Abstract
Many software tools have been developed for the automated identification of peptides from tandem mass spectra. The accuracy and sensitivity of the identification software via database search are critical for successful proteomics experiments. A new database search tool, PEAKS DB, has been developed by incorporating the de novo sequencing results into the database search. PEAKS DB achieves significantly improved accuracy and sensitivity over two other commonly used software packages. Additionally, a new result validation method, decoy fusion, has been introduced to solve the issue of overconfidence that exists in the conventional target decoy method for certain types of peptide identification software.
Peptide identification from tandem mass spectrometry (MS/MS)1 data is a central task in proteomics. The accuracy and sensitivity of this task directly impacts the performance of protein identification from peptide hits, as well as other downstream analyses. Many software tools have been developed for peptide identification; these tools can be broadly divided into two categories: de novo sequencing and database search.
De novo sequencing derives the peptide sequence directly from the MS/MS spectrum, whereas a database search queries a sequence database for the best peptide to explain the peaks in the MS/MS spectrum. Representative de novo sequencing software packages include PEAKS (1), PepNovo (2), NovoHMM (3), and Lutefisk (4), and representative database search software packages include Mascot (5), SEQUEST (6), X!Tandem (7), OMSSA (8), ProteinProspector (9), MaxQuant (10) (11) and MS-GFDB (12).
The database search is generally believed to be a simpler approach because the protein sequence database provides a limited space for the software to search. Therefore, when a protein sequence database is available, a database search is the most common method for peptide identification. However, existing database search tools still experience problems of low identification rates (low sensitivity) (13) (14) and high false discovery rates (low accuracy) (15). The improvement of database search performance has always been an active research area in this field.
Two competing objectives are sought in the database search approach: accuracy and sensitivity. The accuracy is usually measured by the false discovery rate (FDR), which is defined as the percentage of the false identifications in all identifications above the score threshold. Accuracy can be accomplished by increasing the score threshold. However, this will at the same time reduce the sensitivity. To improve both accuracy and sensitivity, a new scoring function needs to be developed that more accurately separates the true and false identifications (16, 17). Meanwhile, to maintain an acceptable search speed, database search software often introduces a filtration method to quickly select a shortlist of protein or peptide candidates and will only evaluate those candidates with a more advanced (and usually slower) scoring function (see for example Ref. 7). However, this simple filtration often excludes real peptides and causes reduced sensitivity. A good filtration technique is required to balance sensitivity, accuracy, and speed.
In this paper, the PEAKS DB software is described for peptide identification using the database search approach. However, as opposed to the traditional database search approach, the PEAKS DB software relies heavily upon de novo sequencing results to improve the filtration and the scoring function. This combination results in significantly improved sensitivity and accuracy in comparison to existing database search software.
In addition to the aforementioned two objectives (accuracy and sensitivity), the high throughput generation of proteomics mass spectrometry data requires the automated validation of database search results. Currently, this validation is typically achieved by the target decoy method (18, 19). This method introduces decoy proteins to be searched by the same search engine and uses the engine's outcome on the decoy proteins to estimate the number of false positives. However, the method has to be used with caution because a multi-stage search procedure can make it biased toward underestimating the FDR (20–22). A fix was initially proposed in Ref. 21, but Bern and Kil (22) pointed out that the fix was still biased. They proposed an alternative solution by adding more decoy proteins at the second stage of the search on top of the decoy proteins introduced initially. This requires changes of the search engine at the source code level and may cause FDR overestimation (which is a smaller problem than FDR underestimation). Another drawback of the standard target decoy method is that it was incapable of validating a search engine's results if the protein information is used in the peptide scoring function (23). In this paper, we show that a slight change to the target decoy method will solve these two problems. Instead of adding the decoy proteins as separate entries of the database, we concatenate the target and decoy sequences of the same protein together as a single entry of the database. In this paper, this new strategy is investigated, and an improved target decoy method, decoy fusion, is presented.
EXPERIMENTAL PROCEDURES
The aim of PEAKS DB is to identify peptides from a sequence database with MS/MS data. As such, PEAKS DB belongs to the database search category of peptide identification software. However, PEAKS DB employs de novo sequencing as a subroutine and exploits the de novo sequencing results to improve both the speed and accuracy of the database search. The main algorithmic steps of the PEAKS DB software proceed as follows:
De novo sequencing: The PEAKS algorithm (1) is used to perform de novo sequencing for each input spectrum.
Protein shortlisting: The de novo sequence tags are used to find approximate matches in the protein sequence database. All of the proteins in the database are evaluated according to the sequence tag matches. The 7,000 top ranked proteins form the protein shortlist and are used in future analysis.
Peptide shortlisting: All of the peptides of the protein shortlist are used to match the MS/MS spectra with a rapid scoring function. Only the 512 highest scoring peptide candidates (including those with PTMs) are kept for each MS/MS spectrum.
Peptide scoring: From the 512 candidates calculated in the peptide shortlisting step, a precise scoring function is used to find the best peptide for each spectrum. The similarity between the de novo sequence and the database peptide is an important component in the scoring function. In addition, the score is normalized to ensure it can be compared across different spectra.
Result validation: A modified target decoy approach is used to determine the minimum peptide spectrum matching score threshold to meet the FDR requirement of the user.
Protein inference and grouping: The high confidence peptides identified through the above steps are used to infer the proteins. Those proteins that share the same set of peptide hits are grouped together for a more convenient report.
The details of these steps are discussed in the following sections.
De Novo Sequencing
The PEAKS algorithm is used to perform de novo sequencing for each input spectrum. The same parameters (mass error tolerance and PTMs) specified by the user for database search are also used for de novo sequencing. For each spectrum, only the first de novo sequencing peptide reported by PEAKS is utilized. The PEAKS algorithm also computes a confidence for each amino acid in the de novo sequence; this confidence is a percentage value. The output of PEAKS is converted to a sequence tag by replacing the low confidence amino acids by their mass values. More specifically, each stretch of adjacent amino acid residues with <30% confidence is replaced by a “mass segment” that is equal to the total mass of the residues. See Fig. 1 as an example.
Fig. 1.
A de novo sequence computed with PEAKS has a local confidence score on each amino acid, as represented by the heights of the vertical bars. By using a threshold of 30%, the consecutive amino acids below the confidence threshold are substituted by their total residue mass.
Protein Shortlisting
In this step, the algorithm uses the de novo sequence tags to select a short list of proteins from the protein database. Future steps in the process will only work on this short list to reduce the total computing time.
The matching quality between a de novo sequence tag and a database peptide is measured by the number of common amino acids (the CAA score). In Fig. 2, the computation of the CAA score is illustrated. Note that in this protein shortlisting step, because there is no modification information in the sequence database, a modified residue on the de novo sequence can match an unmodified residue in the sequence database. However, in the later peptide scoring step, a modified residue can only match the same residue with the same modification for the CAA score calculation.
Fig. 2.

A de novo sequence tag is compared with a database peptide. The alignment ensures that the mass of each aligned block (surrounded by square brackets) is equal for both sequences. The CAA score is the number of common amino acids in the alignment, which is 4 in this example.
The proteins are ranked by the highest CAA score achieved by the peptides of each protein. If two proteins have the same highest CAA score, the tie is broken by the second and the third highest CAA scores. Within this ranking, the 7,000 top database proteins are selected as the protein shortlist, which should be a superset of the identifiable proteins in most proteomics experiments. No special treatment is made on handling homologous proteins in the database. Therefore, the number of shortlist proteins may need to be increased if the biological system studied has a larger number of proteins and the search is on a large database (such as NCBInr) without specifying the taxonomy information. This can be adjusted in the configuration file of PEAKS DB.
Peptide Shortlisting
All of the peptide sequences digested in silico from the protein shortlist are compared against the input spectra to find peptide spectrum matches (PSMs). Each peptide sequence may produce multiple modified peptides by enumerating all possible combinations of the user-specified variable PTMs. For each peptide sequence (modified or not), the peptide mass is calculated, and the MS/MS spectra with the matching precursor mass is compared with the sequence. A “quick scorer” is used to compute the score of the PSM. A priority queue data structure is used to keep the top 512 sequence candidates for each spectrum.
The quick scorer is derived from the same de novo sequencing scoring function used in PEAKS de novo sequencing (1). Briefly, a spectrum is converted to two functions fN(m) and fC(m), where fN(m) indicates the odds that the correct peptide has a prefix (a subsequence containing the N terminus) with total residue mass m, and fC(m) indicates the odds that the correct peptide has a suffix (a subsequence containing the C terminus) with total residue mass m. The odds are estimated with the corresponding fragmentation ions. For a collision induced dissociation (CID) spectrum, a, b, c, y, z, b-H2O, y-H2O, and y-NH3 are used (see Ref. 1 for details). For an ETD spectrum, a, b, c, c-H, y, z, and z+H ions are used (see Ref. 24 for details of the calculation). After fN(m) and fC(m) are calculated, the ion match score of a peptide is determined as the sum of the fN(m) and fC(m′) for all the prefix masses m and suffix masses m′. This score can be calculated efficiently by indexing fN(m) and fC(m) in memory. With this simple quick scorer, the correct peptide of a given MS/MS spectrum may not be the top scoring sequence but is most likely among the 512 top scoring sequence candidates kept in the priority queue for this spectrum.
Peptide Scoring
A more sophisticated scoring function is used to rerank the sequence candidates for each spectrum. First, the ion match score sion_match is normalized by the formula s′ion_match = (sion_match − μ)/σ, where μ represents the mean score of the top 10 candidates, and σ represents the standard deviation of the scores of the top 150 candidates. Such normalization against the incorrect peptides is necessary to compare scores across different spectra. A number of other features are used in addition to the normalized ion match score. Several features have been evaluated. However, the following nine features of a peptide candidate were found to be most effective and are now included in PEAKS DB: 1) the number of amino acids matching the de novo sequence tag (CAA score); 2) the protein feature: each protein obtains a score by adding its three highest peptide CAA scores, and the protein feature of a peptide is the maximum score of the proteins containing this peptide; 3) the peptide length; 4) the average sequence length per missed cleavage in the peptide; 5) the average sequence length per PTM in the peptide; 6) the precursor mass error; 7) the charge state; 8) the maximum length of the consecutively matched fragment ion series; and 9) the number of termini violating the enzyme's digesting rule.
Some of these features or similar features were also previously used in the Percolator (16) and PeptideProphet (17) programs. In particular, 6), 7), and 8) were used in Percolator; 6) and 9) were used in PeptideProphet; features similar to 4) and 5) were used in Percolator; and a feature similar to 4) was used in PeptideProphet. Both Percolator and PeptideProphet used more features than listed here.
These nine features, together with the normalized ion match score, are combined with a weighted sum. The weights are trained with an iterative search on a large LC-MS/MS training data set to maximize the area on the left of the 1% FDR curve, as shown in Fig. 3. Once the weights are determined by the training for a particular instrument type, they do not change from experiment to experiment.
Fig. 3.
The FDR curve shows the FDR (y axis) with respect to the number of peptide spectrum matches to be reported (x axis). The training of the weighted sum coefficients in the peptide scoring function maximizes the area on the left of the curve and below the 1% FDR threshold.
The weighted sum score is converted to a p value for easier human interpretation. For a given score, the corresponding p value is defined as the probability that a false identification in the current search achieves the same or better matching score. The p value attempts to predict the false positive rate, i.e. the ratio between the number of false identifications above the given score T and the total number of false identifications. Note that false positive rate is a different concept from FDR. If the p value is P, the final peptide score (called the significance score) output by PEAKS DB is −10lgP. Here lg(●) is the common logarithm with base 10.
Result Validation
A modified target decoy approach, called decoy fusion, is used to estimate the FDR at any given score threshold. The more conventional target decoy approach requires the generation of a decoy protein sequence for each target protein sequence in the database (16). The target and decoy databases are then searched either separately or together by the software, and the FDR is calculated by the ratio between the numbers of the decoy and target matches. However, in PEAKS DB, the target and decoy sequences are not treated as separate entries in the database. Instead, they are concatenated together for each protein. Thus, the newly generated database contains the same number of protein entries, but the length of each protein is doubled. The software searches this newly generated database. After the search, the target and decoy identifications are separated by checking whether they are from the first or the second half of each concatenated sequence. For each user-specified score threshold, the FDR is calculated as the ratio between the number of decoy hits and the number of target hits above the score threshold.
If the C-terminal amino acid of the target protein is not an enzyme cleavage site, then appending a decoy sequence to its end may prevent the search engine from considering the C-terminal peptide of the target protein. To solve this problem, a special letter J is added in between the target and decoy sequences as the separator. Both Mascot and PEAKS DB algorithm can cleave at both sides of the letter J for the in silico digestion, ensuring that the C-terminal peptide from the target protein is considered.
Protein Inference and Grouping
Although protein inference is not the focus of this paper, the following is a brief outline of the protein inference procedure in PEAKS DB. Proteins are grouped according to their shared peptides. Given a score threshold T, a protein (X) is called to dominate another protein (Y) if all of the peptides of Y with a significance score ≥T are also found in X. In the current version of PEAKS DB, T is equal to 15, corresponding to a p value of ∼0.03.
If X dominates Y, then Y is not a confident identification and is therefore added to the X group. After each pair of proteins is examined for domination relations, the proteins are clustered into several groups. Note that there may be a few proteins dominating each other in a group. For each group, the user can choose to display or export only one dominating protein, all dominating proteins, or all proteins from the user interface.
The significance score of each protein is computed from its identified peptides as follows. First, redundant peptides are removed; if the same peptide is identified multiple times from different spectra, only the highest scoring peptide is retained. Two peptides are considered the same if they are identical or differ only by the PTM location, but considered different if the amino acid sequence or PTMs are different. Second, all the nonredundant significance scores of the peptides are sorted as s1 ≥ s1 ≥ … ≥ sk. Finally, the score of the protein is equal to s1 + (1/2)s2 + (1/3)s3 + … + (1/k)sk. The score of a protein group is equal to the score of the dominating protein.
RESULTS
Two public data sets, one fragmented with CID and the other ETD, were used to evaluate the performance of PEAKS DB. Both data sets were generated with LTQ-Orbitrap instruments.
The CID data set came from the trypsin digest of Pseudomonas aeruginosa and was previously used to study the relation between protein and mRNA abundances (25). The data file was downloaded from http://www.marcottelab.org/MSdata/Data_12/DATA/20090115_SMPA14_2.RAW.gz. For the CID data set, the P. aeruginosa PAO1 protein database, downloaded from PseudoCAP (http://www.pseudomonas.com) in April 2011 was used for database search. The database contains 5566 protein entries.
The ETD data set was obtained from the Lys-C digest of a yeast lysate following strong cation exchange peptide fractionation prior to LC-MS. The raw data from fraction 10 was previously used in the 2011 study by the Proteome Informatics Research Group (iPRG) of the Association of Biomolecular Resource Facilities (15). The same data is used here. For the ETD data set, the same protein sequence database provided by the Association of Biomolecular Resource Facilities iPRG 2011 study was used for database search. It was the complete proteome for Saccharomyces cerevisiae with typical laboratory contaminant proteins appended. The database contains 6666 protein entries.
In all of the experiments involving decoy sequences, the decoy sequences were produced by randomly shuffling the amino acids in each protein. Decoy peptides were removed before FDR calculation. That is, FDR = number of decoy hits/number of target hits. When a target decoy method was used to estimate the FDR, the target and decoy databases were searched together.
The Effectiveness of de Novo Sequencing in Database Search
This section demonstrates the relative performance of the de novo sequencing and database search approaches when analyzing the same data set. Their complementary abilities will justify the utilization of the de novo sequencing results in PEAKS DB. With the CID data set, PEAKS 5.3 and Mascot 2.3 were employed for the de novo sequencing and database search analyses, respectively. For each spectrum, only the first de novo sequencing peptide reported by PEAKS was selected. For each peptide reported by Mascot 2.3, the number of matched amino acids with the de novo sequence (the CAA score) is calculated. Fig. 4 shows the distribution of the scores when the P. aeruginosa database is used. It can be seen that the best separation of the target and decoy matches is achieved by a combination of both the database search score and the CAA score, clearly indicating the effectiveness of using de novo sequencing results in the peptide scoring.
Fig. 4.
The comparison of de novo sequencing results (PEAKS 5.3) with database search results (Mascot 2.3). Each data point represents a peptide found by Mascot database search. The x axis is the Mascot score, and the y axis is the number of matching amino acids with the de novo sequencing result (CAA score). For a better view of the data density, a small random number between 0 and 0.8 is added to each CAA score. The best separation of target and decoy matches is achieved by combining the CAA and Mascot scores (dashed line).
For Mascot to confidently identify a peptide, the required spectrum quality is different when databases of different sizes are used. For example, on the CID data set, the 1% FDR corresponds to Mascot scores of 23.6 and 55.1 when the P. aeruginosa and Swissprot databases were employed, respectively. As a result, the relative performance of de novo sequencing and database search varies. When the P. aeruginosa and Swissprot databases are used for the Mascot database search, respectively, the de novo sequencing was able to correctly compute five or more amino acids (CAA score ≥ 5) on 70 and 88% of the PSMs identified by Mascot with 1% FDR.
Comparing the Target Decoy and Decoy Fusion Methods
The basic assumption of the target decoy and the decoy fusion methods is that the score distribution of the false target hits and the decoy hits are similar. Therefore the number of decoy hits can be used to estimate the number of false target hits. Unfortunately there is no effective way to verify this assumption, because it is difficult to assess whether a target hit is true or false. Thus, the following simulated experiment was conducted to verify the assumption.
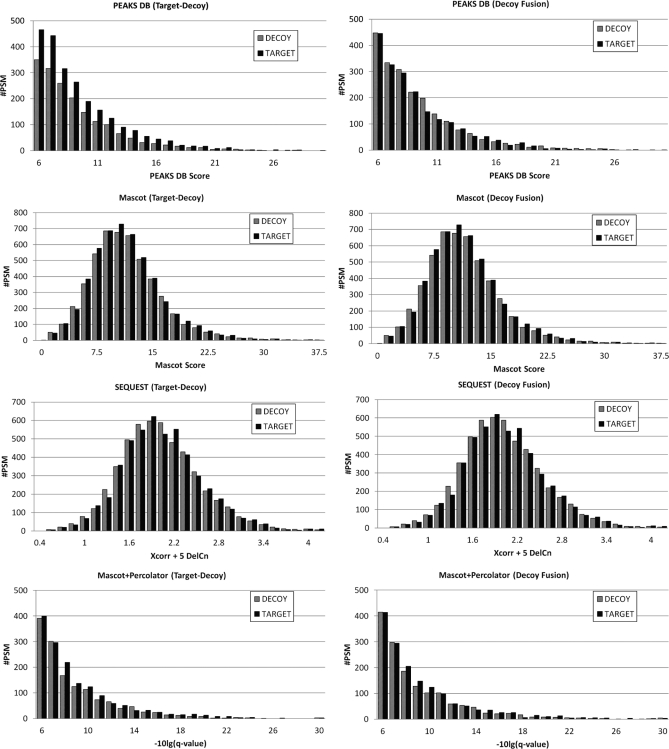
The CID data set was searched against the P. aeruginosa database by Mascot, SEQUEST, and PEAKS DB. The peptides identified by all three engines were considered as correct. A simulated database was created by keeping these peptides unchanged in the P. aeruginosa database, while randomly shuffling all other amino acids in each protein. When a search engine is used to search in this simulated database, the peptides that do not have significant (five or more amino acids) overlap with the unchanged peptides can be safely regarded as false hits. Thus, by using the simulated database as the target, the score distribution of the false target hits and the decoy hits can be compared. Both decoy fusion and target decoy methods were examined, and the results are shown in Fig. 5.
Fig. 5.
The score distribution of the false target hits and the decoy hits when the simulated protein database was used. The height of each bar represents the number of PSMs around the corresponding score. The target decoy method generated fewer decoy hits than the false target hits for the PEAKS DB results, which may lead to FDR underestimation. The decoy fusion method has no such problem.
Fig. 5 illustrates that for the PEAKS DB results, only the decoy fusion method could produce similar score distributions. The target decoy method produced fewer decoy hits than the false target hits, which might cause FDR underestimation. This indicates that decoy fusion is more appropriate for validating the PEAKS DB results. However, the two decoy methods showed no noticeable difference for Mascot, SEQUEST, and Mascot+Percolator results. The result of Fig. 5 is consistent with another experiment aiming to compare the FDR curves estimated by the two decoy methods, respectively (supplement Fig. S1). The two methods produced identical or very similar FDR curves for each of Mascot, SEQUEST, and Mascot+Percolator, whereas the decoy fusion curve of PEAKS DB is noticeably more conservative than the target decoy curve. As such, in all following experiments the decoy fusion method was used to estimate the FDR of PEAKS DB, and the target decoy method was used to estimate the FDR of all other searching methods.
Performance Comparison of PEAKS DB with Other Database Search Tools
Following the general practice, the peptide identification performance of PEAKS DB was compared by FDR curves with two commonly used software packages: Mascot 2.3 and SEQUEST (in Proteome Discoverer 1.2). The search with each of the three engines used the same set of parameters: The parent ion mass error tolerance was 15 ppm, and fragment ion mass error tolerance was 0.8 Da. Up to three missed cleavages were allowed in one peptide, and at most one end of each peptide could violate the enzyme cleavage rule. One fixed PTM: carboxyamidomethylation of Cys, and three variable PTMs: deamidation of Gln and Asn, oxidation of Met, and Pyro-glu from Gln, were used. Trypsin and Lys-C were used as the enzymes for the CID and ETD data sets, respectively. For each peptide spectrum match (PSM), SEQUEST outputs two scores, Xcorr and DelCn. In this experiment Xcorr + 5 DelCn was used as SEQUEST score because this combination produced the optimal FDR curve for SEQUEST.
Recently, the Percolator program has been developed to improve Mascot database search results by rescoring with a rigorous machine learning method (16). It is not a self-contained database search engine. Nevertheless, a comparison with the combination of Mascot and Percolator was also conducted.
Figs. 6 and 7 display the FDR for the CID and ETD data sets, respectively. At a 1% FDR, the numbers of identified target PSMs are PEAKS DB (10668) > Mascot+Percolator (9969) > SEQUEST (8236) > Mascot (7515) from the CID data set; and PEAKS DB (3652) > Mascot+Percolator (2702) > Mascot (2398) > SEQUEST (2233) from the ETD data set.
Fig. 6.
FDR curves of the compared software tools on the CID data set. The x axis represents the number of peptide spectrum matches kept from the target sequences, and the y axis represents the corresponding FDR.
Fig. 7.
FDR curves of the compared software tools on the ETD data set. The x axis represents the number of peptide spectrum matches kept from the target sequences, and the y axis represents the corresponding FDR.
Another recent database search program, MS-GFDB (12), also reported a significant improvement over Mascot. Because the published MS-GFDB does not deal with variable PTMs at the time of this study, we also conducted a special comparison by not specifying any variable PTMs in PEAKS DB (this caused a reduction of the overall performance of PEAKS DB). PEAKS DB also outperformed MS-GFDB by ∼58 and 8% in such a special comparison for CID and ETD, respectively. The detail of this comparison is included in the supplemental materials.
DISCUSSION
Accuracy and Sensitivity
The first conclusion from Figs. 6 and 7 is that PEAKS DB could confidently identify significantly more PSMs than Mascot and SEQUEST. In particular, in comparison to Mascot, at a 1% FDR, PEAKS DB could identify 42% more PSMs for the CID data set and 52% more PSMs for the ETD data set. In fact, PEAKS DB identified more PSMs (9494 for CID and 3299 for ETD) at 0.1% FDR than Mascot (7515 for CID and 2398 for ETD) at 1% FDR. Although Percolator significantly improved the performance of Mascot, PEAKS DB still outperformed Mascot+Percolator by 7% for CID data and by 35% for ETD data at 1% FDR on these data sets.
In terms of the total number of peptides identified, many search engines outperformed Mascot on the ETD data set in the iPRG study mentioned above (15). Among the single-engine results in the iPRG study, the most number of PSMs were reported by the following few engines (in decreasing order): ProteinProspector (9), unnamed in-house software, PEAKS DB, another unnamed in-house software, pFind (26), and Spectrum Mill. However, among these several results, only PEAKS DB and pFind results possessed the accuracy required by the iPRG study (1% FDR). However, it is possible that the FDR estimation method used by the iPRG study and the relative experience of users in operating different software tools might have affected the above ranking. More details are provided in the full report of the iPRG study (15).
Reliable Result Validation
The use of the decoy fusion method is necessary for validation of the PEAKS DB result. As shown under “Results”, the standard target decoy approach may underestimate the FDR of PEAKS DB results and should be avoided. This inaccuracy comes from two sources that are due to the fact that the decoy sequences are introduced as separate entries of the database. First, the protein shortlisting step may select more target proteins than the decoy proteins. This causes the false identifications in later steps to fall in the target proteins with a higher probability. The decoy fusion method avoids this problem by combining the target and decoy sequences in the same protein entry. Second, the “protein feature” is used in the peptide scoring. This increases the scores of the random peptide matches in the highly confident target proteins. Consequently, more false hits will be reported from the target proteins than from the decoy proteins. By fusing the target and decoy sequences together, the score increment is applied equally to the target and decoy peptide hits. Thus, the score distributions of the false target hits and decoy hits remain the same.
There were different opinions in the literature regarding the use of protein information in the peptide scoring function. On one hand, the protein information may compromise the reliability of the target decoy validation method and thus was not used in PeptideProphet (17) and is no longer used in the Mascot Percolator (23). On the other hand, Bern et al. (20) reported significantly improved sensitivity by a second round search on the confidently identified proteins for finding more peptides, which can be regarded as an extreme case of using the protein information in the peptide scoring function. We argue that the use of the protein information is appropriate. By limiting the search on a protein database, a database search engine makes the implicit assumption that each peptide sequence appears in the sample with equal probability, prior to the search. Such prior probability should be updated when another peptide from the same protein is identified with high confidence. This will surely contribute toward the peptide identification sensitivity, but the use of the protein information does require a more robust result validation method than the standard target decoy approach. The decoy fusion method proposed in this paper provides a very simple alternative to solve this problem.
In PEAKS DB, the coefficients for the weighted sum score for peptide scoring are trained only once for each instrument type. This is different from the approach used in Percolator, where the scoring function is retrained for each experiment after the search is completed, and the target and decoy peptides found by the search become known. Although the retraining may further improve the sensitivity, it exposes the decoy information to the scoring function. This creates a risk of impairing the FDR estimation method. To keep the FDR estimation invulnerable, the retraining approach is not used in the current version of PEAKS DB.
De Novo Sequencing and Database Search
De novo sequencing was historically thought to be slow and to require spectra with higher mass accuracy. Therefore it has been mostly used when the protein database was unavailable. Thanks to the recent development in computer algorithms and continuous improvement of computers, the speed is no longer an issue for de novo sequencing. For example, in our experiments the PEAKS algorithm was able to de novo sequence 15 spectra/second on a moderate desktop PC (Intel Core i7 Processor, quad core, 2.8 GHz). The high mass accuracy has also become available because of the development of new mass spectrometers such as the Orbitrap. This makes de novo sequencing a viable choice for every mass spectrometry analysis in proteomics. De novo sequencing and database search should not anymore be regarded as two separate approaches that are used in different circumstances. Instead, they should work together to provide better sensitivity and accuracy in proteomics analysis, as illustrated in this paper. Additionally, the spectra that produce highly confident de novo sequencing tags but no database hits are likely from novel or modified peptides. These “de novo only” peptides may arguably be more interesting than those in the database but are currently rejected in an analysis purely based on database search.
Conclusion
In summary, we described the PEAKS DB software that takes advantages of fast de novo sequencing results and several new features. The net outcome is an increase in both sensitivity and accuracy and an overall superior performance to other commonly used search engines. This is particularly true for mass spectral data obtained by ETD fragmentation, which makes PEAKS DB a particularly useful tool for identifying peptides with PTMs. We also proposed a more robust result validation method, decoy fusion, for controlling the FDR of PEAKS DB results.
Acknowledgments
We are grateful to Dr. Christine Vogel and Dr. Taejoon Kwon for providing the CID data set.
Footnotes
* This work was supported in part by the funds from Natural Sciences and Engineering Research Council of Canada Discovery program (to B. M. and G. L.) and by Bioinformatics Solutions Inc. (to J. Z., L. X., B. S., W. C., M. X., D. Y., W. Z., and Z. Z.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- MS/MS
- tandem mass spectrometry
- PTM
- post-translational modification
- ETD
- electron transfer dissociation
- FDR
- false discovery rate
- PSM
- peptide spectrum match
- iPRG
- Proteome Informatics Research Group.
REFERENCES
- 1. Ma B., Zhang K., Hendrie C., Liang C., Li M., Doherty-Kirby A., Lajoie G. (2003) PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 17, 2337–2342 [DOI] [PubMed] [Google Scholar]
- 2. Frank A., Pevzner P. (2005) PepNovo: De novo peptide sequencing via probabilistic network modeling. Anal. Chem. 77, 964–973 [DOI] [PubMed] [Google Scholar]
- 3. Fischer B., Roth V., Roos F., Grossmann J., Baginsky S., Widmayer P., Gruissem W., Buhmann J. M. (2005) NovoHMM: A hidden Markov model for de novo peptide sequencing. Anal. Chem. 77, 7265–7273 [DOI] [PubMed] [Google Scholar]
- 4. Taylor J. A., Johnson R. S. (1997) Sequence database searches via de novo peptide sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 11, 1067–1075 [DOI] [PubMed] [Google Scholar]
- 5. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 6. Eng J., McCormack A. L., Yates J. R., 3rd (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 [DOI] [PubMed] [Google Scholar]
- 7. Craig R., Beavis R. C. (2004) TANDEM: Matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 8. Geer L. Y., Markey S. P., Kowalak J. A., Wagner L., Xu M., Maynard D. M., Yang X., Shi W., Bryant S. H. (2004) Open mass spectrometry search algorithm. J. Proteome Res. 3, 958–964 [DOI] [PubMed] [Google Scholar]
- 9. Chalkley R. J., Baker P. R., Huang L., Hansen K. C., Allen N. P., Rexach M., Burlingame A. L. (2005) Comprehensive analysis of a multidimensional liquid chromatography mass spectrometry dataset acquired on a quadrupole selecting quadrupole collision cell, time-of-flight mass spectrometer: II. New developments in protein prospector allow for reliable and comprehensive automatic analysis of large datasets. Mol. Cell. Proteomics 4, 1194–1204 [DOI] [PubMed] [Google Scholar]
- 10. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 11. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., Mann M. (2011) Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 12. Kim S., Mischerikow N., Bandeira N., Navarro J. D., Wich L., Mohammed S., Heck A. J., Pevzner P. A. (2010) The generating function of CID, ETD and CID/ETD pairs of tandem mass spectra: Applications to database search. Mol. Cell. Proteomics 9, 2840–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bell A. W., Deutsch E. W., Au C. E., Kearney R. E., Beavis R., Sechi S., Nilsson T., Bergeron J. J. (2009) HUPO Test Sample Working Group: A HUPO test sample study reveals common problems in mass spectrometry-based proteomics. Nat. Methods. 6, 423–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kapp E. A., Schütz F., Connolly L. M., Chakel J. A., Meza J. E., Miller C. A., Fenyo D., Eng J. K., Adkins J. N., Omenn G. S., Simpson R. J. (2005) An evaluation, comparison, and accurate benchmarking of several publicly available MS/MS search algorithms: Sensitivity and specificity analysis. Proteomics 5, 3475–3490 [DOI] [PubMed] [Google Scholar]
- 15. Askenazi M., Bandeira N., Chalkley R. J., Clauser K. R., Deutsch E., Lam H. H. N., McDonald W. H., Neubert T., Rudnick P. A., Martens L. (2011) iPRG 2011: A Study on the Identification of Electron Transfer Dissociation (ETD) Mass Spectra. J Biomol Tech. 22(Supplement), S20 [Google Scholar]
- 16. Brosch M., Yu L., Hubbard T., Choudhary J. (2009) Accurate and sensitive peptide identification with Mascot percolator. J. Proteome Res. 8, 3176–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 18. Elias J. E., Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 19. Käll L., Storey J. D., MacCoss M. J., Noble W. S. (2008) Assigning significance to peptides identified by tandem mass spectrometry using decoy databases. J. Proteome Res. 7, 29–34 [DOI] [PubMed] [Google Scholar]
- 20. Bern M., Phinney B. S., Goldberg D. (2009) Reanalysis of Tyrannosaurus rex mass spectra. J. Proteome Res. 8, 4328–4332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Everett L. J., Bierl C., Master S. R. (2010) Unbiased statistical analysis for multi-stage proteomic search strategies. J. Proteome Res. 9, 700–707 [DOI] [PubMed] [Google Scholar]
- 22. Bern M., Kil Y. J. (2011) Comment on “unbiased statistical analysis for multi-stage proteomic search strategies.” J. Proteome Res. 10, 2123–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Matrix Science Ltd (2010) Mind your P's and Q's: Maximising sensitivity with percolator. Matrix Science ASMS Workshop and User Meeting Salt Lake City, May 23, 2010 [Google Scholar]
- 24. Liu X., Shan B., Xin L., Ma B. (2011) Better score function for peptide identification with ETD MS/MS spectra. BMC Bioinformatics 11, (Suppl 1) 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Laurent J. M., Vogel C., Kwon T., Craig S. A., Boutz D. R., Huse H. K., Nozue K., Walia H., Whiteley M., Ronald P. C., Marcotte E. M. (2010) Protein abundances are more conserved than mRNA abundances across diverse taxa. Proteomics 10, 4209–4212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sun R. X., Dong M. Q., Song C. Q., Chi H., Yang B., Xiu L. Y., Tao L., Jing Z. Y., Liu C., Wang L. H., Fu Y., He S. M. (2010) Improved peptide identification for proteomic analysis based on comprehensive characterization of electron transfer dissociation spectra. J. Proteome Res. 9, 6354–6367 [DOI] [PubMed] [Google Scholar]