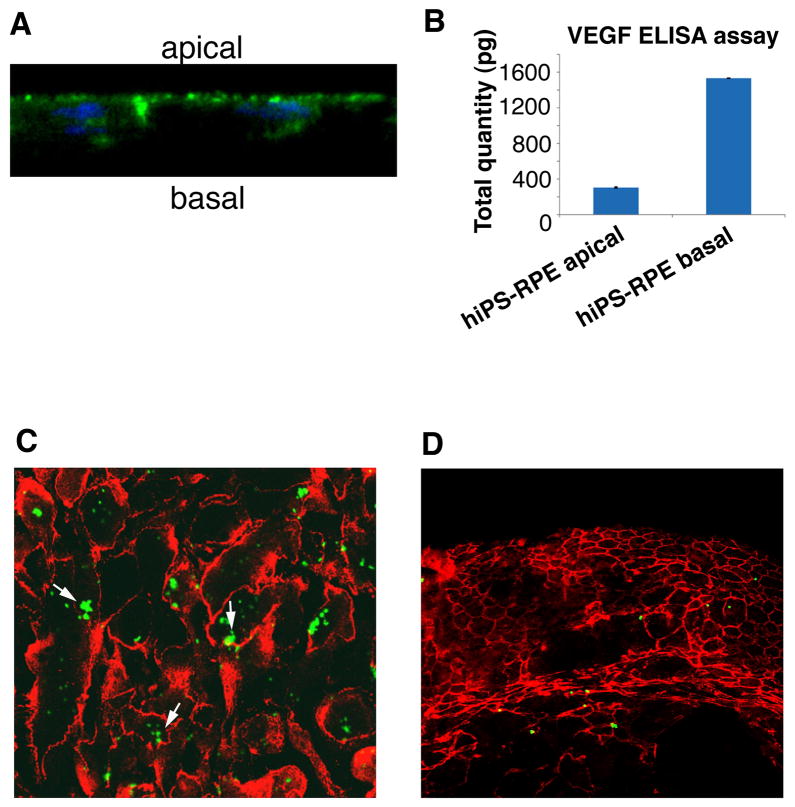
Polarity between apical and basolateral regions and phagocytosis of the outer segments of photoreceptors are important characteristics of RPE. To generate polarized hiPS-RPE, the cells were grown in monolayers on Transwell filters at a density of 50,000 cells / cm2 for 4–6 weeks. When pigmentation was observed, the polarity of the cells was assayed by immunostaining with anti-Na+/K+ ATPase for its localization in apical side and by measuring the amount of secreted VEGF from basolateral side. The Na+/K+ ATPase was localized predominantly at the apical site of the hiPS-RPE (Figure 3A), as previously reported for the RPE[30]. By comparing the morphology of the polarized hiPS-RPE cells to those of non-polarized, we have observed that the polarized hiPS-RPE are also pigmented, as shown in the merged brightfield and fluorescent images of Supplementary Figure 4. In addition, higher expression of Na+/K+ ATPase was observed in the pigmented hiPS-RPE (Supplementary Figure 4). Furthermore, to assay the polarized secretion of VEGF, the hiPS-RPE were grown on Transwells for four weeks and VEGF was detected by ELISA in the culture medium collected from both the upper and lower reservoirs. The ELISA assays showed that mainly the basal site of hiPS-RPE secretes VEGF (Figure 3B), as has been observed for the native RPE [30].
Figure 3. hiPS-RPE are polarized and able to phagocytose.
Pigmented sheets of hiPS-RPE cells after 6 weeks of culture on Transwells show polarized localization of Na+/K+ ATPase at the apical side (A) and polarized secretion of VEGF, from the basal side (B), as detected in the culture medium by ELISA. The bars (B) represent the average total amount of VEGF, detected in duplicate experiments. (C) hiPS-RPE immunostained with anti-ZO-1 have phagocytosed green fluorescent beads (arrows) added in their medium. (D) Less differentiated hiPS-derived cells in clusters, 2 weeks after induction of differentiation, show no phagocytosis capability. Images (A, C and D) were obtained by confocal microscopy with a 60x objective. A z-scan (X-Z scan mode) was performed to obtain optical cross sections, as shown in (A).
To test the ability of the hiPS-RPE to phagocytose, we incubated the cells with green fluorescent Latex 1μm beads before passaging and re-plating, washing them extensively to remove non-internalized beads. The number of cells with internalized beads was assayed by confocal microscopy, after staining with the antibody to tight junction ZO-1 to define the cell boundaries (Figure 3C). Note that the RPE cells in Figure 3C do not show the characteristic epithelial morphology, due to their passaging after incubation with the fluorescent beads, as described in the Materials and Methods. However, the majority of hiPS-RPE cells (80–90%) had phagocytosed fluorescent beads, showing that hiPS-RPE cells are functional. On the contrary, no phagocytosis was observed when less differentiated cells were used for the assay, which correlates the phagocytosis function with late RPE differentiation (Figure 3D).
5. hiPS-RPE have sodium and potassium ion transport activity and membrane potential similar to native RPE
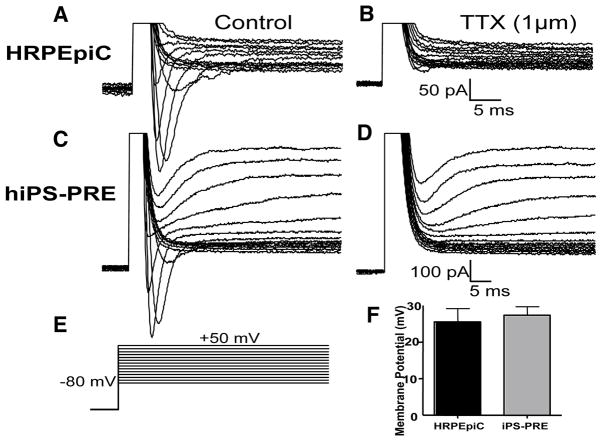
To determine if iPS-RPE are functional, similar to native RPE, we employed whole-cell patch clamp electrophysiology to measure their membrane potential and detect the presence of voltage-gated channels. Since both cell types tended to be large, we targeted single cells covering an area of ~≤40,00μm2 for recording. In all cells studied in response to the voltage-step protocol employed, both sodium and potassium currents were readily activated (Figure 4A–D), suggesting the presence of functional voltage-gated ion transporters. Moreover, bath application of the sodium channel blocker tetrodotoxin (TTX, 1μM) to the perfusate blocked all voltage-activated sodium currents, leaving potassium currents unaffected (Figure 4B & D).
Figure 4. hiPS-RPE have functional Na+ and K+ voltage gated channels and membrane potential similar to that of native RPE.
(A–C), Reprensentative recordings of voltage-gated currents in HRPEpiC (A & B) and hiPS-PRE (C &D). Voltage-activated sodium currents are blocked (B & D) by bath application of tetrodotoxin (TTX). [Note: The presence of potassium currents in both cell types.] (E), The protocol used to activate voltage-gated currents: a prepulse of −170mV (50ms), followed by 14 steps starting at a potential of −80mV and increased in 10 mV increments. (F), Graph showing the relative resting membrane potential of HRPEpiC (n=8) and iPS-PRE (n=12) cells. There was no significant difference in the membrane potential of the two cell types.
By measuring the membrane potential of the two cell types, we found no significant differences between the two. The average membrane potential of the HRPEpiC cells was 25.50+/−3.732 mV (n=8), whereas that of the hiPS-RPE cells was 27.38+/−2.388 (n=12) (Figure 4F).
6. hiPS-RPE have similar signature gene expression to native RPE
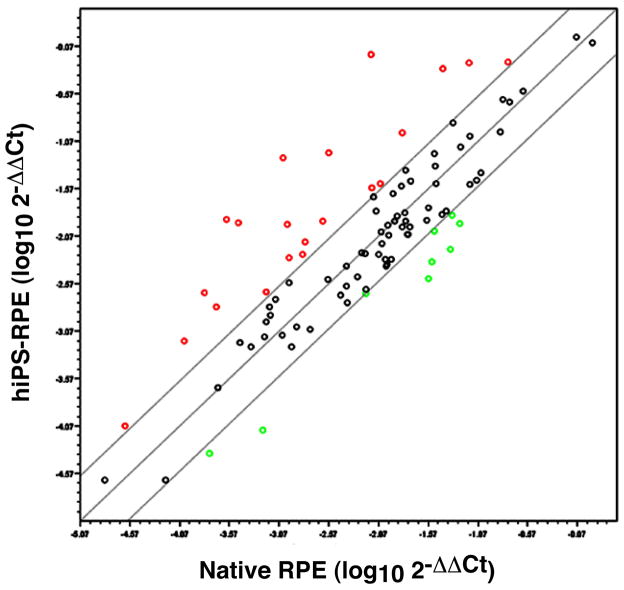
The human RPE molecular signature, consisting of 154 genes, has recently been revealed [31]. We selected 89 of these genes (Table 1), based on their expression values in human fetal and adult RPE, as reported by Strunnikova et al, as well as their association to AMD pathogenesis reviewed in the same paper[31]. We checked the expression of those genes in our pigmented hiPS-RPE cells and compared their levels to those in human native RPE cells (HRPEpiC, ScienCell) by qRT-PCR. The hiPS-RPE that were used for this analysis were the first passage of the expansion phase, growing on monolayers on 6-well dishes for 6–8 weeks, until 80–90% of them were pigmented. Two biological replicas were included in the qRT-PCR analysis. All the 89 signature genes were expressed in the hiPS-RPE as well as in the native RPE. Fifty-six of them showed similar levels of expression (<3x change) between the two cell populations (Figure 5, Table 1). The functions of these genes are discussed in Supplementary Data.
Table 1.
RPE signature gene expression in hiPS-RPE vs native RPE
| Group A
| |
|---|---|
| Gene Symbol | Fold Change |
| COL8A2 * | −2.9 |
| COX15 | −2.9 |
| MET* | −2.8 |
| ARL6IP1 * | −2.7 |
| GPR143 * | −2.6 |
| MED8 ! | −2.4 |
| SLC4A2 * ! | −2.4 |
| SMAD6 * ! | −2.4 |
| LHX2 * | −2.3 |
| SLC16A4 * ! | −2.1 |
| SLC16A1 | −2.1 |
| GULP1 | −2.0 |
| LIN7C | −2.0 |
| IFT74 | −1.9 |
| CLCN4 * ! | −1.9 |
| USP34 * | −1.7 |
| SCAMP1 | −1.7 |
| CRIM1 * ! | −1.6 |
| PHACTR2 | −1.5 |
| RAB38 * ! | −1.5 |
| MAP9 * ! | −1.5 |
| NAV3 * | −1.5 |
| SOSTDC1* | −1.3 |
| RDH11 | −1.2 |
| DMXL1 * ! | −1.2 |
| STAM2 | −1.2 |
| GEM | −1.2 |
| SLC24A1* | −1.2 |
| SORBS2 * | −1.2 |
| BMP4 * ! | −1.1 |
| PITPNA * | −1.1 |
| HSP90B1* | −1.0 |
| BEST1 * | 1.0 |
| DZIP1 * | 1.0 |
| GPM6B | 1.0 |
| SILV * | 1.1 |
| ITGAV * | 1.2 |
| SIX3 * ! | 1.3 |
| SGK3 * ! | 1.4 |
| PLCB4 * | 1.5 |
| WWTR1 * | 1.5 |
| RPE65 * | 1.5 |
| SFRP5 * ! | 1.6 |
| EFHC1 * | 1.6 |
| PDPN | 1.7 |
| LOXL1 * | 1.7 |
| CSPG5 * ! | 1.8 |
| ADCY9 | 1.9 |
| CDH3 | 1.9 |
| ENPP2 * | 2.2 |
| GPNMB * | 2.3 |
| PTPRG | 2.5 |
| EFEMP1 * | 2.8 |
| TRPM1* | 2.9 |
| CDO1 * | 2.9 |
| RRAGD * | 2.9 |
| Group B
| |
|---|---|
| Gene Symbol | Fold Change |
| MAB21L1 * | 3.2 |
| TIMP3 * | 3.4 |
| GAS1 * | 3.9 |
| SDC2 * ! | 4.1 |
| MFAP3L * ! | 4.4 |
| SULF1 * | 5.9 |
| MYRIP * | 6.4 |
| SLC6A20 * | 6.4 |
| SERPINF1 * | 6.7 |
| ASAH1* | 9.4 |
| FGFR2 | 10.6 |
| TFPI2 * | 10.7 |
| TTR * | 20.6 |
| FRZB* | 29.1 |
| DCT* | 42.6 |
| CHRNA3 * | 48.2 |
| TYRP1 * | 81.1 |
| Group C
| |
|---|---|
| Gene Symbol | Fold Change |
| SLC6A15 * | −10.9 |
| PKNOX2 * | −9.7 |
| VEGFA * | −9.2 |
| ALDH1A3 * | −7.8 |
| NRIP1 * | −6.0 |
| FOXD1* | −4.3 |
| FADS1 * | −4.0 |
| WWC2 | −3.8 |
| APLP1 * | −3.7 |
| CRX | −3.5 |
| TTLL4 * ! | −3.3 |
| IGF2BP2 | −3.2 |
| BAT2D1 * | −3.1 |
| DUSP4 * | −3.1 |
| RBP1 * | −3.0 |
| MPHOSPH9 | −3.0 |
Group A: Genes with similar gene expression in hiPS-RPE and native RPE (Fold change between −3 and +3)
Group B: Genes over-expressed in hiPS-RPE vs native RPE (Fold change > +3)
Group C: Genes under-expressed in hiPS-RPE vs native RPE (Fold change < −3)
: Genes included in the ‘core signature’ RPE gene set of 87 genes, as defined by Liao et al (2010).
Genes reported by Liao et al (2010), as expressed in fetal RPE and not in stem-cell derived RPE.
Highlighted in grey are the genes reported by Liao et al (2010) to be commonly expressed by fetal RPE and hES cells.
Figure 5. hiPS-RPE express RPE signature genes.
We assayed the expression of 89 RPE signature genes in hiPS-RPE and compared their expression levels to human native RPE by quantitative RT-PCR, using custom arrays from SABiosciences. In the scatterplot of normalized expression logarithmic values, each gene is represented by a circle, black for the genes that do not change more than 3x between the two cell populations, red for the genes that are over-expressed in hiPS-RPE and green for those that are over-expressed in native RPE.
7. hiPS-RPE undergo rapid telomere shortening and DNA damage
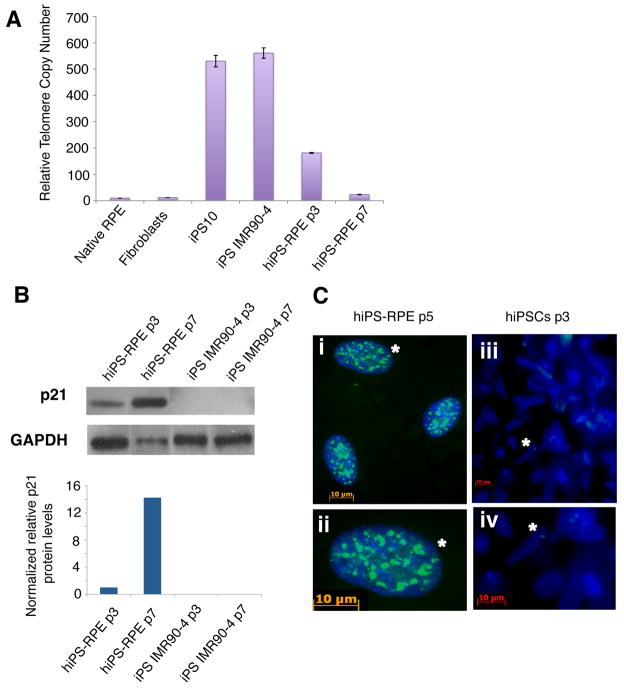
Based on the observation of a significant decrease in cell growth of hiPS-RPE between passages 5–7, we followed the senescence of hiPS-RPE by analyzing their telomere length at different passages (Figure 6A). We showed a rapid shortening of the telomeres that occurs both during differentiation of iPS to RPE and during iPS-RPE culture. At passage 3 (p3), the hiPS-RPE have shortened their telomeres by almost 60% compared to their parental hiPS cells, whereas at passage 7 (p7) their telomere length has dropped to levels similar to native RPE (Figure 6A). Consistently, at p7 we also observed cell growth arrest for the hiPS-RPE, which might suggest that cells derived from hiPS cells by in vitro differentiation exhibit faster senescence. We also investigated the expression of tumor suppressor protein p21 Waf1/Cip1 that acts as an inhibitor of cell cycle progression, in iPS-RPE in order to explain the premature senescence of these cells. Our data show that p21 level increases 14-fold in hiPS-RPE p7 compared to p3, consistent with the shortened telomere observed in the late passage (Figure 6B). Immunostaining using γH2ax antibody, the phosphorylated form of histone H2A in response to double-strand DNA damage, in hiPS-RPE at p5 and p7 showed high level of staining in the nucleus (Figure 6C).
Figure 6. hiPS-RPE exhibit rapid telomere shortening in vitro.
(A) Relative telomere copy numbers were measured by Q-PCR of genomic DNA We used a published assay [28] for telomere size measurement by quantitative PCR. This assay is based on determining relative telomere lengths between DNA samples by measuring the factor by which an experimental sample differs from a control sample in its ratio of telomere repeat copy number to the single-copy gene’s 36B4 copy number. This ratio is proportional to the average telomere length. Therefore, telomere repeats and the single-copy gene were detected in standard qPCR reactions using SYBR green. The quantities of the DNA samples were normalized to 36B4 using the ΔΔCt method. (B) The expression of the cell growth arrest marker protein p21 was analyzed by western blot. Normalized p21 levels shown in the graph were calculated by densitometry analysis of the protein bands for p21 and GAPDH. (C) hiPS-RPE have accumulated DNA damage sites at late passages. (i&ii), Immunostaining of hiPS-RPE at passage 5 with the antibody to phosphorylated histone H2Ax (γH2Ax) (green), showing sites of DNA damage in their nuclei stained with DAPI (blue). The cell marked by an asterisk in Figure 6C-(i) is shown in 6C-(ii) at high magnification. (iii & iv), Immunostaining of IMR90-4 hiPS cells at passage 3 with anti-γH2Ax (green), showing absence of sites of DNA damage in their nuclei stained with DAPI (blue). Figure 6C-(iv) is higher magnification of Figure 6C-(iii).
DISCUSSION
The RPE cells are crucial in maintaining the structure and function of the retina and photoreceptors and their dysfunctions are often related to eye diseases, such as age-related macular degeneration (AMD)[32–34]. Therefore, it is of paramount importance to generate patient-specific RPE cells that can mimic all functions and characteristics of native human RPE. Recent studies have generated RPE from human ES and iPS cells and have tested their functionality by demonstrating the expression of RPE specific proteins, RPE gene transcripts and phagocytosis of outer segment of photoreceptors [23, 35]. However, they do not consider the most important RPE functions that is regulation of ion and fluid transport, resting membrane potential, and polarized secretion of specific growth factors. Recently, the summary of functional tests that should be considered to assess RPE authenticity was published [31]. Here we report the generation of human iPS-derived RPE from the IMR90-4 human iPS cell line that express RPE morphology, specific proteins and genes as reported previously [23] and further demonstrate that hiPS-derived RPE perform similar functions to those of native RPE including ion transport, basal VEGF secretion and exhibit identical membrane potential as that of native RPE.
The ‘safety’ of iPS-derived cells is important for in vivo transplantation and therapeutic applications. We have analyzed the expression of pluripotency factors in differentiated cells derived from IMR90-4 and from two of our iPS cell lines, and have shown that they become irreversibly silent after differentiation, and therefore we called them “safe”. Nonetheless, this will need to be confirmed by transplantation in vivo and long-term observation for the absence of tumor formation. In accordance with our observations, in a few recent transplantation studies of iPS and ES cell-derived RPE, the cells die within 10–15 days [21, 25, 26] without showing any tumor formation confirming that in most cases the differentiation induces irreversible silencing of genes of pluripotency. To confirm the differentiation of hiPS cells to RPE, we have analyzed the gradual expression of RPE marker genes in a timely manner. The expression of the genes of pluripotency gradually decreases after one week of differentiation to a non-detectable level at week 4. Conversely, the expression of RPE genes increases after one week and achieves the highest level at week 6 to 8. Interestingly, the expression of SILV gene increases after 8 weeks of differentiation in accordance with the appearance of RPE pigmentation after 6 weeks. These observations are in agreement with the findings of Buchholz et al. and Lu et al. [21, 23]. In addition, we have shown the expression of RPE specific proteins confirming that the iPS cells have differentiated to RPE cells.
Polarization is one of the most significant characteristics of RPE. The RPE maintains the choriocapillaris in the normal eye and is involved in the pathogenesis of choroidal neovascularization in AMD. Vascular endothelial growth factor-A (VEGF) is secreted by the RPE cells in a polarized manner from the basal membrane [36]. Therefore, it is crucial to verify if the hiPS-RPE are also polarized. We have shown the apical localization of Na+/K+ ATPase by confocal microscopy in our hiPS-RPE confirming the polarization. In addition, our phagocytosis assay revealed that hiPS-RPE are able to phagocytose. More importantly, we demonstrated VEGF secretion from the basal membrane of hiPS-RPE, confirming that these cells not only exhibit the morphology of native RPE but also display adequate functionality comparable to that of native RPE. Although in the previous paper by Buchholz et al. [23] the lateral expression of ZO-1 protein and apical expression of Otx2 was shown, no functional assay related to the polarized cells was performed. Here we have shown for the first time that the hiPS-RPE cells secrete growth factors in a similar manner to that of native RPE. This observation is crucial for the validation of functional RPE for future clinical applications.
RPE form the outer blood-retina barrier and play an important role in ion and fluid transport [37]. In this report we have tested the ability of hiPS-RPE to transport ions and their membrane potential compared to native RPE. Using the patch-clamp technique, we demonstrated the presence of functional voltage-gated sodium and potassium transporters in hiPS-RPE, similar to those of native RPE. In addition, we found no significant differences in the membrane potential of hiPS-RPE and native RPE. These are important markers for the validation of functional hiPS-RPE differentiation and we are the first to report the similarity of hiPS-RPE to native RPE in terms of ion transport and membrane potential. Recently, the molecular signature of human RPE was identified and 154 RPE signature genes were validated by microarray and qRT-PCR analysis in RPE and in an independent set of 78 tissues[31]. The relative expression values (rEx) that were assigned to each gene by this analysis represent the ratio of RPE expression to the median expression of 78 diverse tissues. A gene was included as an RPE signature gene if its mean expression level in all three RPE tissues, native adult and fetal and cultured fetal RPE, was 10-fold or greater than the median expression for that gene in the diverse tissue set. Therefore, these signature genes are good candidates for the validation of iPS and ES-derived. The 89 genes that we analyzed in this study are included in the RPE signature genes. We found that 55 genes of those (~63%) were expressed in a similar pattern to native fetal RPE. Therefore, based on the analysis by Strunnikova et al., our criterion of >3x expression provides a reliable measure to conclude that 63% similarity between native and hiPS-RPE is a significantly high level of similarity. The difference in gene expression pattern in 33 genes might be related to the origin of hiPS cells that are generated from adult tissue as opposed to native RPE that are of fetal origin. While we were preparing this manuscript for submission, a paper was published related to the molecular signature of primary retinal pigment epithelium and stem-sell-derived RPE cells limiting the number of signature genes to 87 out of 154 previously reported [31] and showing that hESC-derived RPE resemble fetal RPE cells more closely than hiPS-RPE [24]. In addition, they show that 87 of the 154 genes are over-expressed in fetal compared to stem-cell derived RPE, including 21 genes that are exclusively expressed in the native fetal RPE. Those genes are involved in lens development and in eye and lens morphogenesis processes. These data further confirm the difference in 33 genes expression pattern that we have observed between fetal RPE and hiPS-RPE and are discussed in detail in Supplementary Data.
It is clear that replacement of RPE by autologous cell-based therapy opens new doors for the treatment of blindness due to retinal degeneration. However, prior to the implementation of this new technology, one would have to generate safe cells that do not form teratomas after transplantation and remain viable and functional in vivo. However, most in vivo studies have shown that the transplanted cells die after two weeks [21, 25, 26]. It is not clear whether the hiPS-RPE cells die because of faster senescence or because they cannot integrate into the existing RPE to form a viable monolayer of cells. Recently, the effect of viral reprogramming on induction of DNA damage and loss of DNA damage-induced G(1)/S cell cycle arrest in iPS cells has been reported [27]. Nonetheless, these reports do not discuss the effect of viral integration on the DNA stability and cell cycle progression of iPS-derived differentiated cells. Recent studies have shown telomere elongation, a common indicator of cell “rejuvenation” in iPS cells compared with that of their parental somatic cells [39–41]. However, these reports do not discuss the telomere shortening in the iPS-derived differentiated cells. To date, it is not yet well established whether the iPS cells will maintain their telomere length after induction of differentiation or the iPS-derived differentiated cells will shorten their telomeres to the length comparable to that of their parental somatic cells [39, 42]. Here we show that hiPS-RPE cells undergo rapid telomere shortening and DNA damage that induce their growth arrest after passage 5. These observations are consistent with our gene expression data showing that the growth arrest specific gene 1 (GAS1) is expressed four times higher in hiPS-RPE compared to native RPE. We have also shown increased level of p21 protein in p7 along with DNA damage accumulation. We show for the first time that although hiPS-RPE exhibit the gene expression, polarity and physiology of native RPE, they undergo rapid telomere shortening, DNA damage and expression of cell cycle arrest protein p21, probably due to reprogramming. Furthermore, since the expression of GAS1 gene is higher in hiPS-RPE compared to native fetal RPE, it might well be that these changes occur well before the growth arrest and therefore we believe that use of these cells after passage 3 should be prohibited for future clinical applications. It is also important to investigate the potential of hiPS-derived RPE for DNA repair and to understand the reason of rapid telomere shortening and elevated DNA damage that induce cell senescence in order to prevent cell death and increase cell survival after transplantation.
CONCLUSION
We have generated hiPS-RPE possessing the specific functions of native RPE, including ion transport, membrane potential, polarized VEGF secretion and proper gene expression pattern. These findings suggest that hiPS-RPE are promising candidates for retinal regeneration therapies in AMD. However, we also demonstrated that the viability of hiPS-RPE is affected by rapid telomere shortening, DNA damage and p21 expression that induce growth arrest. Consequently, further investigations are needed in order to generate viral-free and long-term viable cells before implementing the hiPS-RPE in clinical applications.
Supplementary Material
(A) Validation of pluripotency. (i–ii), Phase contrast images of our homemade iPS cell lines, iPS 10 (i) and iPS 11(ii); (iii–viii), Immunostaining with goat anti-NANOG. Secondary anti-Goat antibody was conjugated to Alexa-546 (red). Positive colonies 10 and 11 are shown in iii and vi, respectively. DAPI nuclear staining (blue) for the same colonies, is shown in iv and vii; v and viii are the merged images; (i–xi), Negative control staining without primary antibody. Objective used is 20X. (B) Validation of chromosomal stability. Normal number of chromosomes was detected in iPS 10 (i) and in iPS 11 (ii).
(A), IMR90-4 iPS cells at day 1 in RPE differentiation medium; (B), Differentiating cell clusters, after 1 week in differentiation medium; (C), clusters appear either transparent or opaque at 2 weeks of differentiation; (D), pigmentation is evident after 4 weeks of differentiation; (E), pigmented cluster at 8 weeks in which the hexagonal shape of the differentiating RPE is also shown; (F), pigmented cluster at 9 weeks; (G), After trypsinization of a pigmented cluster at 12 weeks of differentiation the pigmented cells are transferred to expansion media for adherent culture and pigmentation is lost; (H); Pigmentation becomes evident again after 4 weeks of culture in high cell density (50,000 cells / cm2), starting as pigmentation foci; (I), Passage 3 RPE, grown on 24-well Transwell dishes coated with 2μg/cm2 poly-lysine-D and 4μg/ml laminin for 5 weeks.
Performed in parallel with the immunostainings shown in Figures 2 and 3. FITC (B) or TRITC (E) – conjugated secondary antibodies were incubated with RPE cells in the absence of primary antibody. (A & D), Nuclei stained with DAPI (blue); (C &F), merged images.
(A), Expression of Na+/K+ ATPase (green) on a monolayer of hiPS-RPE grown on Transwells; (B), Brightfield image of the same region of the monolayer, showing hiPS-RPE with different levels of pigmentation; (C), Merged image of (A) and (B), showing that Na+/K+ ATPase higher levels of expression co-localize with pigmentation; (D), Optical cross-sections of 7 cells of the same monolayer of hiPS-RPE, showing the expression of Na+/K+ ATPase (green) in D-(i) and a brightfield image in D-(ii). Cells 1–4 that have polarized Na+/K+ ATPase expression are pigmented, whereas cells 5–7 that are not polarized are not pigmented.
Supplementary Table 1. Primers used in qRT-PCR
Supplementary Table 2. Antibodies used in this study
Acknowledgments
This work was supported by the NIH 1R21EY019383-01.
Footnotes
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
The authors confirm no potential conflicts of interest.
Author contributions:
Maria Kokkinaki: Conception and design, Collection or assembly of data, manuscript writing.
Niaz Sahibzada: Collection or assembly of data, data analysis and interpretation.
Nady Golestaneh: Conception and design, financial support, data analysis and interpretation, manuscript writing and final approval of the manuscript.
References
- 1.Takahashi K, Okita K, Nakagawa M, et al. Induction of pluripotent stem cells from fibroblast cultures. Nat Protoc. 2007;2:3081–3089. doi: 10.1038/nprot.2007.418. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 4.Yamanaka S. Induction of pluripotent stem cells from mouse fibroblasts by four transcription factors. Cell Prolif. 2008;41 (Suppl 1):51–56. doi: 10.1111/j.1365-2184.2008.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okita K, Nakagawa M, Hyenjong H, et al. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- 6.Kaji K, Norrby K, Paca A, et al. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okita K, Hong H, Takahashi K, et al. Generation of mouse-induced pluripotent stem cells with plasmid vectors. Nat Protoc. 2010;5:418–428. doi: 10.1038/nprot.2009.231. [DOI] [PubMed] [Google Scholar]
- 8.Gehrs KM, Anderson DH, Johnson LV, et al. Age-related macular degeneration--emerging pathogenetic and therapeutic concepts. Ann Med. 2006;38:450–471. doi: 10.1080/07853890600946724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bok D. The retinal pigment epithelium: a versatile partner in vision. J Cell Sci Suppl. 1993;17:189–195. doi: 10.1242/jcs.1993.supplement_17.27. [DOI] [PubMed] [Google Scholar]
- 10.Boulton M, Dayhaw-Barker P. The role of the retinal pigment epithelium: topographical variation and ageing changes. Eye (Lond) 2001;15:384–389. doi: 10.1038/eye.2001.141. [DOI] [PubMed] [Google Scholar]
- 11.Strauss O. The retinal pigment epithelium in visual function. Physiol Rev. 2005;85:845–881. doi: 10.1152/physrev.00021.2004. [DOI] [PubMed] [Google Scholar]
- 12.Wimmers S, Karl MO, Strauss O. Ion channels in the RPE. Prog Retin Eye Res. 2007;26:263–301. doi: 10.1016/j.preteyeres.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 13.Treumer F, Bunse A, Klatt C, et al. Autologous retinal pigment epithelium-choroid sheet transplantation in age related macular degeneration: morphological and functional results. Br J Ophthalmol. 2007;91:349–353. doi: 10.1136/bjo.2006.102152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.MacLaren RE, Uppal GS, Balaggan KS, et al. Autologous transplantation of the retinal pigment epithelium and choroid in the treatment of neovascular age-related macular degeneration. Ophthalmology. 2007;114:561–570. doi: 10.1016/j.ophtha.2006.06.049. [DOI] [PubMed] [Google Scholar]
- 15.Heussen FM, Fawzy NF, Joeres S, et al. Autologous translocation of the choroid and RPE in age-related macular degeneration: 1-year follow-up in 30 patients and recommendations for patient selection. Eye (Lond) 2008;22:799–807. doi: 10.1038/sj.eye.6702823. [DOI] [PubMed] [Google Scholar]
- 16.Chen FK, Uppal GS, Rubin GS, et al. Evidence of retinal function using microperimetry following autologous retinal pigment epithelium-choroid graft in macular dystrophy. Invest Ophthalmol Vis Sci. 2008;49:3143–3150. doi: 10.1167/iovs.07-1648. [DOI] [PubMed] [Google Scholar]
- 17.Chen FK, Patel PJ, Uppal GS, et al. A comparison of macular translocation with patch graft in neovascular age-related macular degeneration. Invest Ophthalmol Vis Sci. 2009;50:1848–1855. doi: 10.1167/iovs.08-2845. [DOI] [PubMed] [Google Scholar]
- 18.Chen FK, Uppal GS, MacLaren RE, et al. Long-term visual and microperimetry outcomes following autologous retinal pigment epithelium choroid graft for neovascular age-related macular degeneration. Clin Experiment Ophthalmol. 2009;37:275–285. doi: 10.1111/j.1442-9071.2009.01915.x. [DOI] [PubMed] [Google Scholar]
- 19.Klimanskaya I, Hipp J, Rezai KA, et al. Derivation and comparative assessment of retinal pigment epithelium from human embryonic stem cells using transcriptomics. Cloning Stem Cells. 2004;6:217–245. doi: 10.1089/clo.2004.6.217. [DOI] [PubMed] [Google Scholar]
- 20.Lund RD, Wang S, Klimanskaya I, et al. Human embryonic stem cell-derived cells rescue visual function in dystrophic RCS rats. Cloning Stem Cells. 2006;8:189–199. doi: 10.1089/clo.2006.8.189. [DOI] [PubMed] [Google Scholar]
- 21.Lu B, Malcuit C, Wang S, et al. Long-term safety and function of RPE from human embryonic stem cells in preclinical models of macular degeneration. Stem Cells. 2009;27:2126–2135. doi: 10.1002/stem.149. [DOI] [PubMed] [Google Scholar]
- 22.Hirami Y, Osakada F, Takahashi K, et al. Generation of retinal cells from mouse and human induced pluripotent stem cells. Neurosci Lett. 2009;458:126–131. doi: 10.1016/j.neulet.2009.04.035. [DOI] [PubMed] [Google Scholar]
- 23.Buchholz DE, Hikita ST, Rowland TJ, et al. Derivation of functional retinal pigmented epithelium from induced pluripotent stem cells. Stem Cells. 2009;27:2427–2434. doi: 10.1002/stem.189. [DOI] [PubMed] [Google Scholar]
- 24.Liao JL, Yu J, Huang K, et al. Molecular signature of primary retinal pigment epithelium and stem-cell-derived RPE cells. Hum Mol Genet. 2010 doi: 10.1093/hmg/ddq341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carr AJ, Vugler AA, Hikita ST, et al. Protective effects of human iPS-derived retinal pigment epithelium cell transplantation in the retinal dystrophic rat. PLoS One. 2009;4:e8152. doi: 10.1371/journal.pone.0008152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Idelson M, Alper R, Obolensky A, et al. Directed differentiation of human embryonic stem cells into functional retinal pigment epithelium cells. Cell Stem Cell. 2009;5:396–408. doi: 10.1016/j.stem.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 27.Marion RM, Strati K, Li H, et al. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cawthon RM. Telomere measurement by quantitative PCR. Nucleic Acids Res. 2002;30:e47. doi: 10.1093/nar/30.10.e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced Pluripotent Stem Cell Lines Derived from Human Somatic Cells. Science. 2007 doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 30.Maminishkis A, Chen S, Jalickee S, et al. Confluent monolayers of cultured human fetal retinal pigment epithelium exhibit morphology and physiology of native tissue. Invest Ophthalmol Vis Sci. 2006;47:3612–3624. doi: 10.1167/iovs.05-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strunnikova NV, Maminishkis A, Barb JJ, et al. Transcriptome analysis and molecular signature of human retinal pigment epithelium. Hum Mol Genet. 2010 doi: 10.1093/hmg/ddq129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Longbottom R, Fruttiger M, Douglas RH, et al. Genetic ablation of retinal pigment epithelial cells reveals the adaptive response of the epithelium and impact on photoreceptors. Proc Natl Acad Sci U S A. 2009;106:18728–18733. doi: 10.1073/pnas.0902593106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nussenblatt RB, Liu B, Li Z. Age-related macular degeneration: an immunologically driven disease. Curr Opin Investig Drugs. 2009;10:434–442. [PubMed] [Google Scholar]
- 34.Swaroop A, Chew EY, Rickman CB, et al. Unraveling a multifactorial late-onset disease: from genetic susceptibility to disease mechanisms for age-related macular degeneration. Annu Rev Genomics Hum Genet. 2009;10:19–43. doi: 10.1146/annurev.genom.9.081307.164350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lamba DA, Karl MO, Ware CB, et al. Efficient generation of retinal progenitor cells from human embryonic stem cells. Proc Natl Acad Sci U S A. 2006;103:12769–12774. doi: 10.1073/pnas.0601990103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blaauwgeers HG, Holtkamp GM, Rutten H, et al. Polarized vascular endothelial growth factor secretion by human retinal pigment epithelium and localization of vascular endothelial growth factor receptors on the inner choriocapillaris. Evidence for a trophic paracrine relation. Am J Pathol. 1999;155:421–428. doi: 10.1016/S0002-9440(10)65138-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marmorstein AD, Finnemann SC, Bonilha VL, et al. Morphogenesis of the retinal pigment epithelium: toward understanding retinal degenerative diseases. Ann N Y Acad Sci. 1998;857:1–12. doi: 10.1111/j.1749-6632.1998.tb10102.x. [DOI] [PubMed] [Google Scholar]
- 38.Momcilovic O, Knobloch L, Fornsaglio J, et al. DNA damage responses in human induced pluripotent stem cells and embryonic stem cells. PLoS One. 2010;5:e13410. doi: 10.1371/journal.pone.0013410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suhr ST, Chang EA, Rodriguez RM, et al. Telomere dynamics in human cells reprogrammed to pluripotency. PLoS One. 2009;4:e8124. doi: 10.1371/journal.pone.0008124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Agarwal S, Loh YH, McLoughlin EM, et al. Telomere elongation in induced pluripotent stem cells from dyskeratosis congenita patients. Nature. 2010;464:292–296. doi: 10.1038/nature08792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marion RM, Strati K, Li H, et al. Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell Stem Cell. 2009;4:141–154. doi: 10.1016/j.stem.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 42.Vaziri H, Chapman KB, Guigova A, et al. Spontaneous reversal of the developmental aging of normal human cells following transcriptional reprogramming. Regen Med. 2010;5:345–363. doi: 10.2217/rme.10.21. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Validation of pluripotency. (i–ii), Phase contrast images of our homemade iPS cell lines, iPS 10 (i) and iPS 11(ii); (iii–viii), Immunostaining with goat anti-NANOG. Secondary anti-Goat antibody was conjugated to Alexa-546 (red). Positive colonies 10 and 11 are shown in iii and vi, respectively. DAPI nuclear staining (blue) for the same colonies, is shown in iv and vii; v and viii are the merged images; (i–xi), Negative control staining without primary antibody. Objective used is 20X. (B) Validation of chromosomal stability. Normal number of chromosomes was detected in iPS 10 (i) and in iPS 11 (ii).
(A), IMR90-4 iPS cells at day 1 in RPE differentiation medium; (B), Differentiating cell clusters, after 1 week in differentiation medium; (C), clusters appear either transparent or opaque at 2 weeks of differentiation; (D), pigmentation is evident after 4 weeks of differentiation; (E), pigmented cluster at 8 weeks in which the hexagonal shape of the differentiating RPE is also shown; (F), pigmented cluster at 9 weeks; (G), After trypsinization of a pigmented cluster at 12 weeks of differentiation the pigmented cells are transferred to expansion media for adherent culture and pigmentation is lost; (H); Pigmentation becomes evident again after 4 weeks of culture in high cell density (50,000 cells / cm2), starting as pigmentation foci; (I), Passage 3 RPE, grown on 24-well Transwell dishes coated with 2μg/cm2 poly-lysine-D and 4μg/ml laminin for 5 weeks.
Performed in parallel with the immunostainings shown in Figures 2 and 3. FITC (B) or TRITC (E) – conjugated secondary antibodies were incubated with RPE cells in the absence of primary antibody. (A & D), Nuclei stained with DAPI (blue); (C &F), merged images.
(A), Expression of Na+/K+ ATPase (green) on a monolayer of hiPS-RPE grown on Transwells; (B), Brightfield image of the same region of the monolayer, showing hiPS-RPE with different levels of pigmentation; (C), Merged image of (A) and (B), showing that Na+/K+ ATPase higher levels of expression co-localize with pigmentation; (D), Optical cross-sections of 7 cells of the same monolayer of hiPS-RPE, showing the expression of Na+/K+ ATPase (green) in D-(i) and a brightfield image in D-(ii). Cells 1–4 that have polarized Na+/K+ ATPase expression are pigmented, whereas cells 5–7 that are not polarized are not pigmented.
Supplementary Table 1. Primers used in qRT-PCR
Supplementary Table 2. Antibodies used in this study