Abstract
Two studies recently identified a GGGGCC hexanucleotide repeat expansion in a non-coding region of the chromosome 9 open reading frame 72 gene (C9ORF72) as the cause of chromosome 9p-linked amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). In a cohort of 231 probands with ALS, we identified the C9ORF72 mutation in 17 familial (27.4 %) and six sporadic (3.6%) cases. Patients with the mutation presented with typical motor features of ALS, although subjects with the C9ORF72 mutation had more frequent bulbar onset, compared to those without this mutation. Dementia was significantly more common in ALS patients and families with the C9ORF72 mutation and was usually early-onset FTD. There was striking clinical heterogeneity among the members of individual families with the mutation. The associated neuropathology was a combination of ALS with TDP-ir inclusions and FTLD-TDP. In addition to TDP-43-immunoreactive pathology, a consistent and specific feature of cases with the C9ORF72 mutation was the presence of ubiquitin-positive, TDP-43-negative inclusions in a variety of neuroanatomical regions, such as the cerebellar cortex. These findings support the C9ORF72 mutation as an important newly-recognized cause of ALS, provide a more detailed characterization of the associated clinical and pathological features and further demonstrate the clinical and molecular overlap between ALS and FTD.
Keywords: amyotrophic lateral sclerosis, frontotemporal dementia, frontotemporal lobar degeneration, C9ORF72, TDP-43, chromosome 9p
Introduction
Amyotrophic lateral sclerosis (ALS) is a common neurodegenerative disorder in which the relentless destruction of motor neurons results in progressive weakness, typically leading to death within a few years. Although most cases are sporadic (sALS), approximately 10% of ALS patients have a positive family history (fALS) [24]. Until very recently, the most common known genetic cause was mutations in the Cu/Zn superoxide dismutase gene (SOD1), which explain 15-20% of fALS. A number of other genes have been identified that cause autosomal dominant adult-onset ALS (TARDBP, FUS, VAPB, ANG, OPTN); however, each of these only accounts for a small percentage of cases and the genetic basis of most fALS remains unknown [24].
ALS is a multisystem disorder with dementia as a recognized clinical complication and evidence of frontotemporal lobar dysfunction in up to 50% of patients [11, 21]. The concept that ALS and frontotemporal dementia (FTD) represent a clinicopathological spectrum of disease is strongly supported by the discovery of the transactive response DNA binding protein with Mr 43 kD (TDP-43) as the protein that accumulates abnormally in the vast majority of ALS cases and in the most common pathological subtype of FTD [4, 12, 19], now referred to as frontotemporal lobar degeneration with TDP-43 pathology (FTLD-TDP) [14]. In addition, a number of families have been reported with an autosomal dominant pattern of disease in which affected members develop either ALS or FTD or both (ALS-FTD). Several of these families have shown genetic linkage to a region on chromosome 9p21 [2, 7, 9, 10, 15, 16, 20, 25, 26] and the same chromosomal region has been identified in a number of large genome-wide association studies of both ALS and FTD [8, 23, 27, 28].
Recently, two independent studies reported identification of the FTD/ALS gene defect on chromosome 9p as being a massively expanded GGGGCC hexanucleotide repeat in a non-coding region of the chromosome 9 open reading frame 72 gene (C9ORF72) [5, 22]. The mutation was shown to result in the loss of one alternatively spliced C9ORF72 transcript and the formation of nuclear RNA foci, suggesting multiple pathogenic mechanisms [5]. In these two studies, the C9ORF72 mutation was found to be the most common genetic abnormality in familial and sporadic forms of both ALS and of FTD and was particularly frequent in patients and families with both conditions. However, in these two papers [5, 22], and in more recent studies [1, 17], descriptions of the associated clinical features have been limited to basic demographic information and broad phenotypes. Moreover, the frequency of the C9ORF72 mutation in the different populations reported thus far has varied by more than 100% [5, 22].
In the present study, we describe the frequency, demographic, clinical and neuropathological features associated with the C9ORF72 mutation in a large cohort of patients from our ALS clinic. Important aspects of this study include; (i) determining the mutation frequency in a new, well-characterized ALS patient population, (ii) providing a more detailed description of the clinical presentation associated with the C9ORF72 mutation than previously published, (iii) determining whether the clinical features of ALS patients with the C9ORF72 mutation distinguish them from non-mutation carriers and from the other common fALS genetic subgroup (with SOD1 mutations), (iv) establishing the degree of clinical variation within individual families with the C9ORF72 mutation, (v) evaluating clinicopathological correlations within the C9ORF72 mutation group, (vi) confirming the unique pathological features that have previously been described [1, 2, 5, 17] and (vii) investigating the distribution of C9ORF72 protein in tissue. We believe our findings significantly expand the current knowledge of this newly discovered and important genetic subtype of ALS.
Materials and methods
Subjects
The initial cohort consisted of 231 probands of unrelated families who had been prospectively diagnosed with probable or definite ALS [3], at the ALS Clinic of Vancouver Coastal Health and the University of British Columbia (Vancouver General Hospital and GF Strong Rehabilitation Centre sites). Those suspected of developing dementia during the course of their disease were further investigated at the University of British Columbia Dementia Clinic. Patients were enrolled in an ongoing research study investigating the genetic basis of ALS that had been approved by the local Institutional Research Ethics Board with all subjects signing informed consent. Sixty-two (26.8 %) probands had at least one first or second degree relatives with ALS (fALS) while the remaining 169 (73.2 %) appeared to be sporadic (sALS). Subjects had already been screened for SOD1 mutations and 19 found to be positive. The demographic and clinical features of most of those with SOD1 mutations have been reported previously [6]. Screening for less common ALS-associated mutations (TARDBP, FUS, VAPB, ANG, OPTN) had not been performed in a comprehensive fashion (data not shown). Clinical information was obtained from the study database and through retrospective review of clinic charts. Demographic and clinical features of probands found to carry the C9ORF72 mutation were compared to those with SOD1 mutations and those in which C9ORF72 and SOD1 mutations were excluded (9p/SOD1-negative). In addition to the probands, limited clinical information was available for 11 other affected members of seven families with C9ORF72 mutation and in four of these families, DNA was available from multiple affected members.
Molecular genetics
All samples in which SOD1 mutations had been excluded were screened for the C9ORF72 mutation (N = 212). Genomic DNA was extracted from samples using standard procedures. For each case, the presence of an expanded GGGGCC hexanucleotide repeat in C9ORF72 was detected using a two-step protocol. First, in all samples, the hexanucleotide repeat was PCR amplified using one fluorescently labeled primer followed by fragment length analysis on an automated ABI3730 DNA-analyzer as described [5]. All patients that appeared homozygous in this assay were further analyzed using the repeat primed PCR method as described [5]. A characteristic stutter amplification pattern on the electropherogram was considered evidence of a pathogenic repeat expansion.
Neuropathology
Post mortem brain and spinal cord tissue was available from eight probands with the C9ORF72 mutation and five other affected members from their families (total, N = 13). Their final clinical diagnoses were ALS (N = 4), FTD (N =1) and ALS-FTD (N = 8). Pathological changes were graded in a wide range of neuroanatomical regions, using a semiquantitative system (0, absent; 1, mild/few; 2, moderate; 3, severe/abundant).
Sections from selected anatomical regions from 9p/SOD1-negative sALS (N = 10), fALS with SOD1 mutations (N = 5) and neurologically normal controls (N = 5) were also evaluated to determine the specificity of certain unusual pathological changes found in the C9ORF72 mutation group (specifically, the presence of ubiquitin/p62-positive, TDP-43-negative inclusions) and to investigate the staining pattern with the C9ORF72 antibody.
Histochemistry and immunohistochemistry
Sections of formalin fixed, paraffin embedded tissue were stained with hematoxylin and eosin (HE), HE combined with Luxol fast blue (HE/LFB) and Bielschowsky silver method. Immunohistochemistry (IHC) was performed on 5 μm thick sections using the Ventana BenchMark® XT automated staining system (Ventana, Tuscon, AZ) and developed with aminoethylcarbizole (AEC). The primary antibodies employed recognized ubiquitin (DAKO anti-ubiquitin; 1:500, following microwave antigen retrieval), p62 (BD Transduction Laboratories p62 Lck ligand; 1:500 following microwave antigen retrieval), hyperphosphorylated tau (Innogenetics AT-8; 1:2,000 following microwave antigen retrieval and Sigma TAU-2; 1:1,000 with 3 h initial incubation at room temperature), α-synuclein (Zymed anti-α-synuclein; 1:10,000, following microwave antigen retrieval), Aβ (DAKO anti-beta amyloid; 1:100 with initial incubation for 3 h at room temperature), TDP-43 (ProteinTech Group anti-TARDBP; 1:1,000 following microwave antigen retrieval), FUS (Sigma-Aldrich anti-FUS, 1:200, initial overnight incubation at room temperature, following microwave antigen retrieval) and C9ORF72 (Sigma-Aldrich, anti-C9orf72; 1:50 overnight incubation following microwave antigen retrieval).
Statistical analysis
Data were analyzed with IBM SPSS Statistics Version 19, Release 19.0.0 (IBM Corporation). Mean age at onset of first ALS symptoms, as well as mean survival from first ALS symptoms until death, were compared among genotype groups using one-way between subjects ANOVA. Median survival was also compared using Kruskal-Wallis test for independent samples. Individuals who had received a tracheostomy for permanent assisted ventilation (N = 6) were excluded from survival analyses. Pearson's chi-squared tests were used to compare frequencies of demographic and clinical features among groups. Significance level was set at P = 0.05.
Results
Mutation frequencies
Twenty-three (10.0 %) of the probands were found to carry alleles with abnormally expanded GGGGCC hexanucleotide repeats in intron 1 of C9ORF72, including 17 (27.4 %) cases of fALS and six (3.6%) sALS (Table 1). In the four families where DNA was available from multiple affected members, the mutation was found to segregate with clinical disease. The frequency of C9ORF72 and SOD1 mutations in the fALS group was the same. Although the frequency of the C9ORF72 mutation in sALS was three times greater than SOD1 mutations, the number of cases in both groups was small and the difference did not reach statistical significance.
Table 1.
Mutation frequency, demographics and clinical course of study subjects.
| C9ORF72 | SOD1 | 9p/SOD-neg | P value | |
|---|---|---|---|---|
| Proportion of total cohort (%) | 23/231 (10.0)* | 19/231 (8.2)* | 189/231 (81.8) | NS* |
| Proportion fALS (%) | 17/62 (27.4)* | 17/62 (27.4)* | 28/62 (45.2) | NS* |
| Proportion sALS (%) | 6/169 (3.6)* | 2/169 (1.2)* | 161/169 (95.3) | NS* |
| Sex (male, %) | 11/23 (47.8) | 10/19 (52.6) | 116/189 (61.4) | NS |
| Mean age at ALS onset (years) | 58.2 ± 9.9 | 54.9 ± 12.1 | 57.4 ± 14.7 | NS |
| Range of ALS onset age (years) | 39.5 - 82.7 | 36.8 - 78.3 | 9.3 - 88.4 | - |
| Mean survival (months)† | 34.3 ± 22.8 | 71.2 ± 70.5 | 54.0 ± 48.6 | NS |
| Median survival (months)† | 29.4 | 32.5 | 35.8 | NS |
| Range of survival (months)† | 12.2 - 96.3 | 12.9 - 204.8 | 12.8 - 298.8 | - |
| Dementia‡ in proband (%) | 7/22 (31.8) | 0/19 (0) | 6/178 (3.4) | < 0.001 |
| FTD in proband (%) | 5/22 (22.7) | 0/19 (0) | 2/178 (1.1) | < 0.001 |
| Dementia‡ in relative (%) | 9/22 (40.9) | 1/19 (5.2) | 11/175 (6.3) | < 0.001 |
| Dementia‡ in family• (%) | 11/22 (50.0) | 1/19 (5.2) | 17/173 (9.8) | < 0.001 |
fALS = familial amyotrophic lateral sclerosis; FTD = frontotemporal dementia; NS = not significant; sALS = sporadic amyotrophic lateral sclerosis. Denominator for dementia frequencies based on available clinical information.
Excludes patients maintained on permanent ventilatory assistance via tracheostomy.
Any dementia subtype, including FTD.
Proband or other family member.
Comparison between C9ORF72 and SOD1 groups only.
Demographics
All probands with the C9ORF72 mutation were of European ethnic descent, with the exception of one of Middle Eastern origin (data not shown). A trend towards male sex predominance was only present in the 9p/SOD1-negative cases. Cases with the C9ORF72 mutation had a broad range in age at disease onset (39.5 - 82.7 years) and survival (12.2 - 96.3 months). The mean age of onset for those with the C9ORF72 mutation (58.2 ± 9.9 years) was similar to cases with SOD1 mutations and the 9p/SOD1-negative group. However, because only the 9p/SOD1-negative group included a few cases with juvenile onset, this group had a broader onset range (due to lower minimum value). Although there was a trend for the mean survival of the C9ORF72 mutation carriers to be shorter (34.3 ± 22.8 months), the difference was not significant (ANOVA, P = 0.08) and was primarily due to the presence of a small number of patients with much longer survival in the other two groups.
Clinical features
Dementia was significantly more common in probands (31.8 %) and families (50.0 %) with the C9ORF72 mutation. In these cases, dementia was often present at the time of the initial ALS diagnosis and most often (9/11 cases, 81.8 %) fulfilled clinical diagnostic criteria for FTD [18]. In contrast, only two 9p/SOD1-negative probands developed FTD-type dementia and only one of these had FTD as a presenting feature. All the other 9p/SOD1-negative probands and family members with dementia, and the one demented relative of a SOD1 mutation carrier, were diagnosed with Alzheimer-type dementia (not FTD), late in their disease course.
The spectrum and frequency of motor features at the time of initial assessment was similar in the C9ORF72 mutation carriers and the 9p/SOD-negative group, with most patients in both groups presenting with a combination of both upper motor neuron (UMN) and lower motor neuron (LMN) features (Table 2). The only difference was that bulbar dysfunction was more common in patients with the C9ORF72 mutation (43.5 %), particularly in those with early dementia (data not shown). In contrast, patients with SOD1 mutations showed significant differences in presentation, with symptom onset occurring exclusively in the extremities and a striking predominance of LMN features.
Table 2.
Motor features at time of initial assessment.
| C9ORF72 | SOD1 | 9p/SOD-neg | P value | |
|---|---|---|---|---|
| Site of symptom onset | ||||
| • Extremity (%) | 12/23 (52.2) | 19/19 (100) | 117/174 (67.2) | 0.003 |
| • Bulbar (%) | 10/23 (43.5) | 0/19 (0) | 46/174 (26.4) | 0.005 |
| • Axial (%) | 0/23 (0) | 0/19 (0) | 6/174 (3.4) | NS |
| • Generalized (%) | 1/23 (4.3) | 0/19 (0) | 5/174 (2.9) | NS |
| Motor features | ||||
| LMN | ||||
| • Weakness (%) | 18/22 (81.8) | 18/18 (100) | 167/178 (93.8) | NS |
| • Atrophy (%) | 19/22 (86.4) | 18/18 (100) | 154/178 (86.5) | NS |
| • Fasciculations (%) | 14/22 (63.6) | 15/18 (83.3) | 138/178 (77.5) | NS |
| • Cramps (%) | 5/22 (22.7) | 13/18 (72.2) | 78/178 (43.8) | 0.04 |
| • No LMN signs (%) | 2/22 (9.1) | 0/18 (0) | 3/178 (1.9) | NS |
| UMN | ||||
| • Hyperreflexia (%) | 21/22 (95.5) | 11/17 (64.8) | 148/177 (83.6) | NS |
| • Spasticity (%) | 10/22 (45.5) | 2/17 (11.8) | 66/177 (37.3) | NS |
| • Babinski reflex (%) | 4/22 (18.2) | 2/17 (11.8) | 46/177 (26.0) | NS |
| • Hoffman sign (%) | 5/22 (22.7) | 6/17 (35.3) | 24/177(13.6) | NS |
| • Clonus (%) | 2/22 (9.1) | 0/17 (0.0) | 17/177 (9.6) | NS |
| • No UMN signs (%) | 1/22 (4.5) | 5/17 (29.4) | 14/177 (8.0) | 0.05 |
| initial MND diagnosis | ||||
| • ALS (%) | 20/23 (87.0) | 13/19 (68.4) | 155/187 (83.4) | NS |
| • PLS (%) | 2/23 (8.7) | 0/19 (0) | 13/187 (7.0) | NS |
| • PMA(%) | 1/23 (4.3) | 6/19 (31.6) | 14/187 (7.5) | 0.01 |
| • PBP (%) | 0/23 (0) | 0/19 (0) | 5/187 (2.7) | NS |
ALS = amyotrophic lateral sclerosis; LMN = lower motor neuron; MND= motor neuron disease; PBP = progressive bulbar palsy; PLS = primary lateral sclerosis; PMA = progressive muscular atrophy; UMN = upper motor neuron. Denominator based on available clinical information. All significant differences are due to the SOD1 mutation group.
There was considerable clinical heterogeneity among the members of individual families carrying the C9ORF72 mutation (Table 3). The age of disease onset varied by as much as 18 years and survival showed three-fold variation. In each of the four families for which data was available on members from multiple generations, the ages at onset were consistently earlier in the younger generation. The initial presentation and final diagnoses within a family could include pure ALS, pure FTD or ALS-FTD, and the FTD phenotype could be progressive non-fluent aphasia, behavioural variant FTD or both.
Table 3.
Clinical heterogeneity within families with C9ORF72 mutation
| Family | Member | Onset age (years) | Survival (years) | Initial clinical diagnosis | Final clinical diagnosis |
|---|---|---|---|---|---|
| 1 | A1 | 58 | 1 | ALS | ALS |
| B1* | 47 | 2 | ALS | ALS | |
| B2 | 40 | NA | bvFTD | bvFTD | |
| 2 | A1* | 59 | 4 | ALS | ALS |
| A2 | 56 | 6 | bvFTD | bvFTD + ALS | |
| A3 | 74 | 2 | ALS | ALS | |
| 3 | A1* | 72 | 3 | ALS | ALS |
| B1 | 55 | 1 | ALS | ALS + PNFA | |
| 4 | A1* | 56 | 19 | ALS | ALS + dementia |
| A2 | 57 | NA | ALS + dementia | ALS + dementia | |
| 5 | A1 | 54 | 4 | PNFA | PNFA/bvFTD + ALS |
| A2 | 67 | 9 | PNFA | PNFA/bvFTD | |
| B1* | 52 | 3 | PNFA | PNFA/bvFTD + ALS | |
| B2 | 53 | 3 | ALS | ALS | |
| 6 | A1* | 56 | 3 | PNFA | PNFA/bvFTD + ALS |
| A2 | 53 | 2 | ALS | ALS | |
| 7 | A1 | 61 | 1 | ALS | ALS |
| B1* | 49 | 7 | bvFTD | bvFTD + ALS |
A = older generation; ALS = amyotrophic lateral sclerosis; B = younger generation; bvFTD = behavioural variant frontotemporal dementia; NA = not available; PNFA = progressive non-fluent aphasia.
Proband.
Neuropathology
The post mortem neuropathology of eight probands with the C9ORF72 mutation and five additional affected members from their families (total, N = 13) were all characterized by TDP-43 immunoreative (TDP-ir) neuronal and glial cytoplasmic inclusions in a wide range of neuroanatomical regions (Table 4). Cases with a clinical diagnosis that included ALS (N =12) showed degeneration of the corticospinal tracts and loss of LMN from both brainstem and spinal cord. Many of the remaining LMN contained Bunina bodies and TDP-ir neuronal cytoplasmic inclusions (NCI) of various morphologies (Fig. 1a, b). The only case that did not show pathology of the pyramidal tracts or LMN was the one with a pure FTD clinical diagnosis (case 4). The nine cases with clinical FTD all had TDP-ir NCI in the extramotor neocortex and hippocampus (FTLD-TDP) (Fig. 1c, d). The extramotor cortex was also involved in all four cases with a clinical diagnosis of pure ALS, but usually to a milder degree. In all but one case, the pattern of TDP-ir pathology in the neocortex was consistent with FTLD-TDP type B [13], with NCI in all cortical layers but few dystrophic neurites (DN). The one exception was a case with clinical FTD-ALS (case 1) which, in addition to showing NCI in a type B distribution, also had a laminar concentration of NCI and short DN in layer 2 (consistent with FTLD-TDP type A). Many other neuroanatomical regions demonstrated TDP-ir pathology, with the striatum and substantia nigra consistently affected.
Table 4.
Neuropathological findings of study subjects with C9ORF72 mutation.
| Case | Clinical diagnosis | FC | MtC | HC dent | BG | SN | CST | LMN | cer ctx ub | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| deg | TDP | deg | TDP | TDP | deg | TDP | deg | TDP | deg | deg | TDP | |||
| 1 | FTD-ALS | 1 | 2 | 1 | 0 | 2 | 2 | 2 | 0 | 1 | 2 | 3 | 3 | 3 |
| 2 | ALS | 0 | 1 | 2 | 1 | 3 | 1 | 2 | 1 | 3 | 3 | 3 | 3 | 3 |
| 3 | FTD-ALS | 1 | 2 | NA | NA | 3 | 1 | 2 | NA | 2 | NA | NA | NA | NA |
| 4 | FTD | 1 | 1 | NA | NA | 3 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 2 |
| 5 | FTD-ALS | 1 | 2 | NA | NA | 3 | 0 | 3 | 1 | 2 | 2 | 3 | 3 | 2 |
| 6 | FTD-ALS | 1 | 2 | NA | NA | 3 | 0 | 1 | 2 | 2 | 1 | 2 | 3 | NA |
| 7 | FTD-ALS | 3 | 3 | NA | NA | 3 | 1 | 2 | 1 | 2 | 3 | 3 | 3 | 3 |
| 8 | FTD-ALS | 1 | 2 | NA | NA | 3 | NA | NA | NA | NA | 2 | 1* | 2* | 2 |
| 9 | ALS | 1 | 2 | 2 | 1 | 3 | 1 | 2 | 1 | 2 | 3 | 3 | 3 | 3 |
| 10 | FTD-ALS | 2 | 3 | 1 | 1 | 3 | 2 | 2 | 2 | 3 | 2 | 2 | 3 | 2 |
| 11 | FTD-ALS | 1 | 2 | NA | NA | 3 | 1 | 2 | 1 | 2 | 2 | 2 | 2 | 3 |
| 12 | ALS | 0 | 1 | 1 | 1 | 2 | 1 | 1 | 0 | 2 | 2 | 2 | 3 | 2 |
| 13 | ALS | 0 | 2 | 1 | 2 | 3 | 0 | 2 | 0 | 3 | 1 | 2 | 3 | 3 |
Semiquantitative grading: 0 = absent; 1 = mild/few; 2 = moderate; 3 = severe/abundant; NA = not available. ALS = amyotrophic lateral sclerosis; BG = basal ganglia; cer ctx = cerebellar cortex; CST = corticospinal tracts; deg = degenerative changes; dent = dentate granule cell layer; FC = premotor frontal cortex; FTD = frontotemporal dementia; HC = hippocampus; LMN = lower motor nuclei (hypoglossal nucleus and spinal cord ventral grey matter); MtC = primary motor cortex; SN, substantia nigra; TDP, TDP-43 immunoreactive pathology; ub = ubiquitin immunoreactive pathology.
Only medulla, no spinal cord available.
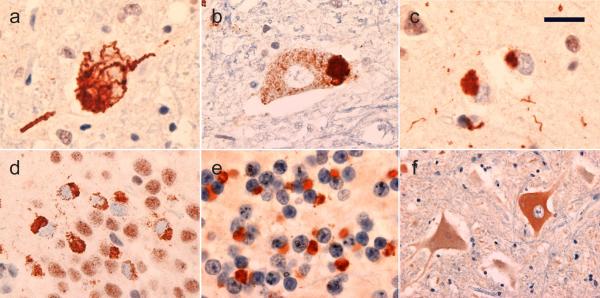
Fig. 1.
Neuropathological features in patients with the C9ORF72 mutation. All cases with a clinical diagnosis of amyotrophic lateral sclerosis had TDP-43 immunoreactive cytoplasmic inclusions in lower motor neurons that were filamentous (a), granular or compact (b). All cases, both with and without clinical frontotemporal dementia, had small TDP-43 immunoreactive neuronal cytoplasmic inclusions in the extramotor neocortex (c) and the hippocampal dentate granule cells (d). A consistent feature was small neuronal cytoplasmic inclusions and short neurites in the granule cell layer of the cerebellar cortex that were immunoreactive for ubiquitin and p62, but negative for TDP-43 (e). Immunohistochemistry from C9ORF72 showed increased cytoplasmic staining of some lower motor neurons in cases of ALS, both with and without the C9ORF72 mutation (f). Immunohistochemistry for TDP-43 (a – d), ubiquitin (e) and C9ORF72 (f). Scale bar 25 μm (a, b, d), 15 μm (c), 12 μm (e), 60 μm (f).
In addition to TDP-ir pathology, all cases with the C9ORF72 mutation were found to have several types of inclusions, in a variety of neuroanatomical regions, which were immunoreactive for ubiquitin and p62 but negative for TDP-43. In the cerebral neocortex, ubiquitin and p62 IHC demonstrated many more cytoplasmic inclusions and swollen DN than were labeled for TDP-43. These included small round compact cytoplasmic bodies in cells with small oval nuclei that could be either neurons or glia, and larger star-shaped NCI in some pyramidal neurons. Similarly, more ubiquitin/p62-positve NCI were present in neurons of the hippocampal dentate layer than were seen with TDP-43 immunostaining. Moreover, small round neuronal intranuclear inclusions (NII) in dentate granule cells and hippocampal pyramidal neurons and large star-shaped NCI in CA3/4 pyramidal neurons were only demonstrated with ubiquitin and p62 IHC, not with TDP-43 IHC. However, the most striking findings were in the cerebellum, where none of the cases showed any TDP-ir pathology, but all had moderate or abundant compact NCI, short DN and occasional small round NII in the granule cell layer that were p62/ubiquitin-positive (Fig. 1e). Occasional small round ubiquitin/p62-ir cytoplasmic and intranuclear inclusions were also present in the molecular layer in cells that could be either small neurons or glia; however, no cytoplasmic or intranuclear inclusions were present in Purkinje cells. All of these changes were demonstrated with IHC for both ubiquitin and p62; however, ubiquitin tended to be more sensitive for demonstrating the inclusions in the cerebellar granule cell layer and the neocortical DN, while p62 labelled more NCI and NII in the neocortex, hippocampus and cerebellar molecular layer. The presence of these types of ubiquitin/p62-positive, TDP-negative inclusions were specific for cases with the C9ORF72 mutation as they were not present in any of the other ALS cases or neurological control cases examined.
Immunohistochemistry using a commercially available antibody for C9ORF72 showed weak cytoplasmic reactivity but no nuclear staining of pyramidal neurons in sections of neocortex and spinal cord from neurologically normal controls (data not shown); a staining pattern consistent with data published on the Human Protein Atlas (http://www.proteinatlas.org/ENSG00000147894/summary). In cases with ALS, LMN that appeared to be chromatolytic showed more intense cytoplasmic reactivity and occasional swollen axons in the spinal cord ventral grey matter were strongly reactive (Fig. 1f); however, this staining pattern was seen in ALS cases both with and without the C9ORF72 mutation. No discrete cellular inclusions labelled for C9ORF72.
Discussion
The frequency of the C9ORF72 mutation in our cohort was very similar to that previously reported in a series of ALS patients from the Mayo Clinic, Jacksonville (23.5% of fALS and 4.1% of sALS) [5]. Much higher frequencies have been described in selected European populations, accounting for 46.0% of fALS and 21.1% of sALS in Finland [22]. In both the Mayo Clinic series and the Finnish population, the C9ORF72 mutation was found to be twice as common as SOD1 mutations [5, 22]; whereas, in our cohort it was only more common in sporadic cases. This difference is partially explained by the relatively high frequency of SOD1 mutations in our series. Together, C9ORF72 and SOD1 mutations accounted for more than half of our fALS population.
The motor features displayed by our patients with the C9ORF72 mutation were typical of ALS and similar to those of the 9p/SOD1-negative group, with the exception of more frequent bulbar presentation in C9ORF72 mutation carriers. This was in contrast to the SOD1 mutation group which most often presented with a predominance of LMN features. The most striking clinical difference in the C9ORF72 mutation group was the higher frequency of dementia in probands and their relatives. Moreover, the dementia subtype associated with the C9ORF72 mutation was most often early-onset FTD, while those without the mutation were usually diagnosed with Alzheimer-type dementia late in their disease. The clinical heterogeneity within many of our families was similar to what has been reported previously in chromosome 9p-linked families [2, 7, 9, 10, 15, 16, 20, 25, 26]. Although the finding of earlier age of onset in subsequent generations is intriguing, data on larger numbers of family members from multiple generations is required to properly evaluate genetic anticipation.
The neuropathology associated with the C9ORF72 mutation was found to be a combination of ALS with TDP-ir inclusions and FTLD-TDP type B [13]. Only one case showed a different pattern of cortical TDP-ir pathology that appeared to represent a combination of type B and type A. Although a recent study reported much greater heterogeneity in the cortical types of FTLD-TDP associated with the C9ORF72 mutation, this may be explained by the higher proportion of their cases with pure FTD (most of which had FTLD-TDP type A); whereas, the majority (71 %) of their cases with combined FTD and ALS had type B pathology, similar to ours [17]. The anatomical distribution of TDP-ir pathology showed even greater overlap than the clinical phenotypes, with all patients having involvement of extramotor neocortex and hippocampus, including those without a clinical diagnosis of dementia.
Similar to several recent studies [1, 2, 5, 17]; we found ubiquitin/p62-ir, TDP-negative inclusions in a variety of neuroanatomical regions, to be a highly consistent and specific feature of the C9ORF72 mutation. The presence of inclusions in the cerebellar granule cell layer was particularly useful for distinguishing cases with the mutation, since this region was devoid of other ubiquitin/p62-positive pathology. Minor differences in our findings compared with previous studies included the absence of inclusions in the Purkinje cell layer in our cases and the fact that we could not always be certain whether small inclusion-bearing cells in the cerebral neocortex and cerebellar molecular layer were neurons or glia [1]. The consistent presence of this ubiquitin/p62-positive pathology implies the mis-metabolism of some protein other than TDP-43 in cases with the C9ORF72 mutation.
Using a commercially available antibody for C9ORF72, we found similar changes in LMN all cases of ALS, both with and without the C9ORF72 mutation, that likely represent a non-specific feature of impaired axonal transport. No discrete cellular inclusions labelled for C9ORF72, including the ubiquitin/p62-postive, TDP-negative ones. Although there was some variation in the intensity of neuronal cytoplasmic staining among cases within the C9ORF72 mutation group and the control groups, no overall trend was apparent to suggest the mutation was associated with an altered level of protein expression. However, it is important to note that C9ORF72 is an uncharacterized protein and that nothing is currently known about its normal function or cellular distribution [5, 22]. Although a number of antibodies against C9ORF72 are commercially available, none has been fully validated. Future studies, using better characterized antibodies are needed to determine whether there are abnormalities in the level of expression or cellular distribution of the protein in cases with the C9ORF72 mutation.
In summary, our findings confirm that expansion of a GGGGCC hexanucleotide repeat in intron 1 of C9ORF72 is a major genetic cause of ALS. The C9ORF72 mutation appears to be at least as common as SOD1 mutations and likely accounts for most familial cases of ALS combined with FTD. The neuropathology in these cases includes some unusual features that appear to be highly sensitive and specific for the C9ORF72 mutation. These findings provide further support for the concept that ALS and FTD represent a clinical spectrum of disease with overlapping molecular basis.
Acknowledgements
We are grateful to the patients and family members who participated in this research. We thank Margaret Luk (research technologist, Vancouver General Hospital) for her excellent technical assistance. This work was supported by grants from the Canadian Institutes of Health Research [operating grants #179009, #74580 to IM], the Pacific Alzheimer's Research Foundation [center grant C06-01 to IM], the ALS Association [to RR] and the National Institutes of Health [grants # R01 NS065782, R01 AG026251 to RR].
Contributor Information
Heather Stewart, Division of Neurology, University of British Columbia, Vancouver, BC, Canada.
Nicola J. Rutherford, Department of Neuroscience, Mayo Clinic, Jacksonville, FL, USA
Hannah Briemberg, Division of Neurology, University of British Columbia, Vancouver, BC, Canada.
Charles Krieger, Division of Neurology, University of British Columbia, Vancouver, BC, Canada.
Neil Cashman, Division of Neurology, University of British Columbia, Vancouver, BC, Canada.
Marife Fabros, Division of Neurology, University of British Columbia, Vancouver, BC, Canada.
Matt Baker, Department of Neuroscience, Mayo Clinic, Jacksonville, FL, USA.
Alice Fok, Division of Neurology, University of British Columbia, Vancouver, BC, Canada.
Mariely DeJesus-Hernandez, Department of Neuroscience, Mayo Clinic, Jacksonville, FL, USA.
Andrew Eisen, Division of Neurology, University of British Columbia, Vancouver, BC, Canada.
Rosa Rademakers, Department of Neuroscience, Mayo Clinic, Jacksonville, FL, USA.
Ian R. A. Mackenzie, Department of Pathology, University of British Columbia, Vancouver, BC, Canada
References
- 1.Al-Sarraj S, King A, Troakes C, et al. p62 positive, TDP-43 negative, neuronal and cytoplasmic and intranuclear inclusions in the cerebellum and hippocampus define the pathology of C9orf72-linked FTLD and MND/ALS. Acta Neuropathol. 2011;22:691–702. doi: 10.1007/s00401-011-0911-2. [DOI] [PubMed] [Google Scholar]
- 2.Boxer AL, Mackenzie IR, Boeve BF, et al. Clinical, neuroimaging and neuropathological variability in a chromosome 9p-linked FTD-ALS family. J Neurol Neurosurg Psychiatr. 2011;82:196–203. doi: 10.1136/jnnp.2009.204081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brooks BR, Miller RG, Swash M, Munsat TL, World Federation of Neurology Research Group on Motor Neuron Diseases El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Motor Neuron Disord. 2000;1:293–299. doi: 10.1080/146608200300079536. [DOI] [PubMed] [Google Scholar]
- 4.Cairns NJ, Neumann M, Bigio EH, et al. TDP-43 in familial and sporadic frontotemporal lobar degeneration with ubiquitin inclusions. Am J Pathol. 2007;171:227–240. doi: 10.2353/ajpath.2007.070182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, et al. Expanded GGGGCC Hexanucleotide Repeat in Noncoding Region of C9ORF72 Causes Chromosome 9p-Linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eisen A, Mezei MM, Stewart HG, Fabros M, Gibson G, Andersen PM. SOD1 gene mutations in ALS patients from British Columbia, Canada: clinical features, neurophysiology and ethical issues in management. Amyotroph Lateral Scler. 2008;9:108–119. doi: 10.1080/17482960801900073. [DOI] [PubMed] [Google Scholar]
- 7.Gijselinck I, Engelborghs S, Maes G, et al. Identification of 2 Loci at chromosomes 9 and 14 in a multiplex family with frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Arch Neurol. 2010;67:606–616. doi: 10.1001/archneurol.2010.82. [DOI] [PubMed] [Google Scholar]
- 8.Laaksovirta H, Peuralinna T, Schymick JC, et al. Chromosome 9p21 in amyotrophic lateral sclerosis in Finland: a genome-wide association study. Lancet Neurol. 2010;9:978–985. doi: 10.1016/S1474-4422(10)70184-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Le Ber I, Camuzat A, Berger E, et al. Chromosome 9p-linked families with frontotemporal dementia associated with motor neuron disease. Neurology. 2009;72:1669–1676. doi: 10.1212/WNL.0b013e3181a55f1c. [DOI] [PubMed] [Google Scholar]
- 10.Luty AA, Kwok JB, Thompson EM, Blumbergs P, et al. Pedigree with frontotemporal lobar degeneration--motor neuron disease and Tar DNA binding protein-43 positive neuropathology: genetic linkage to chromosome 9. BMC Neurol. 2008;8:32. doi: 10.1186/1471-2377-8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lomen-Hoerth C, Murphy J, Langmore S, Kramer JH, Olney RK, Miller B. Are amyotrophic lateral sclerosis patients cognitively normal? Neurology. 2003;60:1094–1097. doi: 10.1212/01.wnl.0000055861.95202.8d. [DOI] [PubMed] [Google Scholar]
- 12.Mackenzie IRA, Bigio EH, Ince PG, et al. Pathological TDP-43 distinguishes sporadic ALS from ALS with SOD-1 mutations. Ann Neurol. 2007;61:427–434. doi: 10.1002/ana.21147. [DOI] [PubMed] [Google Scholar]
- 13.Mackenzie IRA, Neumann M, Baborie A, et al. A harmonized classification system for FTLD-TDP pathology. Acta Neuropathol. 2011;122:111–113. doi: 10.1007/s00401-011-0845-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mackenzie IR, Neumann M, Bigio EH, et al. Nomenclature for neuropathologic subtypes of frontotemporal lobar degeneration: consensus recommendations. Acta Neuropathol. 2009;117:15–18. doi: 10.1007/s00401-008-0460-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Momeni P, Schymick J, Jain S, et al. Analysis of IFT74 as a candidate gene for chromosome 9p-linked ALS-FTD. BMC Neurol. 2006;6:44. doi: 10.1186/1471-2377-6-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morita M, Al-Chalabi A, Andersen PM, et al. A locus on chromosome 9p confers susceptibility to ALS and frontotemporal dementia. Neurology. 2006;66:839–844. doi: 10.1212/01.wnl.0000200048.53766.b4. [DOI] [PubMed] [Google Scholar]
- 17.Murray ME, DeJesus-Hernandez, Rutherford NJ, et al. Clinical and neuropathologic heterogeneity of c9FTD/ALS associated with hexanucleotide repeat expansion in C9ORF72. Acta Neuropathol. 2011;122:673–690. doi: 10.1007/s00401-011-0907-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neary D, Snowden JS, Gustafson L, et al. Frontotemporal lobar degeneration: a consensus on clinical diagnostic criteria. Neurology. 1998;51:1546–1554. doi: 10.1212/wnl.51.6.1546. [DOI] [PubMed] [Google Scholar]
- 19.Neumann M, Sampathu DM, Kwong LK, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 20.Pearson JP, Williams NM, Majounie E, et al. Familial frontotemporal dementia with amyotrophic lateral sclerosis and a shared haplotype on chromosome 9p. J Neurol. 2011;258:647–655. doi: 10.1007/s00415-010-5815-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phukan J, Pender NP, Hardiman O. Cognitive impairment in amyotrophic lateral sclerosis. Lancet Neurol. 2007;6:994–1003. doi: 10.1016/S1474-4422(07)70265-X. [DOI] [PubMed] [Google Scholar]
- 22.Renton AE, Majounie E, Waite A, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shatunov A, Mok K, Newhouse S, et al. Chromosome 9p21 in sporadic amyotrophic lateral sclerosis in the UK and seven other countries: a genome-wide association study. Lancet Neurol. 2010;9:986–994. doi: 10.1016/S1474-4422(10)70197-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valdmanis PN, Daoud H, Dion PA, Rouleau GA. Recent Advances in the genetics of amyotrophic lateral sclerosis. Curr Neurol Neurosci Rep. 2009;9:198–205. doi: 10.1007/s11910-009-0030-9. [DOI] [PubMed] [Google Scholar]
- 25.Valdmanis PN, Dupre N, Bouchard JP, et al. Three families with amyotrophic lateral sclerosis and frontotemporal dementia with evidence of linkage to chromosome 9p. Arch Neurol. 2007;64:240–245. doi: 10.1001/archneur.64.2.240. [DOI] [PubMed] [Google Scholar]
- 26.Vance C, Al-Chalabi A, Ruddy D, et al. Familial amyotrophic lateral sclerosis with frontotemporal dementia is linked to a locus on chromosome 9p13.2-21.3. Brain. 2006;129:868–876. doi: 10.1093/brain/awl030. [DOI] [PubMed] [Google Scholar]
- 27.Van Deerlin VM, Sleiman PM, Martinez-Lage M, et al. Common variants at 7p21 are associated with frontotemporal lobar degeneration with TDP-43 inclusions. Nat Genet. 2010;42:234–239. doi: 10.1038/ng.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Es MA, Veldink JH, Saris CG, et al. Genome-wide association study identifies 19p13.3 (UNC13A) and 9p21.2 as susceptibility loci for sporadic amyotrophic lateral sclerosis. Nat Genet. 2009;41:1083–1087. doi: 10.1038/ng.442. [DOI] [PubMed] [Google Scholar]