Abstract
With great biological interest in post-translational modifications (PTMs), various approaches have been introduced to identify PTMs using MS/MS. Recent developments for PTM identification have focused on an unrestrictive approach that searches MS/MS spectra for all known and possibly even unknown types of PTMs at once. However, the resulting expanded search space requires much longer search time and also increases the number of false positives (incorrect identifications) and false negatives (missed true identifications), thus creating a bottleneck in high throughput analysis. Here we introduce MODa, a novel “multi-blind” spectral alignment algorithm that allows for fast unrestrictive PTM searches with no limitation on the number of modifications per peptide while featuring over an order of magnitude speedup in relation to existing approaches. We demonstrate the sensitivity of MODa on human shotgun proteomics data where it reveals multiple mutations, a wide range of modifications (including glycosylation), and evidence for several putative novel modifications. Based on the reported findings, we argue that the efficiency and sensitivity of MODa make it the first unrestrictive search tool with the potential to fully replace conventional restrictive identification of proteomics mass spectrometry data.
Post-translational modifications (PTMs)1 regulate protein function, localization, and interactions inside a cell (1). Hundreds of PTM types are known so far, and yet a lot more may remain to be discovered (2, 3). The identification of PTMs is critical to gaining insight into biological functions but remains a formidable challenge. Tandem mass spectrometry (MS/MS) has emerged as a powerful tool for rapid identification of PTMs (4, 5), which can be detected by PTM-related diagnostic mass shifts of fragment ions in MS/MS spectra. However accurate computational identification of modified peptides remains a difficult problem often addressed with restrictive approaches that require “guessed” lists of possible PTMs to be provided in advance (6–8). Such an approach may overlook potentially important PTMs if they are not guessed in advance. In recent PTM identification algorithms, peptide sequence tag approaches have been proposed to search for more types of PTMs and to speed up the search (9–12). A small set of short sequence tags (2–4 amino acids long) are derived from an MS/MS spectrum and used to screen for matching peptides in a protein database; possible modifications are then inferred from the difference between the precursor ion mass of the experimental spectrum and the theoretically calculated mass of the matched peptide.
In contrast with restrictive approaches, unrestrictive or blind approaches search MS/MS spectra for all known and even possibly unknown types of PTMs at once and derive the list of modifications directly from MS/MS data (13–22). OpenSea (13) and SPIDER (14) compared de novo sequencing results with peptide sequences from a protein database. The differences between fragment ion masses of de novo and database sequences can be used to localize the modification or substitution. TagRecon (15) and MODmap (16) used partial sequence tags. MS-Alignment (17, 18) first proposed a spectral alignment between spectra and database sequences using dynamic programming, and ModifiComb (19) introduced a ΔM histogram between unassigned spectra and unmodified peptides with similar retention times. Spectral networks (20) derived possible modifications from spectrum/spectrum alignments.
Although unrestrictive searches can improve sensitivity in the detection of peptides with unexpected modifications, these searches traditionally face serious challenges in increased run time and substantially decreased identification of unmodified peptides. Allowing arbitrary modification masses on any residue leads to an exponential growth in the size of the virtual database of possible modified peptide sequences that need to be matched against each and every MS/MS spectrum. This creates a serious bottleneck in high throughput proteomics, where the data analysis step often takes 10–50 times longer than the data acquisition. The first challenge is the increase in both false positives (incorrect identifications) and false negatives (missed identifications of peptides). Allowing many modifications increases false positives because of the combinatorial increase in the number of possible matches (23, 24) because even bad matches can be “tweaked” by introducing arbitrary modifications that increase the match score. Consequently, maintaining a fixed 1% false discovery rate (FDR) usually requires a much higher score threshold for peptide identification, which conversely results in a large number of false negatives. Both of these effects worsen as the protein sequence database gets larger. In our analysis of existing unrestrictive search tools, the number of false positives (determined by matches to a decoy database (25)) with high match score increased for larger sequence databases, and as a result, the number of identifications at the same FDR was dramatically reduced by ∼53%.
The second challenge addressed in this manuscript is the number of modified sites allowed per peptide—a critical analytical capability with severe implications in the software performance. In the case of localizing only one modification Δ to one position on a peptide P (where Δ is determined by the precursor mass of the MS/MS spectrum minus the mass of the unmodified peptide), the modified site can be determined in time proportional to the length of P (i.e. by trying all possible site assignments). However, for multiple modifications, the time complexity grows exponentially. For example, in the case of two modifications, there are many ways to split Δ into Δ1 and Δ2, such that Δ = Δ1 + Δ2, and then one has to combinatorially search for pairs of sites in P for each possible Δ1 and Δ2. This explosion in the size of the virtual database of possible peptide matches is the fundamental reason why increasing numbers of modified sites per peptide significantly deteriorates the efficiency of database search while generating many more false positives and substantially fewer true positives. Although many unrestrictive approaches have been developed, none has fully addressed these issues. Most approaches allow only a single unknown, variable modification per peptide and either limit the search database to a small set of proteins or require that the unmodified peptides be identified in advance.
In this work, we propose a novel spectral alignment approach, MODa (MODification via alignment), enabling fast “multi-blind ” unrestrictive PTM search with an order of magnitude speedup over existing approaches while allowing, for the first time, no limitation on the number of variable modifications per peptide. Different from alternative approaches, MODa simultaneously uses multiple sequence tags from each MS/MS spectrum, and a dynamic programming algorithm is used to identify modifications between sequence tags matched to a database peptide. The sequence tag approach provides various advantages in identifying modifications. First, sequence tags dramatically reduce the number of database peptides matched to each spectrum and thus alleviate the impact of database size on an unrestrictive search. Second, sequence tags effectively localize modified regions within a spectrum; the mass difference (Δ mass) between the flanking mass of a tag (e.g. the lowest mass of a peak in the tag) and the mass of the corresponding subsequence from the database peptide suggests that the region is modified by Δ mass. Expanding this notion, if several different Δ mass values are discovered from multiple matched tags, then this suggests multiple modifications whose sites can be restricted to the regions between the tags (see Fig. 6). Thus, an unrestrictive search can be freed from the limitation on the number of modifications per peptide. As a result, MODa is able to remove key limiting factors affecting the complexity of traditional spectral alignment algorithms (17, 18), such as the number of modifications per peptide and mass range of modifications, while significantly improving the speed of unrestrictive searches. Most importantly, MODa nearly eliminates the increases in false positives and false negatives by using the multiple-tag approach to filter out many useless and incorrect peptide matches. The effective performance of MODa is demonstrated by comparison with established restrictive and unrestrictive modification search tools.
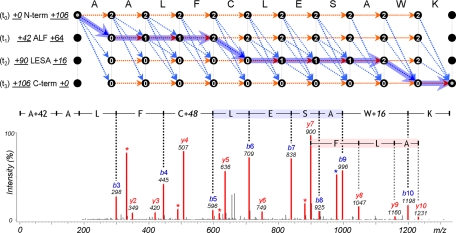
Fig. 6.
Dynamic programming to identify triply modified peptide, A+42ALFC+48LESAW+16K, and its MS/MS spectrum. First, two sequence tags, ALF (t1) and LESA (t2), were derived from the spectrum. The t0 and t3 tags are special tags of length zero that define the start and end nodes of dynamic programming. Numeric figures at both ends of a tag represent mass differences (Δ mass) between the tag's flanking mass and the mass of subsequence corresponding to the flanking mass region: the left figure for N terminus and the right figure for C terminus. They represent the total modified mass of the region. To restrict jumps between nodes, nodes are labeled according to the position of matched tag: 0 for before the tag, 1 for inside the tag, and 2 for after the tag. Dashed arrows represent all possible jumps satisfying constraints on the sequence tags and the highlighted path represents the optimal alignment. For a modification jump, its modified mass is defined as a difference between N-terminal Δ mass values of two tags.
EXPERIMENTAL PROCEDURES
LC-MS/MS-based Analysis of Human Proteome
We analyzed three proteomics data sets acquired from human plasma, HEK293 cell line, and human lens; the detailed description and availability of the data are provided in previous publications. The human plasma data set consists of 67,648 MS/MS spectra, acquired on a ThermoFinnigan (San Jose, CA) LTQ instrument (26). This data set was used to demonstrate the performance of MODa against the established search engines SEQUEST (6), Mascot (7), InsPecT (11), and MS-Alignment (18). The HEK293 data set consists of 363,807 MS/MS spectra, acquired on a Thermo LTQ instrument (27) and represents an analysis of a large scale complex mixture from whole cell lysate. The human lens data set consists of 381,224 MS/MS spectra acquired on a Thermo LCQ Classic instrument (13, 28, 29) from seven different samples: 3-day and 2-, 18-, 35-, and 70-year-old normal lens and 70- and 93-year-old cataract lens. Because many lens proteins do not turn over and tend to become substantially modified over time, PTM identification for this sample has been extensively studied by others, and their results were compared with the results from MODa analysis. The peak lists for plasma and HEK293 data were generated by converting the raw data into mzXML format and, for lens data, Bioworks software (version 3.1SR1, ThermoFinnigan).
MODa Search
MODa searches were conducted against human plasma, HEK293, and lens data sets with the following parameters: ±2.5 and ±0.5 Da mass tolerances for peptide and fragment ions, respectively, no enzyme specificity, ±200 Da for modification mass size, “multi-mod” mode (allowing arbitrary number of modifications per peptide). All of the data sets were searched against IPI human database (version 3.41, 72,155 entries) appended with the shuffled sequences. From all of the search results, peptide identifications were obtained at FDR 1% using target-decoy approach. FDR was calculated as (2 × D)/(T + D), where T and D are the number of target and decoy hits above score threshold, respectively (25). For human plasma data, additional MODa search was conducted allowing one modification/peptide (“one-mod” mode). This was to assess the effect of the number of modifications/peptide in search condition and to test the performance.
Established Restrictive/Unrestrictive Searches for High Throughput Complex Mixture Data
We conducted several searches to test MODa performance in various aspects. All of the searches for this experiment were conducted against human plasma data using IPI human database (version 3.41, 72,155 entries) appended with its shuffled sequences.
First, to compare MODa performance with the identification performance of standard database search tools, SEQUEST (version 28, rev. 12; ThermoFinnigan) and InsPecT searches were conducted with the following parameters: no enzyme specificity, precursor mass tolerance = ±2.5 Da, fragment mass tolerance = ±0.5 Da, fixed modification = carbamidomethyl (Cys), no variable modifications. The peptide identifications were obtained at FDR 1% using target-decoy. XCorr and DeltaCN for SEQUEST, and F-Score for InsPecT were used for thresholding.
Next, a Mascot error-tolerant search was conducted to test the identification performance including modified peptides. The Mascot error-tolerant search was selected because other database search tools have a limitation on the number of modifications as an input parameter. The Mascot (version 2.2.07; Matrix Science) search was conducted using the following parameters: enzyme = trypsin, missed cleavage = 1, precursor mass tolerance = ±2.5 Da, fragment mass tolerance = ±0.5 Da, variable modifications = oxidation (Met) and carbamidomethyl (Cys). The Mascot error-tolerant search consists of two stages. A standard, first pass search is performed using the search parameters specified. The proteins identified from the results of the first pass search are selected for an error-tolerant, second pass search. Then the second pass search is performed with relaxed enzyme specificity, while iterating through a comprehensive list of chemical and post-translational modifications, together with residue substitutions. The peptide identifications were obtained at target-decoy FDR 1% using ion score and homology threshold.
Finally, for the performance comparison with an existing unrestrictive search tool, MS-Alignment (InsPecT with blind option allowing one modification per peptide) search was conducted with the same parameters as MODa search. The identifications were obtained below p value of 0.01 using its p value script.
Unrestrictive Searches for PTM-rich Data
The human ocular lens tissue consists of a small number of crystallin proteins, which often become substantially modified post-translationally as this tissue ages. Related studies have reported a wide variety of modifications, and to fully analyze these data, it is necessary to allow for identification of multiply modified peptides. Thus, for this PTM-rich data, the performance of MODa was compared with those of Mascot error-tolerant search and Protein Prospector (21), all of which support different types of unrestrictive searches and are also able to identify multiply modified peptides. Protein Prospector permits the identification of multiply modified peptides by a combination of one or more predefined modifications (i.e. known in advance) and at most one unknown modification, whereas Mascot error-tolerant search allows identifying multiply modified peptides by a combination of predefined modifications and at most one modification listed in Unimod (3).
The three searches were conducted using 30,575 MS/MS spectra (obtained with LCQ Classic ion trap mass spectrometry) from 93-year-old cataract lens (13) and searching against the Swiss-Prot human protein database (release 2011_01) appended with its randomly shuffled sequences, where the shuffled sequences that Protein Prospector used were different from those that MODa and Mascot used (because it was not possible to reproduce the shuffled sequences generated by the Protein Prospector web server). However, it is expected that different decoy databases generated in the same manner (randomly shuffled) would not significantly change the results (25).
For Protein Prospector, an initial search was first conducted using the following parameters: fully tryptic specificity, up to one missed cleavage, ±2.5 Da and ±0.5 Da mass tolerances for peptide and fragment ions, respectively, acetyl (protein N terminus), acetyl + oxidation (protein N-terminal Met), Met loss (protein N-terminal Met), Met loss + acetyl (protein N-terminal Met), oxidation (Met), Gln → pyro-Glu (N-terminal Gln), and carbamidomethyl (Cys) as variable modifications, up to two modifications per peptide. Next, the extensive search was performed for 88 proteins identified in the initial search, where in addition to predefined variable modifications, a single mass modification between −200 and +200 Da was allowed on any amino acid, and partially tryptic specificity was allowed.
MODa and Mascot searches were conducted using the same parameters as described in previous sections. The final peptide identifications were obtained at FDR 1% using target-decoy. Expectation value for Protein Prospector, ion score and homology threshold for Mascot, and probability for MODa were used for FDR thresholding.
Simulation Test for Modified Peptides
A simulation test was performed to evaluate how sensitive MODa is to a modified region in an MS/MS spectrum. First, of 2,423 peptide sequences from 14,623 SEQUEST peptide-spectrum matches (PSMs) identified in human plasma data (more details in previous section), ∼50% of their corresponding database sequences were mutated by changing one residue of each sequence to a random residue in protein database. As a result, the 1,213 peptide sequences from 7,353 spectra were mutated from the original database, whereas the 1,210 peptide sequences from the remaining 7,270 spectra were kept intact. We retained the mutated rates at the levels of peptides and spectra to 50% to prevent a particular peptide from being identified too many times and thus affecting the overall performance significantly (there can be many spectra belonging to a single peptide). Then we checked how many spectra could be identified as the original peptides with one amino acid mutation when MODa search is conducted against the mutated database. MS-Alignment search was also conducted to compare the performance (search parameters in the previous section). Final mutated database (mutDB) consisted of mutated sequences (of version 3.41 IPI human proteins, 72,155 entries) and their shuffled sequences.
Dynamic Programming Based on Multiple Tags
Let S be an MS/MS spectrum, P = a1 … an be a peptide selected from the database, and T be a set of tags matched to P. A tag t of length n is defined by n + 1 masses; start(t)/end(t) indicates the position in P where the tag match starts/ends (e.g. start(t) = 0 and end(t) = 1 mean that only a1 is matched).
Let M[p][t][s] be the maximum score path at position p (on the peptide sequence) on the diagonal defined by a tag t; s indicates whether the position on the diagonal is before/after the tag using values 0/1, respectively. Intuitively, the main goal of the third dimension (spanned by s) is to avoid having to decompose each tag into multiple “subtags.” As defined, one can jump inside a tag at any point and jump out again as soon as at least one amino acid is used in the tag (branch labeled as “jumps inside the tag”).
M[p][t][s] is initialized to 0 and defined recursively as score(p,t) plus the maximum of the following options:
- Amino acid jumps
- Before the tag: M[p−1][t][0], iff p ≤ start(t)
- Inside the tag: max(M[p−1][t][0], M[p−1][t][1]), iff s = 1, start(t) < p ≤ end(t)
- After the tag: M[p−1][t][1], iff p > end(t)
- Modification jump
- M[p−1][q][1] + pf(Δ,ap), iff s = 0, p < end(t), over all tags q such that start(q) < start(t), where pf(Δ,aa) ≤ 0 is a penalty function for a modification Δ on the amino acid (aa). Given the frequent and common modification list in advance, the function can be adjusted. In this work, we considered pf(Δ,aa) as a constant C for all of the modifications.
To avoid special cases, we consider that T always contains two special tags (t0 and tv) of length 0, one ending at position zero on P (t0) and the other starting at n (tv), to define the zero/parent mass diagonals. The overall best candidate score is obtained from M[n][tv][0].
The time and space complexity of the algorithm above are O(max|S|, |T|2 × |P|); one only needs to scan the spectrum once at the start to compute every score(p,t).
MODa Score and Probability Computation
To compute the score during dynamic programming, an experimental spectrum S is converted into prefix residue mass (PRM) spectrum (11). First, S is separated into windows of 100 m/z units. Within each window, top 10 peaks are retained according to their intensity, and each peak is given a weight according to its ranking. For every retained peak's mass m, nodes of masses (m − 1) and (PrecursorMass(S) − m + 1) are added to the PRM spectrum. The score of the PRM node is defined as the sum of weights of expected ion peaks from the PRM. The expected ion peaks include ion types b, b-H2O, b-NH3, y, y-H2O, and y-NH3 and isotopes. In case of y-ion, only peaks with less intensity are considered as neutral losses. If any peak is assumed as the secondary isotopic peak (this is determined by whether there is the parent peak at −1 Da position), it is not given additional weight from its supporting peaks. For the spectrum of charge states more than or equal to 3, doubly charged ion types are also included. Then the score of a candidate peptide is computed as the sum of scores of PRM nodes for the masses of prefixes of the candidate peptide.
The PRM score described above is to rank the candidate identifications from a single spectrum during dynamic programming and is not sufficient to assess the quality of the top scoring match. Thus, we compute the probability that the top identification is correct. The probability is evaluated by taking into account various properties representing the quality of match between the peptide and the MS/MS spectrum: 1) PRM score, 2) mass errors of matched fragment ions, 3) the fractions of b and y ions found, and 4) the propensity to a particular ion type: tryptic peptide features a stronger y-ion ladder than b-ion ladder. The four components are combined by a logistic regression, the result of which represents the probability of correct match using a logistic function over a weighted linear sum of the components. The weights were trained and validated over correct and incorrect matches from the Institute for Systems Biology's standard protein mixture data set (30). For LTQ and Q-TOF MS/MS data obtained from mixtures 2 and 3, SEQUEST and Mascot searches were done against the 18 standard proteins and 15 contaminants database appended with the reverse sequences of IPI human. To construct the training data set, top-ranked matches to one of the standard proteins, and contaminants were classified as correct, and their second-ranked matches were classified as incorrect (31). Finally, the weights were obtained separately according to instrument types (ion trap and Q-TOF) and charge states (2+ and 3+) of precursor ions.
Sequence Tag Generation
We generated tags of length 3 and used 100 top scoring tags per spectrum for subsequent processes. The tag generation algorithm was described in our previous work (12).
Software
MODa was implemented in Java programming language and will be available for download from our website (http://prix.uos.ac.kr).
RESULTS
Overall PTM Identification
MODa search was applied to three data sets obtained on ion trap mass spectrometers as follows: 1) 67,648 spectra from human plasma; 2) 363,807 spectra from human HEK293 cell line; and 3) 381,224 spectra from six human aged lenses. The identifications were obtained at FDR 1%. 18,419, 83,554, and 53,724 PSMs for plasma, HEK293, and lens data were identified, respectively. The peptide identifications are listed in supplemental Table 1. Of them, modified PSMs were 3,698, 10,400, and 13,042 for plasma, HEK293, and lens data, respectively. Fig. 1 summarizes the types of modification found in these three samples. Artifacts and chemical derivatives were commonly found, and their presence seemed to vary with experimental conditions. For example, carbamidomethyl DTT formed a major population in plasma but was not observed in the other two samples. In contrast, other modifications are more commonly observed: oxidations and N-terminal pyroglutamate were frequently observed in all samples and are naturally often used as variable modifications in most database search tools. Besides these, N-terminal S-carbamoylmethylcysteine was also commonly observed in all data sets. It is caused by losing NH3 from S-alkylated cysteine, and as a result, a mass shift of +40 Da (57 Da for carbamidomethyl minus 17 Da for NH3) appears on an N-terminal cysteine. It was known that the occurrence of this modification is similar to the rate at which N-terminal pyroglutamate forms from N-terminal glutamine (32), and our analysis confirms that its frequency is not negligible and is thus worthy of being included as a common search parameter. Detecting and allowing for these artifacts would be necessary for maximum coverage of proteins from proteomic experiments. Supporting spectra for modifications in Fig. 1 are provided in supplemental data 1. The analysis of the lens samples resulted in more modifications than the other samples because lens proteins often become substantially modified over time. The observed modifications were in accordance with previous studies (23, 28), and the identified modified peptides with age are shown in supplemental Table 2.
Fig. 1.
Summary of frequent modification types observed from human plasma (a), human HEK293 (b), and human lens data sets (c). +28 and +42 Da on Lys could be also explained as formylation and trimethylation, respectively.
In addition to S-carbamoylmethylcysteine, MODa highlighted modifications on alkylated cysteine. Although the oxidation of methionine residue is most commonly observed, MODa also found the oxidized form of an alkylated cysteine residue. Peptides and fragment ions containing oxidized Met are almost always accompanied by satellite ions by losing methane sulfenic acid (33). Similarly, peptides and fragment ions with alkylated and oxidized Cys were accompanied by distinguishing satellite ions by the loss of ROSH (R = alkylation derivative) (34). As shown in Fig. 2, it is difficult for search algorithms to identify those peptides because these losses become dominant alternative fragmentation pathways, resulting in a reduction of the information in MS/MS spectrum (supplemental data 2). The detection of these modifications demonstrates that MODa is robust to deficiency of fragmentation and is sensitive to modifications.
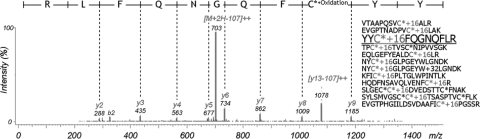
Fig. 2.
Peptides containing alkylated and oxidized cysteine, identified in plasma data, and an MS/MS spectrum. C* represents carbamidomethylated cysteine. In the spectrum, ions that are not generated by peptide backbone fragmentation are observed at 703 and 1078 m/z. The ions are produced by the loss of ROSH (R = alkylation derivative) and are spaced by 107 Da in case of R = carbamidomethyl. These are the evidence for the existence of alkylated and oxidized cysteine, but it becomes the dominant fragmentation pathway, resulting in a low quality MS/MS spectrum.
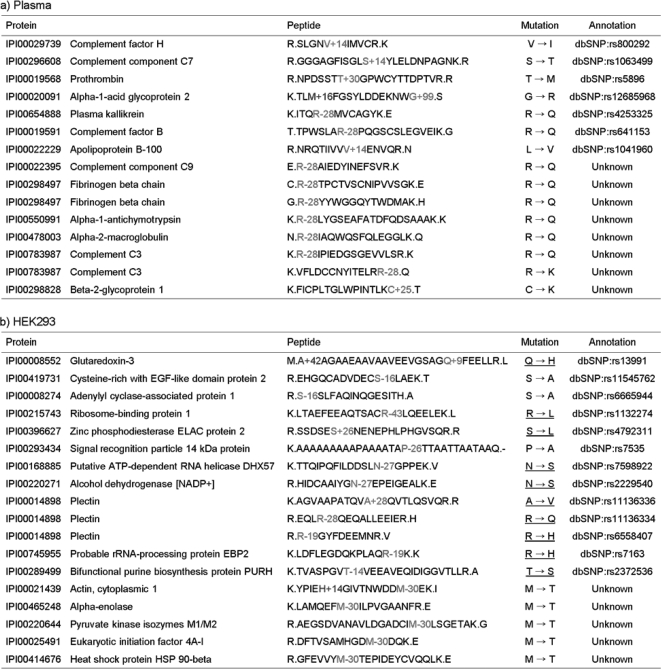
Mutation Analysis
Fig. 3 shows the mutations discovered by MODa analysis of plasma and HEK293 data. Approximately half of these mutations are listed in dbSNP, and known mutations identified from HEK293 are in agreement with those (underlined) from a previous analysis of the same data (27). MODa further found 13 previously unreported mutations, 12 of which could possibly be explained by a single nucleotide polymorphism (SNP). In particular, mutations of Arg → Gln or Lys and Met → Thr were found in plasma and HEK293 data, respectively. In this analysis, ambiguous mutations with no confident peak assignment were rejected and are not reported here. The annotated MS/MS spectra of mutated peptides in Fig. 3 are shown in supplemental data 3.
Fig. 3.
Mutations identified in plasma (a) and HEK293 data sets (b). M+16, A+42, and H+14 represent oxidized methionine, acetylated alanine, and methylated histidine, respectively. The mutations from HEK293 data are underlined if they agree with previous results from gene annotation (27). R-28 could be explained as either Arg → Gln or Arg → Lys substitution. One of the two was selected, assuming that identified peptides are tryptic. Consequently, only in the case where R-28 was detected at C terminus of a peptide was it regarded as an Arg → Lys substitution, and in other cases where Arg → Lys substitution resulted in missed cleavage, it was regarded as Arg → Gln substitution.
Although most discovered mutations were found as single mutation in a peptide, we argue for the necessity of the identification of multiply mutated (or modified) peptides and suggest the potential of MODa for mutation identification. First, from HEK293 analysis, Gln → His substitution was discovered in glutaredoxin-3 by the peptide A+42AGAAEAAVAAVEEVGSAGQ+9FEELLR, which was also acetylated at its N terminus. This peptide was found only in its multiply modified form, which is supported by the UniProt annotation of N-acetylation and the substitution (dbSNP:rs13991). This identification is unique to MODa and cannot be obtained by other unrestrictive tools, which searched for modifications against identified unmodified peptides or considered only one modification/peptide. Similarly, in plasma analysis, the peptide TLM+16FGSYLDDEKNWG+99 in α1-acid glycoprotein was identified as a multiply modified form with oxidized Met and Gly → Arg substitution (dbSNP:rs12685968). Nontryptic cleavage at C terminus of this peptide was also supported by substituting Gly to Arg.
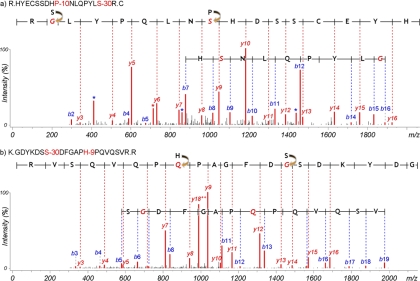
Fig. 4 shows identified peptides with two mutations. HYECSSDHP−10NLQPYLS−30R was identified in γd-crystallin and was found in addition to the wild type (unmodified) form. The Pro → Ser substitution is listed in dbSNP as rs28931605 and is known as a mutation associated with polymorphic congenital cataract (35), and the Ser → Gly substitution is novel. The presence of such mutations was confidently supported by the annotated MS/MS spectrum in Fig. 4. The Pro → Ser substitution of this peptide was not observed alone but only jointly with the Ser → Gly substitution. The coexistence of two mutations could be interpreted as a haplotype, caused by a particular combination of a SNP allele at one site and a specific allele at other nearby variant sites (36). The determination of haplotypes is an important aspect of disease association because altered phenotypes often result from a combination of multiple factors. Interestingly, the mutated peptide was found only in 70- and 93-year cataract lens samples, and another peptide GDYKDSS−30DFGAPH−9PQVQSVR in βB2-crystallin was also identified with two unknown substitutions (Ser → Gly and His → Gln) only in 70- and 93-year cataract lens samples. Altogether these results indicate that further study of genetic variants related with cataract disease may be justified.
Fig. 4.
MS/MS spectra of multiply mutated peptides identified in cataract lens sample, HYECSSDHP−10NLQPYLS−30R in γd-crystallin (a) and GDYKDSS−30DFGAPH−9PQVQSVR (b) in βB2-crystallin. Both peptides were only identified in samples from patients with cataract lens.
Rare and Unknown Modifications
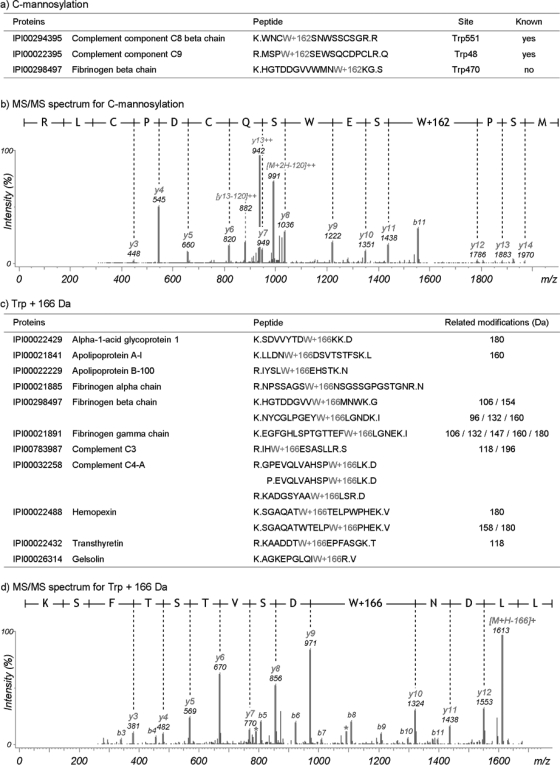
Glycosylation is the enzymatic process that attaches glycans to proteins (37), and at least 22% of human proteins are glycosylated according to current Swiss-Prot annotations. Although N- and O-linked types of glycosylations are well known, MODa was able to further detect a rare form of glycosylation, C-linked glycosylation or C-mannosylation, which indicates the covalent attachment of a mannose residue to a tryptophan residue in an extracellular protein (38). Fig. 5a shows three mannosylated peptides found in human plasma data and containing the recognition motif WXXW; mannosylation of either tryptophan results in a nominal mass increase of 162 Da. Fig. 5b shows an annotated spectrum with prominent neutral losses of 120 Da from precursor and fragment ions, a known characteristic neutral loss from C-mannosylation (38). Interestingly, MODa identified a modification with a form that is analogous to C-mannosylation. The modification was characterized by a mass shift of 166 Da at Trp and was supported by 15 unique peptides listed in Fig. 5c. The comparison of MS/MS spectra of the modified peptides and the corresponding unmodified peptides clearly showed the mass shift of 166 Da at Trp (annotated MS/MS spectra in supplemental data 4). This modification can be explained as kynurenine (+4 Da) plus C-mannosylation (+162 Da). It was known that the reducing sugars like glucose and mannose react with an aromatic amine of kynurenine to form glycosylamines (39). In addition, the other types of modifications were observed at the same Trp residue (related modifications column of Fig. 5c). Fig. 5d shows an annotated MS/MS spectrum for the 166 Da modification. In this spectrum, the neutral loss of the modification from the precursor was clearly visible, but it did not form a dominant fragmentation pathway like the neutral losses from oxidized or phosphorylated peptides (40).
Fig. 5.
Glycosylation on tryptophan residue. a, C-mannosylation sites identified in plasma data. b, MS/MS spectrum of C-mannosylation. MODa identification of C-mannosylated peptides is corroborated by the C-mannosylation recognition motif WXXW and by the characteristic neutral losses of 120 Da from precursor and fragment ions. c, +166 Da modification on Trp. Identified peptides are listed. Extra type column represents the other types of unknown modifications on the corresponding Trp residue. d, MS/MS spectrum of Trp+166 Da. In the MS/MS spectrum of the modified peptide with Trp+166 Da, we observed the ion losing the modification from precursor ion, and consequently the ion corresponds to its unmodified peptide ion.
The MODa Algorithm
The MODa workflow is summarized as follows: 1) candidate peptides are selected from a database using sequence tags derived from each MS/MS spectrum, and 2) a dynamic programming algorithm is used to find an optimal spectrum/peptide to identify modifications between matched sequence tags. Fig. 6 shows the improved spectral alignment algorithm using dynamic programming based on multiple sequence tags. When a mismatch occurs while matching N-terminal flanking mass of a sequence tag, the Δ mass was computed from its corresponding database sequence. Each row represents the theoretical spectrum shifted by N-terminal Δ mass of each sequence tag (or modified by the Δ mass at N terminus of a peptide). Nodes in each row are first classified according to the position of the matched tag and labeled as follows: 0 for before the tag including the start node of the tag; 1 for inside the tag; 2 for after the tag including the end node of the tag (note that nodes at both ends of a tag are not inside the tag), which are used to restrict jumps between nodes. Amino acid jump is allowed only in the same row. Modification jumps between different rows follow the following rules: jump from 0-labeled node is never allowed; jumps from 1- and 2-labeled nodes are allowed only to 0- and 1-labeled nodes; two consecutive modification jumps are not allowed without at least one tag in-between (at least one amino acid jump is required inside the tag). These rules ensure that dynamic programming must use at least one tag out of matched tags, but does not require using all the matched tags. Generally, matched tags are not always correct because during collision activated dissociation, many supplementary fragment ions can be produced by unexpected dissociation pathways, and as a result, polymorphous (having the same partial sequence but shifted by mass) sequence tags can be obtained from continuous internal ions or neutral loss ions (33). Under these constraints, dashed arrows of Fig. 6 represent all possible jumps between nodes. Here, the spectral alignment problem is finding the highest scoring path, where the score of a path is the sum of node scores on the path and each node's score is calculated based on the intensity of the corresponding peak in the experimental spectrum. In improved spectral alignment, its time complexity is O(T2N), where T is the number of tags matched to the peptide (usually less than 5), and N is the peptide length (more details under “Dynamic Programming Based on Multiple Tags” under “Experimental Procedures”).
In contrast, traditional spectral alignments were computed using a two-dimensional matrix with rows representing masses in an MS/MS spectrum and columns representing masses from a database peptide (17, 18). The time complexity depends on spectrum size (M), peptide length (N), the number of modifications (k) per peptide, and mass range (d) for modifications, resulting in O(MNkd). When compared with a representative spectral alignment algorithm, MS-Alignment, our method was 40 times faster under the same condition. Notably, the most prominent difference between MODa and others is in the use of the parameter k. Although other tools would require a known fixed k in advance, MODa determines k automatically during dynamic programming, where k is related to how many tags contributed to its path.
Competence in Peptide Identifications
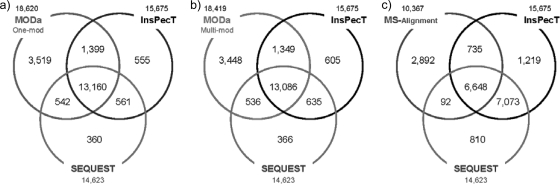
To test MODa performance in various aspects, several searches were conducted against human plasma data. Overall, for used search tools, the numbers of identified PSMs were as follows: 1) SEQUEST: 14,623; 2) InsPecT: 15,675; 3) Mascot/error-tolerant: 17,131; 4) MS-Alignment: 10,367; 5) MODa/one-mod: 18,620; and 6) MODa/multi-mod: 18,419 (more details under “Experimental Procedures”).
To test the identification sensitivity of unmodified peptides, MODa results were first compared with the identification performance of standard database search algorithms, SEQUEST and InsPecT. SEQUEST and InsPecT searches were conducted with no variable modifications to evaluate how well MODa maintains its standard identification performance while dealing with the expected increase in false positives. Fig. 7 shows the identification comparisons between search tools. MODa (Fig. 7a for one-mod and Fig. 7b for multi-mod) retained 93 and 92% of SEQUEST and InsPecT identifications, respectively. This is not likely to be regarded as the reduced performance of MODa for unmodified peptides because identifications between search tools can vary depending on their algorithms, as can be seen by the fact that 13,721 PSMs (94% of SEQUEST and 88% of InsPecT identifications) were overlapped between SEQUEST and InsPecT. In contrast, MS-Alignment retained 46 and 47% of SEQUEST and InsPecT identifications, respectively (Fig. 7c), showing the significant drop in identification performance for unmodified peptides. These results demonstrated that MODa is very robust and does not lose unmodified peptide identifications, even when it is run against a much larger search space of multiply modified peptides. Furthermore, MODa was 40 times faster than MS-Alignment under the same search condition. Additionally, MODa and MS-Alignment results were compared with Mascot error-tolerant search results to test the identification performance including modified peptides. The results are described in supplemental data 5.
Fig. 7.
Comparison of identifications by MODa/one-mod (a), MODa/multi-mod (b), and MS-Alignment (c) with identifications from SEQUEST and InsPecT searches against human plasma data. All of the identifications were obtained at FDR 1%. The numeric figure above or below each tool represents the number of its overall identifications.
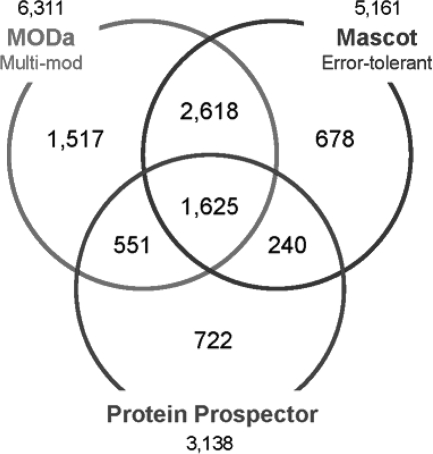
Next, for PTM-rich 93-year-old cataract lens data, we compared three tools, MODa/multi-mod, Mascot error-tolerant search, and Protein Prospector, all of which support different types of unrestrictive searches and are also able to identify multiply modified peptides. Fig. 8 shows the identification comparison and the numbers of PSMs obtained at FDR 1% were as follows: 1) MODa/multi-mod: 6,311; 2) Mascot/error-tolerant: 5,161; and 3) Protein Prospector: 3,138 (search parameters details under “Experimental Procedures”). It should be noted that MODa search was conducted against whole Swiss-Prot human proteins, whereas Mascot and Protein Prospector searches were conducted against a subset of proteins identified from their initial searches (i.e. multi-stage searches). MODa resulted in 22 and 100% more PSMs than Mascot and Protein Prospector, respectively, even though the multi-stage strategy is expected to alleviate the increase of false negatives/positives and thus expected to yield more identifications in unrestrictive searches (41, 42). We also compared the modification types detected by each tool, and supplemental Table 3 shows the top 20 modification types in the PTM frequency matrix of each tool. Most types detected by MODa agreed with previous reports (13, 18), whereas Protein Prospector reported many artifact modifications common in unrestrictive searches (23). Noticeably, hits of known types in the MODa PTM frequency matrix are more clearly distinguishable from the background, when compared with other tools. Unrestrictive searches predict modifications to be present in a sample using any forms of profiles (matrix or histogram). Such profiles commonly contain noise and have difficulty in telling real modifications from noise. For example, in PTM frequency matrixes in supplemental Table 3, Mascot cannot confidently tell deamidation sites, but MODa shows the high bias toward the expected Asn and Gln sites, thus further showing the effectiveness of the MODa algorithm.
Fig. 8.
Comparison among identifications from MODa/multi-mod, Protein Prospector, and Mascot/error-tolerant searches against human 93-year-old cataract lens data. All of the identifications were obtained at FDR 1%. The numeric figure above or below each tool represents the number of its overall identifications.
Sensitivity over Modified Peptides
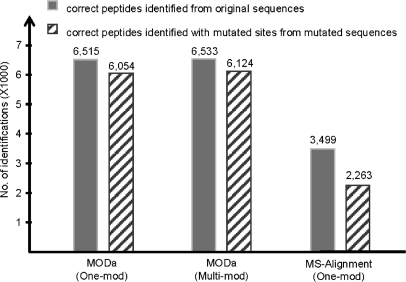
We performed a simulation test to evaluate how sensitive MODa is to a modified region in an MS/MS spectrum. For 14,623 PSMs confidently identified from plasma data, database sequences corresponding to ∼50% were mutated by changing one residue of each sequence to a random residue (mutDB; see “Experimental Procedures”). Then it was measured how many spectra could be identified as the original peptides with one amino acid mutation when searched against mutDB.
Two (one- and multi-mod) MODa searches for whole spectra from human plasma data were conducted against the mutated database (of IPI human proteins) appended with its shuffled sequences, and MS-Alignment search was also conducted to compare the performance. MODa identifications were obtained at FDR 1%, and MS-Alignment identification was obtained below p value of 0.01. Finally, 15,809, 16,320, and 8,339 PSMs for MODa/one-mod, MODa/multi-mod, and MS-Alignment were obtained, respectively. Fig. 9 shows the numbers of PSMs matched with the 14,623 PSMs of SEQUEST. MODa correctly identified 90% of nonmutated PSMs and 83% of mutated PSMs, whereas MS-Alignment identified 48% of nonmutated PSMs and 31% of mutated PSMs.
Fig. 9.
MODa performance on mutated database.
In this experiment with the mutated database, MODa/multi-mod search identified more spectra than MODa/one-mod search. This is due to the existence of originally modified peptides. For example, assume the search against the normal database had identified an unmodified peptide MVFAIPFSFR and, at the same time, its modified peptide M+16VFAIPFSFR. If we mutated the sequence of the unmodified peptide to MVFAIPPSFR (7th F→P), the spectrum corresponding to the unmodified peptide MVFAIPFSFR must be identified as MVFAIPP+40SFR, and the spectrum corresponding to the modified peptide M+16VFAIPFSFR must be identified as M+16VFAIPP+40SFR in the search against the mutated database. However, MODa/one-mod search could not identify the latter spectrum as M+16VFAIPP+40SFR, and the spectrum would result in an incorrect identification matching target or decoy proteins. If the spectrum is identified as a decoy peptide, it would make a score threshold for FDR calculation higher. Actually, in MODa/one-mod search against the mutated database, the score threshold at FDR 1% was higher than in the search against the normal database, and as a result the identification number was decreased.
DISCUSSION
MODa is the first practical multi-blind unrestrictive approach for the identification of multiply modified peptides. Some tools such as TagRecon (15), MODmap (16), and Protein Prospector (21) permitted the identification of multiply modified peptides, but it is possible only by a combination of one or more predefined modifications (i.e. known in advance) and at most one unknown modification allowed per peptide (single-blind). MODa efficiently addresses the computational limitations of current spectral alignment algorithms for unrestrictive PTM searches and achieves over an order of magnitude speedup while delivering significant improvements in identification accuracy. The utility of MODa was demonstrated by generating more peptide identifications and, at the same time, discovering mutated and rare modified peptides from high throughput complex mixture data. In plasma data analysis, MODa increased whole identifications by 78% (18,419 versus 10,367) over MS-Alignment and modified ones by 26% (3,698 versus 2,920); the increase in common modifications was more dramatic: identifications modified by methionine oxidation or N-terminal pyroglutamate were increased by 400% (1,079 versus 213), for example. In this work, unrestrictive PTM search of multiply modified peptides was for the first time conducted against large data sets. Approximately 10–25% of all identified peptides were modified (12, 20, and 24% for HEK293, plasma, and lens, respectively). As samples become more complex, the observed fraction of modified peptides was lower, possibly because of the low stoichiometry of modified proteins. Nevertheless, we found that the rate of multiply modified peptides was ∼5% for all the data sets, and modified peptides with more than three modifications were rarely found.
Computational analysis of post-translational modifications can be divided into three separate subproblems: 1) discovery and identification of unexpected modifications, 2) assignment of post-translationally modified peptide sequences to MS/MS spectra, and 3) localization of modifications to specific sites in the modified peptide sequence of each identified spectrum. MODa was designed to address subproblems 1 and 2 but not subproblem 3. Even though several tools correctly identify peptide sequences and their modifications (subproblem 2), it is often difficult to assign the exact site of modification because of insufficient information in a spectrum (e.g. because of missing ions near the N/C termini). To address this problem, referred to as Δ-correct, we adopted a modified “strength in numbers” strategy (18). For ambiguous site assignments with little difference in score, the sites were reassigned via post-processing based on known modification knowledge and the values in PTM frequency matrix, each entry (Δ,aa) of which represents the number of PSMs with modification mass Δ on amino acid (aa).
Recently, assessing the reliability of a site assignment has become an important issue. PTMFinder post-processed the results from unrestrictive database searches and assessed the accuracy of modification discovery based on multiple witnesses (23). As a special case, AScore assessed the correctness of phosphorylation site localization using site-determining ions (43).
PTMs are a signature for biological events, and their roles in protein functions are still unknown in many cases. MODa revealed that the extent and diversity of modifications in human samples is likely much wider than is currently acceptable to search with existing database search tools. In addition, MODa found substantial support for novel modifications whose structure, mechanism, and significance will require further investigation with additional follow-up experiments.
Footnotes
* The work described in this paper has been conducted while S. Na and E. Paek were at University of Seoul. This work was supported by 21C Frontier Functional Proteomics Project Grant FPR08-A1-020 from the Korean Ministry of Education, Science & Technology (MEST); by National Research Foundation of Korea Grant 2011-0006244 funded by the Korea Government; by the Center for Cell Signaling & Drug Discovery Research at Ewha Womans University; and by the 2010 Research Fund of the University of Seoul. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- PTM
- post-translational modification
- SNP
- single nucleotide polymorphism
- PSM
- peptide-spectrum match
- PRM
- prefix-residue mass
- FDR
- false discovery rate
- IPI
- International Protein Index.
REFERENCES
- 1. Mann M., Jensen O. N. (2003) Proteomic analysis of post-translational modifications. Nat. Biotechnol. 21, 255–261 [DOI] [PubMed] [Google Scholar]
- 2. Nielsen M. L., Savitski M. M., Zubarev R. A. (2006) Extent of modifications in human proteome samples and their effect on dynamic range of analysis in shotgun proteomics. Mol. Cell. Proteomics 5, 2384–2391 [DOI] [PubMed] [Google Scholar]
- 3. Creasy D. M., Cottrell J. S. (2004) Unimod: Protein modifications for mass spectrometry. Proteomics 4, 1534–1536 [DOI] [PubMed] [Google Scholar]
- 4. Cantin G. T., Yates J. R., 3rd (2004) Strategies for shotgun identification of post-translational modifications by mass spectrometry. J. Chromatogr. A 1053, 7–14 [DOI] [PubMed] [Google Scholar]
- 5. Steen H., Mann M. (2004) The ABC's (and XYZ's) of peptide sequencing. Nat. Rev. Mol. Cell Biol. 5, 699–711 [DOI] [PubMed] [Google Scholar]
- 6. Eng J. K., McCormack A. L., Yates J. R. (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 [DOI] [PubMed] [Google Scholar]
- 7. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 8. Craig R., Beavis R. C. (2003) A method for reducing the time required to match protein sequences with tandem mass spectra. Rapid Commun. Mass Spectrom. 17, 2310–2316 [DOI] [PubMed] [Google Scholar]
- 9. Mann M., Wilm M. (1994) Error-tolerant identification of peptides in sequence databases by peptide sequence tags. Anal. Chem. 66, 4390–4399 [DOI] [PubMed] [Google Scholar]
- 10. Tabb D. L., Saraf A., Yates J. R., 3rd (2003) GutenTag: High-throughput sequence tagging via an empirically derived fragmentation model. Anal. Chem. 75, 6415–6421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tanner S., Shu H., Frank A., Wang L. C., Zandi E., Mumby M., Pevzner P. A., Bafna V. (2005) InsPecT: Identification of posttranslationally modified peptides from tandem mass spectra. Anal. Chem. 77, 4626–4639 [DOI] [PubMed] [Google Scholar]
- 12. Na S., Jeong J., Park H., Lee K. J., Paek E. (2008) Unrestrictive identification of multiple post-translational modifications from tandem mass spectrometry using an error-tolerant algorithm based on an extended sequence tag approach. Mol. Cell. Proteomics 7, 2452–2463 [DOI] [PubMed] [Google Scholar]
- 13. Searle B. C., Dasari S., Wilmarth P. A., Turner M., Reddy A. P., David L. L., Nagalla S. R. (2005) Identification of protein modifications using MS/MS de novo sequencing and the OpenSea alignment algorithm. J. Proteome Res. 4, 546–554 [DOI] [PubMed] [Google Scholar]
- 14. Han Y., Ma B., Zhang K. (2005) SPIDER: Software for protein identification from sequence tags with de novo sequencing error. J. Bioinform. Comput. Biol. 3, 697–716 [DOI] [PubMed] [Google Scholar]
- 15. Dasari S., Chambers M. C., Slebos R. J., Zimmerman L. J., Ham A. J., Tabb D. L. (2010) TagRecon: High-throughput mutation identification through sequence tagging. J. Proteome Res. 9, 1716–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Na S., Paek E. (2009) Prediction of novel modifications by unrestrictive search of tandem mass spectra. J. Proteome Res. 8, 4418–4427 [DOI] [PubMed] [Google Scholar]
- 17. Pevzner P. A., Mulyukov Z., Dancik V., Tang C. L. (2001) Efficiency of database search for identification of mutated and modified proteins via mass spectrometry. Genome Res. 11, 290–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tsur D., Tanner S., Zandi E., Bafna V., Pevzner P. A. (2005) Identification of post-translational modifications by blind search of mass spectra. Nat. Biotechnol. 23, 1562–1567 [DOI] [PubMed] [Google Scholar]
- 19. Savitski M. M., Nielsen M. L., Zubarev R. A. (2006) ModifiComb, a new proteomic tool for mapping substoichiometric post-translational modifications, finding novel types of modifications, and fingerprinting complex protein mixtures. Mol. Cell. Proteomics 5, 935–948 [DOI] [PubMed] [Google Scholar]
- 20. Bandeira N., Tsur D., Frank A., Pevzner P. A. (2007) Protein identification by spectral networks analysis. Proc. Natl. Acad. Sci. U.S.A. 104, 6140–6145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chalkley R. J., Baker P. R., Medzihradszky K. F., Lynn A. J., Burlingame A. L. (2008) In-depth analysis of tandem mass spectrometry data from disparate instrument types. Mol. Cell. Proteomics 7, 2386–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen Y., Chen W., Cobb M. H., Zhao Y. (2009) PTMap: A sequence alignment software for unrestricted, accurate, and full-spectrum identification of post-translational modification sites. Proc. Natl. Acad. Sci. U.S.A. 106, 761–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tanner S., Payne S. H., Dasari S., Shen Z., Wilmarth P. A., David L. L., Loomis W. F., Briggs S. P., Bafna V. (2008) Accurate annotation of peptide modifications through unrestrictive database search. J. Proteome Res. 7, 170–181 [DOI] [PubMed] [Google Scholar]
- 24. Ong S. E., Mittler G., Mann M. (2004) Identifying and quantifying in vivo methylation sites by heavy methyl SILAC. Nat. Methods 1, 119–126 [DOI] [PubMed] [Google Scholar]
- 25. Elias J. E., Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 26. Na S., Paek E., Lee C. (2008) CIFTER: Automated charge-state determination for peptide tandem mass spectra. Anal. Chem. 80, 1520–1528 [DOI] [PubMed] [Google Scholar]
- 27. Tanner S., Shen Z., Ng J., Florea L., Guigó R., Briggs S. P., Bafna V. (2007) Improving gene annotation using peptide mass spectrometry. Genome Res. 17, 231–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wilmarth P. A., Tanner S., Dasari S., Nagalla S. R., Riviere M. A., Bafna V., Pevzner P. A., David L. L. (2006) Age-related changes in human crystallins determined from comparative analysis of post-translational modifications in young and aged lens: Does deamidation contribute to crystalline insolubility? J. Proteome Res. 5, 2554–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dasari S., Wilmarth P. A., Rustvold D. L., Riviere M. A., Nagalla S. R., David L. L. (2007) Reliable detection of deamidated peptides from lens crystallin proteins using changes in reversed-phase elution times and parent ion masses. J. Proteome Res. 6, 3819–3826 [DOI] [PubMed] [Google Scholar]
- 30. Klimek J., Eddes J. S., Hohmann L., Jackson J., Peterson A., Letarte S., Gafken P. R., Katz J. E., Mallick P., Lee H., Schmidt A., Ossola R., Eng J. K., Aebersold R., Martin D. B. (2008) The standard protein mix database: A diverse data set to assist in the production of improved peptide and protein identification software tools. J. Proteome Res. 7, 96–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Elias J. E., Gibbons F. D., King O. D., Roth F. P., Gygi S. P. (2004) Intensity-based protein identification by machine learning from a library of tandem mass spectra. Nat. Biotechnol. 22, 214–219 [DOI] [PubMed] [Google Scholar]
- 32. Geoghegan K. F., Hoth L. R., Tan D. H., Borzilleri K. A., Withka J. M., Boyd J. G. (2002) Cyclization of N-terminal S-carbamoylmethylcysteine causing loss of 17 Da from peptides and extra peaks in peptide maps. J. Proteome Res. 1, 181–187 [DOI] [PubMed] [Google Scholar]
- 33. Paizs B., Suhai S. (2005) Fragmentation pathways of protonated peptides. Mass Spectrom. Rev. 24, 508–548 [DOI] [PubMed] [Google Scholar]
- 34. Steen H., Mann M. (2001) Similarity between condensed phase and gas phase chemistry: Fragmentation of peptides containing oxidized cysteine residues and its implications for proteomics. J. Am. Soc. Mass Spectrom. 12, 228–232 [DOI] [PubMed] [Google Scholar]
- 35. Plotnikova O. V., Kondrashov F. A., Vlasov P. K., Grigorenko A. P., Ginter E. K., Rogaev E. I. (2007) Conversion and compensatory evolution of the gamma-crystallin genes and identification of a cataractogenic mutation that reverses the sequence of the human CRYGD gene to an ancestral state. Am. J. Hum. Genet. 81, 32–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. International HapMap Consortium (2005) A haplotype map of the human genome. Nature 437, 1299–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Spiro R. G. (2002) Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptides bonds. Glycobiology 12, 43R–56R [DOI] [PubMed] [Google Scholar]
- 38. Hofsteenge J., Blommers M., Hess D., Furmanek A., Miroshnichenko O. (1999) The four terminal components of the complement system are C-mannosylated on multiple tryptophan residues. J. Biol. Chem. 274, 32786–32794 [DOI] [PubMed] [Google Scholar]
- 39. Gokhale M. Y., Kearney W. R., Kirsch L. E. (2009) Glycosylation of aromatic amines I: Characterization of reaction products and kinetic scheme. AAPS PharmSciTech 10, 317–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tabb D. L., Friedman D. B., Ham A. L. (2006) Verification of automated peptide identifications from proteomic tandem mass spectra. Nat. Protoc. 1, 2213–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Everett L. J., Bierl C., Master S. R. (2010) Unbiased statistical analysis for multi-stage proteomic search strategies. J. Proteome Res. 9, 700–707 [DOI] [PubMed] [Google Scholar]
- 42. Bern M., Kil Y. J. (2011) Comment on “Unbiased statistical analysis for multi-stage proteomic search strategies.” J. Proteome Res. 10, 2123–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Beausoleil S. A., Villén J., Gerber S. A., Rush J., Gygi S. P. (2006) A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 24, 1285–1292 [DOI] [PubMed] [Google Scholar]