Abstract
Oxidation is a double-edged sword for cellular processes and its role in normal physiology, cancer and aging remains only partially understood. Although oxidative stress may disrupt biological function, oxidation-reduction (redox) reactions in a cell are often tightly regulated and play essential physiological roles. Cysteines lie at the interface between these extremes since the chemical properties that make specific thiols exquisitely redox-sensitive also predispose them to oxidative damage by reactive oxygen or nitrogen species during stress. Thus, these modifications can be either under reversible redox regulatory control or, alternatively, a result of reversible or irreversible oxidative damage. In either case, it has become increasingly important to assess the redox status of protein thiols since these modifications often impact such processes as catalytic activity, conformational alterations, or metal binding. To better understand the redox changes that accompany protein cysteine residues in complex biological systems, new experimental approaches have been developed to identify and characterize specific thiol modifications and/or changes in their overall redox status. In this review, we describe the recent technologies in redox proteomics that have pushed the boundaries for detecting and quantifying redox cysteine modifications in a cellular context. While there is no one-size-fits-all analytical solution, we highlight the rationale, strengths, and limitations of each technology in order to effectively apply them to specific biological questions. Several technological limitations still remain unsolved, however these approaches and future developments play an important role toward understanding the interplay between oxidative stress and redox signaling in health and disease.
Cysteine: An Uncommonly Reactive Amino Acid
The nucleophilic sulfur atom allows cysteines to undergo a broad range of chemical modifications. These modifications include redox reactions, lipid acylation, and metal binding motifs that are important for protein structure, localization, regulation, and catalysis. Metal binding and oxidation play a role in protein structure through iron-sulfur (Fe-S) clusters, zinc fingers (ZF)1, and disulfide bonding, among others. Catalytic cysteines are essential to the function of numerous enzymes such as the E1 and E2 ligases of ubiquitin and ubiquitin-like proteins; the HECT domain of ubiquitin E3 ligases; SENP family sumo proteases; the tyrosine phosphatases protein phosphatase 1b (PTP1b) and PTEN; and many others including antioxidants in the thioredoxin, glutaredoxin, and peroxiredoxin families.
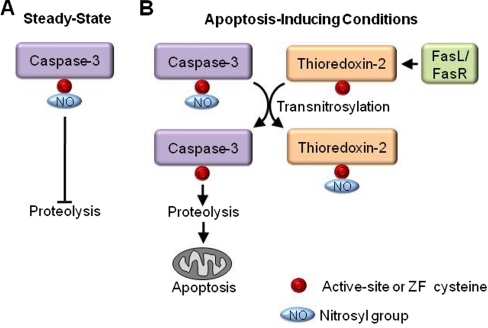
Multiple thiol chemistries can converge to regulate the function of individual cysteines in a biological context. An example of the interconnection between the catalytic and redox properties of a cysteine is found in the cysteine-dependent aspartate-directed protease family of caspases. Essential to apoptosis, caspases are cysteine proteases that utilize the nucleophilicity of their catalytic cysteine for protease activity. Caspase-3 is an executioner caspase that is constitutively S-nitrosylated at its active site cysteine, inhibiting its activity during steady-state conditions (1). When Fas is up-regulated to signal apoptosis, thioredoxin-2 removes the thiol NO group from mitochondrial-associated caspase-3 via transnitrosylation, which derepresses caspase-3 protease activity and promotes apoptosis (2) (Fig. 1).
Fig. 1.
Crosstalk between catalytic activity and redox regulation. Caspase-3 is the terminal protease in the apoptosis cascade and cleaves numerous proteins to complete apoptosis. A, Under steady-state conditions the catalytic cysteine of caspase-3 is nitrosylated which inhibits its protease activity and prevents apoptosis (1). B, When tumor necrosis factor family member FasL binds to its cognate receptor FasR to trigger apoptosis, thioredoxin-2 transnitrosylates mitochondrial-associated caspase-3 derepressing its catalytic activity and promoting apoptosis (2).
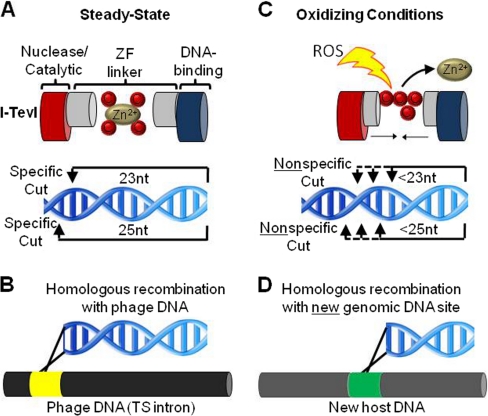
Chemical crosstalk between oxidation and metal binding also regulates individual cysteines. I-TevI is an intron endonuclease located in the thymidylate synthase gene of bacteriophage T4. The nuclease specificity of I-TevI is governed by a four cysteine ZF located between its catalytic and DNA-binding domains that is fully extended and precisely determines the spacing of the two domains (3) (Fig. 2A). Although I-TevI usually cleaves DNA 23 and 25 nucleotides away from the DNA binding site, disruption of the ZF by hydrogen peroxide-induced oxidation leads to cleavage of shorter DNA fragments with no sequence specificity (Fig. 2B). These degenerate DNA sequences are able to recombine at unrelated genomic locations, allowing I-TevI to “jump” into the genome of a new host (Fig. 2C). This is a novel adaptive mechanism by which cysteine oxidation may stimulate I-TevI and other mobile genetic elements to translocate if its host is threatened by oxidative stress (3).
Fig. 2.
Crosstalk between metal binding and redox regulation. A, The intron endonuclease I-TevI has two domains, a DNA-binding domain and a catalytic nuclease domain, separated by a linker region that uses a zinc finger (ZF) to stabilize the extended structure. Under steady-state conditions the linker is fully extended and the nuclease cleaves 23 and 25 nucleotides from the DNA-binding site. B, This allows maintenance of the endonuclease in an intron of the thymidylate synthase gene (TS intron) of the bacteriophage T4. C, Hydrogen peroxide-induced oxidation disrupts the ZF, shortening the linker between the DNA-binding domain and the nuclease domain leading to shorter, nonspecific DNA cleavage (3). D, Although I-TevI typically recombines within an intronless TS gene, the nonspecifically cleaved DNA sequences which result due to oxidation of I-TevI can homologously recombine at a new genomic site or host.
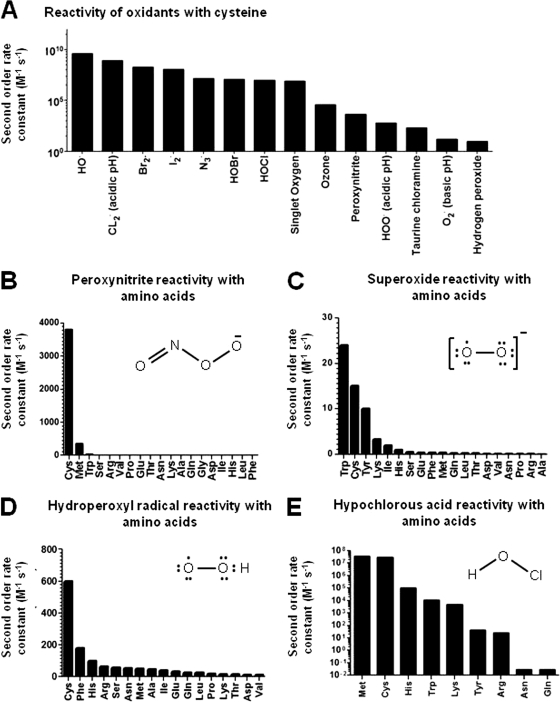
Cysteines lie at the interface between essential redox signaling and the chronic effects of oxidative stress. They can participate in numerous mechanistically distinct redox reactions, including thiol/disulfide exchange, oxygen transfer redox couples, and thiol/thiyl radical transfer reactions, all of which occur during steady-state cellular conditions (4). Cysteine oxidation is prevalent even during steady state conditions, with 5.8% and 9.5% of the cysteines in the proteins of HEK and HeLa cells oxidized, respectively (5). Oxidation can be formed catalytically, such as by the thiol oxidases ErvI (6) and EroI (7) that generate disulfide bonds to facilitate oxidative folding in the endoplasmic reticulum (ER). Alternatively, many redox modifications are mediated by nonenzymatic reactions with reactive oxygen and nitrogen species (abbreviated here as ROS for simplicity). ROS are a diverse set of reactive radical and nonradical oxidants, which include in part: superoxide (O2·−), hydroxyl radical (OH·), hydrogen peroxide (H2O2), nitric oxide (NO·), hypochlorous acid (HOCl), peroxynitrite (ONOO−), nitroxyl (HNO), and a variety of lipid peroxide electrophiles (e.g. 4-hydroxynonenal). Each ROS has unique reactivities and specificities with various cellular components, and for many of them, but not all, the cysteine thiol group is one of the most reactive, especially among amino acid side chains (8–17) (Fig. 3). Endogenously generated nitric oxide and hydrogen peroxide are at low levels and are essential to physiology (18). These act as signaling molecules and react almost exclusively with cysteines (18, 19). Cysteine is also a major target of the ROS associated with oxidative stress such as peroxynitrite, the product of nitric oxide and superoxide (12, 20). HOCl produced by phagocytes as part of a host response to kill invading microorganisms reacts with cysteine and methionine residues two orders of magnitude faster than with other amino acids (21), and cysteine and tyrosine react fastest with singlet oxygen (22). Highly reactive oxidants such as OH· are under diffusion control and thought to have limited specificity. However, even in these cases it is likely that cysteines are an important cellular target. Proteins, such as collagen and albumin, have a rate constant with OH· that is roughly 100 times faster than RNA and DNA (23). Among amino acids, cysteines have the highest rate constant with OH·, threefold higher than tryptophan, tyrosine, and histidine (23). It was recently determined that in the mitochondria the concentration of solvent exposed cysteines is 26-fold higher than the cysteine-containing tripeptide glutathione, typically thought of as the primary cellular antioxidant (24). This suggests that cysteines in proteins are likely an important sink for ROS produced during respiration, including those that are highly reactive and less specific.
Fig. 3.
Reactivity of oxidants with cysteine and other amino acids. A, Second order rate constants (M−1 s−1) spanning nine orders of magnitude for radical and nonradical oxidants with cysteine. From (8–17, 20, 21) with selected data from NDRL/NIST database (http://kinetics.nist.gov/solution/). Rate constants are in water at ∼pH 7.0 except where indicated. Rate constants for biologically relevant oxidants B, peroxynitrite at pH 7.4 (20), C, superoxide at pH 10.0 (8), D, hydroperoxyl radical at pH 1.6 (8), and E, hypochlorous acid at pH 7.4 (21) with all tested amino acids in each study to compare the relative reactivity of cysteine.
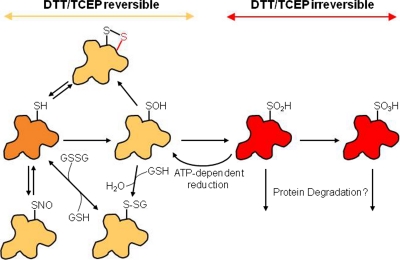
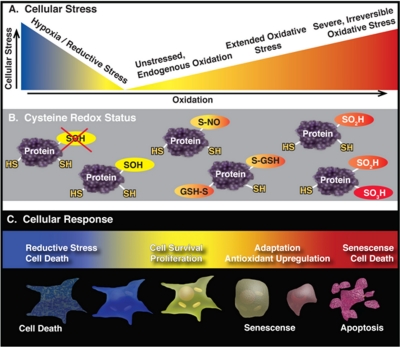
The oxidation state of a free sulfhydryl is −2 and cysteines can form up to ten different oxidation states in vivo (4). Widespread cysteine oxoforms include (oxidation states listed in brackets): S-nitrosyl [0], glutathionyl [−1], disulfide [−1], sulfenylamide [0], sulfenic acid [0], sulfinic acid [+2], sulfonic acid [+4], and cysteinyl [−1] (Fig. 4). Recently cysteine sulfhydration [−1], formed via reaction with hydrogen sulfide, was demonstrated to be prevalent as well (25). The diverse array of cysteine oxoforms differs widely in their reactivity, origin, stability, and reducibility. Constitutive disulfide bonds, typically found in nonreducing environments such as the endoplasmic reticulum or extracellular domains, are highly stable and generally not redox-regulated. On the other hand, the sulfenic acid modification is generally thought to be a transient intermediate, often formed en route to more stable oxoforms. In general, redox-regulated cysteine modifications are transient and can be readily removed upon reduction. Reversibility is an essential property of any regulatory signaling network, but also allows cysteine modifications such as glutathionylation to act as a temporary buffer to oxidative stress which can in turn be removed enzymatically. If sulfhydryls are not recycled back to a lower oxidation state they can continue on to higher oxidation states such as sulfinic acid (–SO2) and sulfonic acid (–SO3). These oxoforms are hallmarks of oxidative stress and in most cases are terminal, “over-oxidized ” modifications that are irreversible by chemical or enzymatic means (Fig. 4). In a unique instance, the sulfinic acid modified peroxiredoxin family members can be enzymatically reduced by sulfiredoxin (26, 27). Cysteines, therefore, play a central role in a redox continuum in which oxoforms associated with low levels of oxidation are required for cellular function, where the cell can adapt to moderate levels of thiol oxidation and respond through antioxidant enzymes, and where overoxidation leads to protein dysfunction and cell death (Fig. 5). Conversely, oxygen deprivation leads to hypoxia which in turn leads to reductive stress. Reductive stress is a buildup of reducing equivalents, NADPH and NADH, that occurs when oxidative phosphorylation is inhibited, such as during hypoxia, when reducing equivalents cannot be transferred to oxygen (28). Both hypoxia and reductive stress can lead to cell death (Fig. 5).
Fig. 4.
Common cysteine oxoforms and their chemical reversibility. Cysteines (orange) can be oxidized to a diverse set of oxidized species, including S-nitrosylation (SNO), glutathionylation (S-SG), disulfide (S-S), sulfenic acid (SOH), sulfinic acid (SO2H), and sulfonic acid (SO3H). Sulfenic acid is often an intermediate to other cysteine oxoforms. Oxidized cysteines in a yellow shade are chemically reversible by DTT and TCEP, whereas those shaded in red are chemically irreversible. In the case of peroxiredoxins, sulfinic acid is reducible through and ATP-dependent process catalyzed by sulfiredoxins (26, 27). There is no known repair process for sulfonic acids which are likely degraded.
Fig. 5.
Oxidation levels affect the redox status of cysteines in vivo and lead to divergent cellular responses. A, The redox continuum of oxidation in the cell ranges from reductive stress to oxidative stress with low levels of oxidation present in unstressed cells. B, C, Cysteines can be oxidized to different states depending on the oxidation level in the cell. While oxoforms essential to cell survival and proliferation (yellow) are the primary type of cysteine modification found in unstressed cells, a moderate increase in oxidation leads to an adaptive response and glutathionylation (S-GSH) of cysteines. Severe oxidative stress leads to senescence or cell death and is accompanied by overoxidized cysteine oxoforms such as sulfinic (orange) and sulfonic acid (red) which are dysfunctional. Hypoxia and reductive stress disrupts essential redox reactions (red X) and can result in cell death.
Predicting which cysteines are oxidized, the type of modifications that are present, and the stability of the thiol modification is very difficult (29, 30). Redox modifications are governed by complex kinetics and are dependent upon which cellular compartment the protein is located in, which ROS are present, and the levels and localization of various antioxidant enzymes (29, 31). To complicate matters, antioxidant enzymes are known to act as oxidants under certain conditions (32, 33). Two chemical properties that primarily govern redox-activity are solvent accessibility and low thiol pKa. The pKa of the cysteine sulfhydryl is heavily influenced by the local milieu and nearby amino acids. Therefore the thiol redox-activity is in part an intrinsic property of the protein structure, however, the environment also influences reactivity via changes in pH. Changing the thiol pKa affects the equilibrium constant between the free thiol (Cys-SH) and the redox-active deprotonated thiolate anion (Cys-S−). The Ka of cysteines can vary over 6 orders of magnitude in a cell. Typically, the pKa of a cysteine sulfhydryl is ∼8.6, yet it can be as low as 3.5 (34, 35) or as high as 10 in some cases. The wide range of factors that contribute to a cysteine's reactivity and susceptibility to oxidation therefore are major determinants of the specificity of redox reactions in cellular systems.
The role that oxidation plays in cancer, aging, and other biological processes remains unclear and at times controversial (36). For example, there is limited correlation between resistance to oxidative stress and increased lifespan in mouse knockout models of antioxidant genes (36). Although sod2 knockout mice exhibit neonatal lethality (37) and sod1 knockout mice have moderate to severe disease pathologies (38, 39), most other antioxidant enzymes do not have obvious lifespan phenotypes (36). Therefore, in isolation, antioxidant enzymes do not play a functional role in aging, though the overall antioxidant system may remain largely intact in these models (36). ROS play a paradoxical role in cancer as well, associated with both proliferative and apoptotic pathways (40).
The conflicting role of oxidation in complex phenotypes may also lie in the fact that numerous markers of oxidative stress, such as 3-nitrotyrosine and carbonyl formation, are often used interchangeably. However, these oxidative modifications are not equally reactive and are the result of different oxidation chemistries. Markers for oxidative stress assays are often chosen based on convenience or ease of analysis rather than because they are the appropriate assay. Due to the high reactivity of cysteines, measuring their oxidation requires thoughtful sample preparation and specialized approaches to preserve the endogenous oxidation state and prevent oxidation artifacts. The technical difficulties associated with cysteine oxidation analysis have hampered our understanding of their role in complex biological questions to some extent. Here we review some of the recent redox proteomics approaches that have improved the analytical sensitivity, depth of coverage, throughput, and selectivity for specific cysteine oxoforms. Protein-level methods includes fluorescent- or Western blot-based approaches in which the measured signal is a composite of the redox changes to every cysteine in the protein. Alternatively, protease digestion and peptide-level analysis by mass spectrometry can detect and quantify oxidation at the site-specific level (41, 42). Among these techniques tradeoffs exist, and when appropriate we will attempt to point out how each approach can be integrated with protein- or peptide-level analysis to effectively characterize oxidized cysteines in a dynamic cellular environment.
Redox Proteomics
Sample Preparation
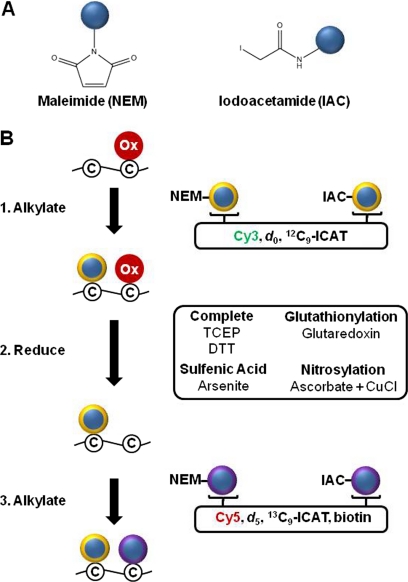
Redox proteomics relies on specialized sample preparation workflows to carefully preserve the endogenous oxidation state of cysteines. Many cysteine oxoforms are potentially labile, and upon cell disruption oxidation can be artificially introduced by air or diminished by unregulated activity of antioxidant enzymes. An additional concern is disulfide bond “shuffling” in which two sets of disulfide-linked cysteines, CysA-CysB and CysC-CysD for example, rearrange through thiol-disulfide exchange reactions to become CysA-CysC and CysB-CysD. Cell lysis using trichloroacetic acid (TCA) is the gold standard to limit oxidation artifacts during cell or tissue disruption. TCA quenching is fast with a rate constant of 109 sec−1 M−1 (43) and has two primary functions: (1) protonating redox-active thiolate anions (S−) by lowering the pH below their pKa and (2) limiting disulfide shuffling by precipitating and denaturing proteins (44). Alternatively, cysteine-specific alkylating reagents can be added to the lysis buffer to trap cysteines in their in vivo redox state for sample preparation procedures not compatible with protein denaturation such as organelle enrichment. N-ethylmaleimide (NEM) and iodoacetamide (IAM) (Fig. 6A) are often used interchangeably for this purpose, however these two reagents have distinct chemical properties and react with thiols by different mechanisms. NEM reacts based on a Michael-type addition with the deprotonated thiolate anion as the Michael donor. Iodoacetamide reacts via nucleophilic substitution with the thiolate acting as the nucleophile and iodine as the leaving group. NEM is faster by 1–2 orders of magnitude and is fast enough to even trap transient protein folding intermediates in structural studies. NEM is also less pH-dependent (45) and more cysteine-specific (43) but is less reactive with buried or partially exposed thiols under nondenaturing conditions (43) and can undergo hydrolysis of the maleimide ring above pH 9.5 (46).
Fig. 6.
Differential alkylation indirectly detects oxidation and is modular. A, Structures of the maleimide and iodoacetamide functional groups. Bifunctional thiol specific reagents combine these two groups with fluorescent, stable isotope, or epitope tags (blue circles). B, The differential alkylation procedure labels nonoxidized cysteine thiols before (yellow ring) and after (purple ring) reduction with two different alkylation reagents to determine the percent of the cysteine that is reversibly alkylated. Fluorescent reagents include Cy3 and Cy5 tags and stable isotope labeled regents include d0 and d5 NEM or 12C9-ICAT and 13C9-ICAT reagents. Alkylation with an untagged alkylation reagent followed by a biotinylated thiol-specific reagent allows enrichment of reversibly oxidized peptides or proteins. Complete reduction of all reversibly oxidized cysteines can be achieved with TCEP or DTT. Selective reduction of specific cysteine oxoforms is possible using ascorbate plus CuCl to reduce S-nitrosylation, the enzyme glutaredoxin to reduce glutathionylation, and arsenite to reduce sulfenic acid.
Differential Alkylation
Differential alkylation is a flexible, modular redox analysis technique that labels cysteines before and after reduction with coded cysteine-specific reagents (Fig. 6B). Alkylating reagents only react with free cysteines and can be used combinatorially to code for non-oxidized cysteines at any step in a sample preparation procedure. Thiol-specific alkylation reagents conjugated to a wide variety of fluorophores, epitope tags, and/or stable-isotope labels are available for enrichment or detection of labeled cysteines. Reducing reagents include the chemicals dithiothreitol (DTT) and tris(2-carboxyethyl)phosphine (TCEP) which reduce most cysteine oxoforms except for sulfinic acid and sulfonic acid (Fig. 4). Although the specificity of ascorbate to reduce nitrosothiols has been controversial, most notably in the context of the biotin switch technique, a recent study established its copper-dependent selectivity (47). Several phosphine derivatives have also shown excellent potential to selectively reduce SNO (48, 49). Arsenite can selectively reduce sulfenic acids (50). There are no chemical reductants available for selective reduction of glutathione (GSH) adducts, although it has been reported that treatment of samples with the C14S, C65Y double mutant of E. coli glutaredoxin-3 (GRX3) preferentially reduces GSH-protein mixed disulfides (51). Combining selective GSH reduction and alkylation with NEM-biotin after reduction led to detection of 43 putative glutathionylated proteins (51). It has been reported more recently that the wild-type mammalian glutaredoxin GRX1 selectively reduces GSH-protein adducts in situ (52) and may also prove to be useful for redox proteomics applications.
S-Nitrosylation: The Biotin Switch Technique
Development of the biotin switch technique to measure cysteine S-nitrosylation by Jaffrey et al. in 2001 (53) first established the utility of the three-step differential alkylation procedure to characterize oxidized proteins. As first reported, this method used methyl methanethiosulfonate under denaturing conditions to first trap free thiols in disulfide bonds, then ascorbate to selectively reduce nitrosothiols, followed by incubation with the cysteine specific biotin-HPDP to label the reduced thiols with a biotin tag. S-nitrosylated cysteines are at very low levels, such as nanomolar concentrations in plasma (54), and additional epitope handles such as His-tag (55) have been employed to maximize their enrichment and identification. Alternatively, a resin capture strategy has recently been developed using thiopropyl Sepharose to capture S-nitrosylated peptides after ascorbate reduction (56).
Redox-DIGE: Redox Differential Gel Electrophoresis
Redox-DIGE is a gel-based fluorescence approach that compares the relative levels of oxidation between two samples. Each sample is initially alkylated with either unlabeled NEM or IAC, reduced, and fluorescently labeled with either green Cy3- or red Cy5-maleimide to code each sample (57). Infrared maleimide-based probes DY-680 and DY-780 have also been used to improve the sensitivity of detection (58). After labeling, the samples are combined and separated by 2D-gel electrophoresis. Protein spots which have equal levels of oxidation in each sample show up as yellow due to an equal mixture of green and red fluorescent signals. Spots that are either green or red are potentially differentially oxidized and are typically identified by in-gel protease digestion and mass spectrometry. Although the protein is often identifiable, the oxidized cysteine is not because of the difficulty of detecting and quantifying the large fluorescent moiety linked to each cysteine. Hurd et al. used this approach to measure the oxidation of mitochondria-enriched rat hearts and detected ∼50 proteins, most of which were differentially oxidized after treatment with hydrogen peroxide or the NO donor S-Nitroso-N-Acetyl-D,l-Penicillamine (SNAP) (57).
An important caveat when comparing two samples that are labeled only after reduction is that differences in signal may solely reflect changes in protein abundance between samples rather than oxidation. This limits the applicability of this approach for long-term studies, for instance, in which widespread changes to protein expression are expected. Differentially alkylating each sample before and after reduction does account for differences in protein levels and overcomes this limitation (59). In addition, unless a single protein is identified per spot it is difficult to prove conclusively which of the proteins is oxidized. Despite the limited dynamic range of 2D-gels and their bias against large or hydrophobic proteins, Redox-DIGE has shown excellent utility to identify oxidized proteins in mitochondria after organelle enrichment (57, 60). However, experimental designs which use pooled samples and biological variance analysis (BVA) are an important experimental design strategy to minimize false positives with this approach (60).
A related differential gel-based approach that uses radiolabeled [14C]-iodoacetamide after reduction has also been used to identify oxidized proteins in 2D-gels (61). In this study, the overall cysteine oxidation state of ∼100 E. coli proteins was determined per 2D-gel. The level of oxidation was assessed by normalizing the level of radioactivity in each spot to the level of total protein as determined by Coomassie staining. A second gel is run in parallel without radiolabeled IAM for analysis by mass-spectrometry to identify the oxidized proteins. As in the Redox-DIGE, this method also relies on final 2D gel separation and analysis, a technique that is waning in use in proteomics due to its inherent limits of protein resolution and overall sampling efficiency.
ICAT: Isotope-coded Affinity Tags
The iodoacetamide-based 12C9 or 13C9 stable isotope labeled pair of ICAT reagents were originally designed to quantify the relative changes in global protein expression between two samples (62). However, these reagents have also become widely used in redox proteomics because they label cysteines, have a biotin handle for enrichment, and are readily quantifiable by mass spectrometry. The first cysteine oxidation studies using ICAT chemistry were reported in the Costello laboratory (63, 64) where free cysteines were first alkylated in each sample with IAM, followed by reduction. Each sample was then labeled with a one of the two nonisobaric ICAT reagents and the two samples were mixed. As discussed for the Redox-DIGE approach, this has the important caveat that differences in peptide levels are potentially due only to differences in protein expression rather than oxidation. Nonetheless, these initial ICAT studies were the first to simultaneously identify and quantify the oxidation state of specific cysteines in an unbiased manner, quantifying the oxidation state of 18 cysteines in a rabbit heart membrane preparation (63). As many as 71 oxidized cysteine-containing peptides have been uncovered in subsequent studies (65).
To address the confounding issue of protein expression levels, Leichert et al. differentially alkylated a single sample with both ICAT reagents, one reagent before reduction to code for the level of the cysteine that is nonoxidized and the second reagent after reduction to label the oxidized form (66). This “OxICAT” approach takes advantage of the fact that because the stable isotope labeled peptides have equivalent ionization efficiencies by mass spectrometry, the sum of the intensity of the two labeled alkylated peptides equals the total amount of cysteine. Therefore, the ratio of the oxidized signal to the total signal is equivalent to the percent oxidation of the cysteine and is not influenced by differences in protein expression. The original OxICAT study in E. coli detected ∼120 ICAT labeled peptides per sample with the oxidation of 27 increasing over 1.5-fold after hypochlorite treatment (66). The OxICAT approach has detected as many as 42 cysteines with over a 1.5-fold increase in oxidation in Caenorhabditis elegans treated with hydrogen peroxide, representing ∼20% of the total cysteines detected in the sample (67).
One of the potential explanations for why there are fewer cysteines detected in OxICAT experiments than typical ICAT-based quantitation experiments is that the highly reducing cellular environment of the nucleus, cytoplasm, and mitochondria leads to low levels of oxidized cysteines, typically under 10% (5). This means that after reduction and second ICAT labeling step, the formerly oxidized cysteines are an order of magnitude less abundant than the nonoxidized cysteines. Because ICAT quantitation is performed at the MS level, rather than MS/MS level, which has a dynamic range of three orders of magnitude, there is less potential to detect and accurately quantify the oxidized form of the ICAT-labeled peptides that are at lower levels. Quantitation at the MS level also limits ICAT-based studies from distinguishing the oxidation status of individual cysteines in peptides with two or more cysteines. Finally, ICAT workflows are difficult to scale down and have had limited applicability to quantify targeted proteins from endogenous sources. ICAT-based studies that have focused on targeted proteins, such as Hsp33 (66), protein disulfide isomerase (68), p21 HRas (69), and heme oxygenase (70), have all relied on analysis of purified, recombinant proteins instead of the endogenous protein.
OxMRM: Quantitative Cysteine Oxidation Analysis by MRM
Multiple reaction monitoring (MRM) is a targeted quantitative technique with arguably the highest sensitivity and reproducibility among mass spectrometry-based approaches. Our laboratory has developed an approach, termed OxMRM, which combines: (1) differential alkylation of samples with unlabeled d0 and a generic d5 stable isotope labeled NEM, (2) affinity purification of the endogenous protein or proteins of interest, and (3) analysis by MRM to quantify the percent oxidation of virtually any targeted cysteine or protein from a cellular source (71). In addition, because MRM quantifies at the MS/MS level, it can distinguish between the oxidation of two cysteines within a peptide if a fragment ion between the two cysteines is quantified.
Redox regulation is a dynamic process and its in-depth characterization requires routine, inexpensive, and sensitive targeted assays downstream to unbiased discovery workflows for follow-up time course analyses or comparing a variety of experimental conditions. OxMRM assays meet these requirements and can be developed from any LC-MS/MS dataset in which the cysteine-containing peptides of interest are identified. Although the OxMRM approach is a targeted assay, it is possible to measure many cysteines simultaneously. We recently quantified the reversible oxidation status of 34 cysteines in the 45 subunit Complex I of the electron transport chain using the OxMRM approach and discovered 6 of those to be oxidized in a mouse model of Parkinson's disease (72). Because triple quadrupole mass spectrometers are widely used and software for developing MRM assays has greatly improved (73), studies measuring hundreds of peptides by MRM is becoming routine (74, 75). However, developing and validating MRM assays for all these peptides can be time consuming and some peptides may prove refractory to this approach.
The sensitivity of OxMRM analysis also allows a unique workflow to quantify the redox status of low-abundance target proteins that are undetectable by unbiased mass spectrometry approaches with limited dynamic range. Our laboratory used OxMRM to quantify the percent oxidation of seven of the 10 cysteines of endogenous p53, which is a highly negatively regulated protein with a half-life of 6–30 min (76), in both cancer cell lines and primary fibroblasts (71). We found that Cys182 of p53 was particularly susceptible to diamide oxidation intracellularly. In contrast, diamide-induced oxidation of purified, isolated p53 showed very little amino acid selectivity (71). Our findings confirmed that the cellular environment, such as intracellular compartmentalization (77), interaction with other proteins, and/or antioxidant activities, can have a significant effect on the redox status of proteins and underscores the importance of measuring endogenous oxidation when possible. From the same cell lysates we quantified the percent oxidation of eight of 10 cysteines in endogenous PTP1b. In comparison, a recent ICAT-based study reported quantitation of oxidation of only three cysteines in recombinant PTP1b (78), demonstrating the improved utility of OxMRM for analysis of target proteins and cysteines.
Because the OxMRM approach does not rely on epitope-tagged alkylation reagents for enrichment of the protein or protein complex of interest, it is amenable to quantifying sulfinic and sulfonic acids levels which cannot be alkylated. However, the ionization efficiency of these species is potentially not equivalent to that of the unmodified peptide and only a relative comparison of irreversible oxidation levels between samples can be measured (71). Nonetheless, it is an initial step toward comprehensively characterizing the complete oxidation state of specific cysteines.
A limitation of OxMRM, as is the case for all peptide-based mass spectrometry approaches, is that a single protease rarely generates peptides with appropriate LC-MS characteristics to detect the entire protein sequence. Therefore not all cysteines can be detected using a single enzyme. However, use of several enzymes or combinations thereof can greatly improve the sequence coverage of proteins and, in principle, can be used to quantify any cysteine of interest.
Selectively Labeling Reactive Cysteines: Biotin-Conjugated Iodoacetamide (BIAM)
The BIAM approach aims to selectively label cysteines which are found in the reactive, oxidizable thiolate anion form at neutral pH (79). Because most cysteines have a pKa of ∼8.5, cysteines with lower pKa values are both more likely to become oxidized and also react much faster with alkylating agents. By incubating a sample with biotin-conjugated iodoacetamide at a low pH, typically 6.5, which is at least 1–1.5 pH units below the maximum rate of iodoacetamide alkylation, these low pKa cysteines can selectively be biotin-tagged for enrichment or detection. By not labeling the “bulk” cysteines that are not reactive, this approach is able to detect small changes in oxidation because background labeling is minimal. For analysis, samples can be digested into peptides, biotin enriched, and analyzed by mass spectrometry to identify the reactive cysteines. Alternatively, intact proteins can be purified and the change in oxidation of specific proteins between conditions can be assessed by Western blot. Because oxidized cysteines do not react with iodoacetamide, decreased BIAM labeling signal corresponds to increased oxidation in these studies.
It has been reported that a potential limitation of this approach is that the bulky nature of the BIAM reagent may limit its labeling of certain known oxidized cysteines, (79); though this may be true for any of the approaches that use sizeable alkylating agents if alkylation is not performed under denaturing conditions. In addition, because this approach does not label the nonoxidized protein, the total protein levels in the sample must be measured in parallel for normalization.
Labeling Cysteines in the Mitochondria
Mitochondria are a major source of ROS, especially superoxide, and proteins in the mitochondria are expected to undergo a relatively high level of oxidative damage or other redox modifications. Lipophilic cations such as triphenylphosphonium (TPP) readily pass through lipid bilayers and preferentially partition to the mitochondria because of the large membrane potential across the mitochondrial inner membrane (80). When TPP is conjugated to other molecules or probes, one can obtain a 20- to 200-fold enrichment in the mitochondria relative to cytoplasmic concentrations (80). Because TPP-conjugated molecules at low levels are not toxic, they have been exploited as chemical probes to specifically examine mitochondrial processes both in situ and in vivo. 4-iodobutyl)triphenylphosphonium (IBTP), a thiol-specific TPP derivative developed by Murphy and colleagues, allows for selective labeling of mitochondrial cysteines that can be detected by Western blot using an antibody to the TPP moiety (81). This approach is especially amenable to one- and two-dimensional gel-based approaches with differences in IBTP labeling indicating relative changes to the cysteine redox status. In one study, labeling of mitochondria with IBTP after treatment with several exogenous oxidants demonstrated that a subset of mitochondrial proteins are particularly susceptible to oxidation (81). However, since high levels of lipophilic cations such as IBTP can disrupt mitochondrial function and become toxic (80), labeling with IBTP is typically carried out at very low stoichiometries. The low labeling efficiency places limits on its sensitivity for detecting proteins at low abundance as well as limiting its utility for differential alkylation which requires complete labeling. In addition, since the membrane potential drives the localization of TPP-conjugated molecules to the mitochondria, changes in labeling may reflect membrane potential differences between condition rather than redox processes and must be taken into consideration in the experimental design. Nonetheless, the method is unique in its ability to capture redox processes while the cell or organelle is still functional and intact.
Dimedone-based Sulfenic Acid Probes
Sulfenic acid is a transient cysteine oxoform that is an important intermediate often formed en route to more stable regulatory or stress-associated oxidation states (Fig. 4). Although arsenite selectively reduces sulfenic acids and has been utilized for differential alkylation to a limited extent (50), the use of dimedone, 5,5-dimethyl-1,3-cyclohexanedione, to directly react with sulfenic acid is more widespread. The specificity of dimedone results from the partial electrophilic character of sulfenic acid (82). Initial dimedone-based derivatives conjugated to biotin (83) or fluorescent tags (84) were not cell permeable, however more recent analogs of dimedone such as DAz-2 have been used to trap and tag the sulfenic modification in situ (85). One study using the DAz-2 dimedone analog identified 193 protein targets of sulfenic acid oxidation (86). This is a very impressive number given the expected low abundance of this transient modification; however, no specific sites of modification were reported, leaving the identities of the reactive cysteines undefined. To determine the relative levels of sulfenic acid between samples, a d0 and d6 isotope coded pair of dimedone analog was recently reported (87).
Direct Detection of Sulfenic Acids
The 16 Da mass shift which results from sulfenic acid modification can be measured by mass spectrometry and has been detected on a purified peptide from MMP-7 that was oxidized in vitro (88). Because of the high reactivity and limited stability of the modification it is unclear if direct analysis of SOH modified cysteines is amenable to samples derived from cellular sources. However, it has been reported that differences in sulfenic acid content could be measured in commercially prepared tissue lysates spotted into microarray format prior to dimedone treatment (89). This suggests that sulfenic acid does have some inherent stability in vitro.
Direct Detection of Glutathionylated Proteins
Glutathione adducts can be measured by nonreducing Western blots with cysteine-specific glutathione antibodies (90) or pan-glutathione antibodies, though pan-glutathione antibodies may not detect all glutathionylated proteins (91). Glutathionylated proteins can also be directly labeled and purified by the cell membrane permeable biotinylated glutathione ethyl ester, Biotin-GEE (92). N,N-biotinyl glutathione disulfide (biotin-GSSG) has also been used to label glutathionylated proteins in cells and has the potential to mimic an oxidative stress induced shift in the glutathione redox couple as a result of the large influx of GSSG from the reagent into the cell (91).
If preserved, direct measurement of glutathione adducts by mass spectrometry is possible through a mass shift of 305.3 Da. Without affinity enrichment, GSH-modified peptides typically exist at very low levels in protein digests. However, numerous sophisticated approaches to quantify glutathionylated intermediates in xenobiotic metabolism could be applied to peptide-level analysis. These include neutral loss scanning mass spectrometry for a loss of the 129 Da pyroglutamic acid from endogenous glutathione adducts (93), or treatment of cells with glutathione ethyl ester (GEE) combined with precursor ion scanning of 300 m/z to detect the GEE linked analyte (94). In addition, treating cells with stable-isotope labeled GSH improves selective detection of glutathionylated molecules because they appear as a pair of light and heavy ions (95, 96).
Direct Detection of S-nitrosylated Proteins
Because of the labile nature of the S-nitrosylated cysteines, most studies on S-nitrosylation use indirect, differential alkylation-based approaches for analysis. However, antibodies to S-nitrosylation are available and have been used for Western (97) and immunohistochemistry analysis (98). In addition, mass spectrometry can detect the 29 Da mass increase that corresponds to S-nitrosylation and the facile neutral loss of NO under MS/MS conditions can be used to improve the sensitivity and specificity of detection (78, 99). However, in a complex matrix S-nitrosylated peptides may be susceptible to loss through transnitrosylation or denitrosylation enzyme activity (100) or decomposition catalyzed by light during sample preparation. Therefore, this approach is likely to be more amenable for studies in which a purified protein or peptide is in vitro modified.
Quantifying the Total Amount of Disulfide Bonding in a Cell
Hansen et al. have developed an approach to measure the percent oxidation of cysteines in a cell at a global level which can distinguish intra- or interprotein disulfide bonds from protein-bound low-molecular weight thiols such as glutathione (5). They found that cysteines in proteins are oxidized under 10% in HEK and HeLa cells which can increase to 43 and 56% after a brief treatment with high, but non-lethal, levels of the oxidant diamide. Although this likely represents close to the maximum level of oxidation that a cell can tolerate on a temporary basis, it demonstrates the tremendous capacity for the cell to respond to oxidation. This study also determined that glutathione is the most abundant oxidized low molecular weight protein adduct in cells on a molar basis. Even so, it still exists at relatively low levels with less than 0.1% of cysteines in proteins glutathionylated under steady state conditions in HEK and HeLa cells (5). After diamide treatment this can increase ∼300-fold with 23 and 36%, respectively, of the total glutathione pool becoming conjugated to proteins in these cells (5).
Future Directions in Redox Proteomics
Integrating Biochemical and Genetic Tools With Mass Spectrometry for Redox Analysis
Identifying oxidized cysteines and quantifying their levels are only part of their characterization, going hand-in-hand with biochemical and genetic follow-up experiments. As with other post-translational modifications (PTMs), modification of a cysteine by oxidation doesn't necessarily mean that it plays a functional role. Site-directed mutagenesis or other techniques that assess the impact that oxidation has on the protein's function are important; however, cysteine knockout experiments must be interpreted with great care. For example, these experiments often do not show a clear phenotype if the site plays a regulatory, but not essential, role. Furthermore, mutagenesis cannot distinguish the role of oxidation from other potential functional roles (metal binding, disulfide bond) that the cysteine fulfills. Nevertheless, these approaches are especially important to complement redox proteomics studies that focus on the essential or regulatory role of oxidized cysteines in signaling.
Regulatory Crosstalk Between Cysteine Oxidation and Other PTMs
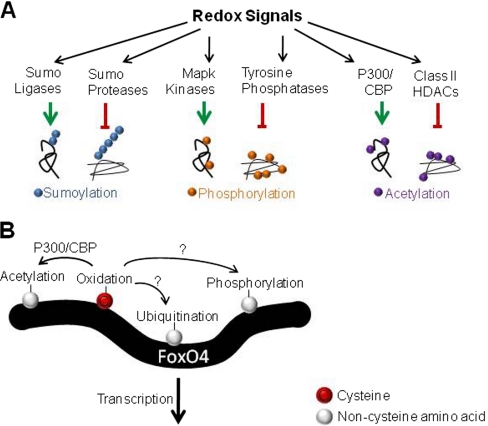
Oxidation affects the function of numerous enzymes which modulate protein PTMs, including the ubiquitin E1 (101) and E3 ligases (102, 103), the E1 ligase of ubiquitin-like modifiers SUMO (104) and ISG15 (105), SUMO proteases (106), histone deacetylases (HDACs) (107), MAP kinase kinases (108), MAPK phosphatases (109) as well as the tyrosine phosphatases PTP1b (110) and PTEN (111) (Fig. 7A). Global analysis of sumoylation, ubiquitination, acetylation, and phosphorylation have become established workflows in proteomics (Reviewed in (112) and (113)) and should allow examination of the crosstalk between redox regulation of these enzymes and subsequent changes to cellular PTM status. A major challenge will be delineating the changes in cellular PTM status that directly result from oxidation of key enzymes from secondary effects.
Fig. 7.
Regulatory crosstalk between cysteine oxidation and other PTMs. A, Redox signals have been implicated in regulating the function of numerous enzymes which affect PTMs such as sumoylation, phosphorylation, acetylation, as well as others. Enzymes that both conjugate and remove PTMs can be affected with redox regulation typically repressing but occasionally enhancing enzyme activity. B, FoxO4 is a paradigm for the crosstalk between cysteine oxidation and other PTMs at the level of a single protein. Cysteine oxidation of FoxO4, via disulfide-mediated interaction with the acetyltransferase p300/CBP, increases acetylation and represses FoxO4 transcriptional activity in a redox-dependent manner (114). FoxO4 is phosphorylated and ubiquitinated as well, however it is not known if these PTMs are redox-regulated.
Crosstalk between cysteine oxidation and other PTMs can also occur within a single protein and is an important signaling node (Fig. 7B). For example, FoxO4 is a transcription factor that plays an important role in complex phenotypes such as aging and tumor suppression. FoxO4 interacts with the acetyltransferase p300/CBP via an intermolecular disulfide link (114). This leads to acetylation of FoxO4 and represses transcriptional activity in a redox-dependent manner (114). In addition, cysteines can be lipid acylated to direct proteins to membranes or modulate protein-protein interactions. Crosstalk between redox regulation and palmitylation has been shown in the case of CD81, in which oxidation decreases CD81 palmitylation and decreases its association with the epsilon isoform of the protein 14–3-3 (115). Identifying PTMs co-associated with oxidized cysteines is an important next step in teasing apart these regulatory interactions.
Determining the Comprehensive Picture of Oxidation of a Cysteine
The use of OxMRM and ICAT-based redox technologies has introduced the ability to measure the extent of thiol oxidation of specific cysteines for the first time and has enabled the ability to distinguish highly oxidized cysteines from those that are minimally oxidized. However, these approaches cannot determine the percentage of each cysteine oxidized to sulfinic and sulfonic acid, which are chemically irreversible and therefore not compatible with differential alkylation or enrichment with biotinylated alkylating reagents. These species are especially important when considering the interface between essential redox signaling and oxidative stress. For example, the protein tyrosine phosphatase PTP1b is an important regulator of redox signaling whose catalytic Cys215 can be overoxidized to sulfinic and sulfonic acid quite readily, inhibiting its phosphatase activity. In our OxMRM studies, PTP1b was enriched by immunopurification and we were able to quantify the relative levels of sulfinic and sulfonic acid modified Cys215 between treatments with various oxidants (71). However, we could not determine the stoichiometry because of potential differences in ionization efficiencies that can affect quantitation by mass spectrometry. Stable isotope dilution analysis with labeled peptides corresponding to the nonoxidized, sulfinic acid, and sulfonic acid forms would allow determination of the molar quantity of these peptides in a sample and an accurate determination of the percentage of the cysteine in various oxidized species could be assessed.
It is known that different cysteine oxoforms can lead to varying functional outcomes. For example, the promoter activity of OxyR differs by over 30-fold depending on the redox modification (NO, GSH, SOH) (116). By selectively reducing NO, GSH, sulfenic acid in a stepwise manner the percent oxidation of each oxoform could, in principle, be determined. Generating a comprehensive picture of site-specific cysteine oxidation would reveal insights into the role of cysteine oxidation in physiology, redox regulation, and oxidative stress.
CONCLUSIONS
A wide range of proteomic techniques are now available for redox analysis, providing researchers with the tools and workflows necessary to assess thiol redox changes in complex biological systems (Table 1). Nonetheless, there is not a one size fits all solution to employing these methods and the limitations and trade-offs for each methodology must be considered. For instance, data-dependent acquisition mass spectrometry or gel-based fluorescent techniques generally have less dynamic range but a higher capacity to identify novel oxidized cysteines than those that target specific proteins or cysteines for analysis, such as Western blot or MRM. Although cysteine redox analysis can be challenging, the application of these approaches and future developments will continue to uncover the unique role that redox regulation of cysteines plays in physiology. One potential application is reconciling conflicting data with regard to the significance of oxidative stress in aging. The long-lived naked mole rat has a maximum lifespan of 28 years but paradoxically has higher levels of protein carbonylation (117) and similar amounts of reactive oxygen species (118) compared with much shorter lived mice. However, compared with mice, naked mole rats are protected from overoxidation of cysteines during aging which associates longer lifespan with decreased cysteine oxidation (119). In any case, it must be kept in mind that ROS and oxidation are essential for physiological processes and should not be considered simply deleterious. The use of redox proteomics technologies combined with biochemical and genetic approaches are positioned to improve our understanding of the unique interaction between oxygen and cysteine in both health and disease.
Table I. List of references containing detailed protocols for the redox analysis techniques covered in this review.
| Technique | References with detailed protocols |
|---|---|
| Redox-DIGE and 2D gel approaches | 57: Alkylation after reduction |
| 59: Differential alkylation | |
| 61: [14C]-iodoacetamide | |
| Redox ICAT | 64: Alkylation after reduction |
| 66: Differential alkylation | |
| OxMRM | 71 |
| Biotin-conjugated iodoacetamide (BIAM) | 79 |
| Organelle specific alkylation | 81 |
| Sulfenic Acid | 86: Dimedone trapping |
| 88: Direct analysis | |
| S-Nitrosylation | 53: Biotin switch |
| 47: Biotin switch with copper | |
| 78: Direct analysis | |
| Total quantitation of disulfide bonding in proteome | 5 |
Footnotes
* This work was supported by grants from the National Cancer Institute (CA138308 to J. M. H.) and the Geroscience Mass Spectrometry and Imaging Core (PL1 AG032118 to B. W. G.).
1 The abbreviations used are:
- ZF
- zinc finger
- IBTP
- (4-iodobutyl)triphenylphosphonium
- ROS
- reactive oxygen species
- MRM
- Multiple reaction monitoring
- PTP1b
- Protein tyrosine phosphatase 1b
- IAM
- iodoacetamide
- NEM
- N-ethymaleimide
- DTT
- dithiotreitol.
REFERENCES
- 1. Mannick J. B., Hausladen A., Liu L., Hess D. T., Zeng M., Miao Q. X., Kane L. S., Gow A. J., Stamler J. S. (1999) Fas-induced caspase denitrosylation. Science 284, 651–654 [DOI] [PubMed] [Google Scholar]
- 2. Benhar M., Forrester M. T., Hess D. T., Stamler J. S. (2008) Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science 320, 1050–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Robbins J. B., Smith D., Belfort M. (2011) Redox-responsive zinc finger fidelity switch in homing endonuclease and intron promiscuity in oxidative stress. Curr. Biol. 21, 243–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Giles N. M., Giles G. I., Jacob C. (2003) Multiple roles of cysteine in biocatalysis. Biochem. Biophys. Res. Commun. 300, 1–4 [DOI] [PubMed] [Google Scholar]
- 5. Hansen R. E., Roth D., Winther J. R. (2009) Quantifying the global cellular thiol-disulfide status. Proc. Natl. Acad. Sci. U.S.A. 106, 422–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mesecke N., Terziyska N., Kozany C., Baumann F., Neupert W., Hell K., Herrmann J. M. (2005) A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell 121, 1059–1069 [DOI] [PubMed] [Google Scholar]
- 7. Tu B. P., Weissman J. S. (2002) The FAD- and O(2)-dependent reaction cycle of Ero1-mediated oxidative protein folding in the endoplasmic reticulum. Mol. Cell 10, 983–994 [DOI] [PubMed] [Google Scholar]
- 8. Bielski B. H., Shiue G. G. (1978) Reaction rates of superoxide radicals with the essential amino acids. Ciba Found Symp 65, 43–56 [DOI] [PubMed] [Google Scholar]
- 9. Peskin A. V., Winterbourn C. C. (2001) Kinetics of the reactions of hypochlorous acid and amino acid chloramines with thiols, methionine, and ascorbate. Free Radic. Biol. Med. 30, 572–579 [DOI] [PubMed] [Google Scholar]
- 10. Pryor W. A., Giamalva D. H., Church D. F. (1984) Kinetics of ozonation. 2. Amino acids and model compounds in water and comparisons to rates in nonpolar solvents. J. Am. Chem. Soc. 106, 7094–7100 [Google Scholar]
- 11. Luo D., Smith S. W., Anderson B. D. (2005) Kinetics and mechanism of the reaction of cysteine and hydrogen peroxide in aqueous solution. J. Pharm. Sci. 94, 304–316 [DOI] [PubMed] [Google Scholar]
- 12. Radi R., Beckman J. S., Bush K. M., Freeman B. A. (1991) Peroxynitrite oxidation of sulfhydryls. The cytotoxic potential of superoxide and nitric oxide. J. Biol. Chem. 266, 4244–4250 [PubMed] [Google Scholar]
- 13. Pattison D. I., Davies M. J. (2004) Kinetic analysis of the reactions of hypobromous acid with protein components: implications for cellular damage and use of 3-bromotyrosine as a marker of oxidative stress. Biochemistry 43, 4799–4809 [DOI] [PubMed] [Google Scholar]
- 14. Land E. J., Prütz W. A. (1979) Reaction of azide radicals with amino acids and proteins. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 36, 75–83 [DOI] [PubMed] [Google Scholar]
- 15. Adams G. E., Aldrich J. E., Bisby R. H., Cundall R. B., Redpath J. L., Willson R. L. (1972) Selective free radical reactions with proteins and enzymes: reactions of inorganic radical anions with amino acids. Radiat. Res. 49, 278–289 [PubMed] [Google Scholar]
- 16. Nucifora G., Smaller B., Remko R., Avery E. C. (1972) Transient radicals of DNA bases by pulse radiolysis. Effects of cysteine and cysteamine as radioprotectors. Radiat. Res. 49, 96–111 [PubMed] [Google Scholar]
- 17. Devasagayam T. P., Sundquist A. R., Di Mascio P., Kaiser S., Sies H. (1991) Activity of thiols as singlet molecular oxygen quenchers. J. Photochem. Photobiol. B 9, 105–116 [DOI] [PubMed] [Google Scholar]
- 18. Rhee S. G. (2006) Cell signaling. H2O2, a necessary evil for cell signaling. Science 312, 1882–1883 [DOI] [PubMed] [Google Scholar]
- 19. Janssen-Heininger Y. M., Mossman B. T., Heintz N. H., Forman H. J., Kalyanaraman B., Finkel T., Stamler J. S., Rhee S. G., van der Vliet A. (2008) Redox-based regulation of signal transduction: principles, pitfalls, and promises. Free Radic. Biol. Med. 45, 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Alvarez B., Ferrer-Sueta G., Freeman B. A., Radi R. (1999) Kinetics of peroxynitrite reaction with amino acids and human serum albumin. J. Biol. Chem. 274, 842–848 [DOI] [PubMed] [Google Scholar]
- 21. Pattison D. I., Davies M. J. (2001) Absolute rate constants for the reaction of hypochlorous acid with protein side chains and peptide bonds. Chem. Res. Toxicol. 14, 1453–1464 [DOI] [PubMed] [Google Scholar]
- 22. Davies M. J. (2004) Reactive species formed on proteins exposed to singlet oxygen. Photochem. Photobiol. Sci. 3, 17–25 [DOI] [PubMed] [Google Scholar]
- 23. Davies M. J. (2005) The oxidative environment and protein damage. Biochim. Biophys. Acta 1703, 93–109 [DOI] [PubMed] [Google Scholar]
- 24. Requejo R., Hurd T. R., Costa N. J., Murphy M. P. (2010) Cysteine residues exposed on protein surfaces are the dominant intramitochondrial thiol and may protect against oxidative damage. FEBS J. 277, 1465–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mustafa A. K., Gadalla M. M., Sen N., Kim S., Mu W., Gazi S. K., Barrow R. K., Yang G., Wang R., Snyder S. H. (2009) H2S signals through protein S-sulfhydration. Sci. Signal. 2, ra72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Biteau B., Labarre J., Toledano M. B. (2003) ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature 425, 980–984 [DOI] [PubMed] [Google Scholar]
- 27. Woo H. A., Chae H. Z., Hwang S. C., Yang K. S., Kang S. W., Kim K., Rhee S. G. (2003) Reversing the inactivation of peroxiredoxins caused by cysteine sulfinic acid formation. Science 300, 653–656 [DOI] [PubMed] [Google Scholar]
- 28. Kehrer J. P., Lund L. G. (1994) Cellular reducing equivalents and oxidative stress. Free Radic. Biol. Med. 17, 65–75 [DOI] [PubMed] [Google Scholar]
- 29. Winterbourn C. C. (2008) Reconciling the chemistry and biology of reactive oxygen species. Nat. Chem. Biol. 4, 278–286 [DOI] [PubMed] [Google Scholar]
- 30. Marino S. M., Gladyshev V. N. (2011) Redox biology: computational approaches to the investigation of functional cysteine residues. Antioxid. Redox. Signal. 15, 135–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Winterbourn C. C., Hampton M. B. (2008) Thiol chemistry and specificity in redox signaling. Free Radic. Biol. Med. 45, 549–561 [DOI] [PubMed] [Google Scholar]
- 32. Delaunay A., Pflieger D., Barrault M. B., Vinh J., Toledano M. B. (2002) A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell 111, 471–481 [DOI] [PubMed] [Google Scholar]
- 33. Vivancos A. P., Castillo E. A., Biteau B., Nicot C., Ayté J., Toledano M. B., Hidalgo E. (2005) A cysteine-sulfinic acid in peroxiredoxin regulates H2O2-sensing by the antioxidant Pap1 pathway. Proc. Natl. Acad. Sci. U.S.A. 102, 8875–8880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gan Z. R., Wells W. W. (1987) Identification and reactivity of the catalytic site of pig liver thioltransferase. J. Biol. Chem. 262, 6704–6707 [PubMed] [Google Scholar]
- 35. Grauschopf U., Winther J. R., Korber P., Zander T., Dallinger P., Bardwell J. C. (1995) Why is DsbA such an oxidizing disulfide catalyst? Cell 83, 947–955 [DOI] [PubMed] [Google Scholar]
- 36. Pérez V. I., Bokov A., Van Remmen H., Mele J., Ran Q., Ikeno Y., Richardson A. (2009) Is the oxidative stress theory of aging dead? Biochim. Biophys. Acta 1790, 1005–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li Y., Huang T. T., Carlson E. J., Melov S., Ursell P. C., Olson J. L., Noble L. J., Yoshimura M. P., Berger C., Chan P. H., Wallace D. C., Epstein C. J. (1995) Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase. Nat. Genet. 11, 376–381 [DOI] [PubMed] [Google Scholar]
- 38. Shefner J. M., Reaume A. G., Flood D. G., Scott R. W., Kowall N. W., Ferrante R. J., Siwek D. F., Upton-Rice M., Brown R. H., Jr. (1999) Mice lacking cytosolic copper/zinc superoxide dismutase display a distinctive motor axonopathy. Neurology 53, 1239–1246 [DOI] [PubMed] [Google Scholar]
- 39. Flood D. G., Reaume A. G., Gruner J. A., Hoffman E. K., Hirsch J. D., Lin Y. G., Dorfman K. S., Scott R. W. (1999) Hindlimb motor neurons require Cu/Zn superoxide dismutase for maintenance of neuromuscular junctions. Am. J. Pathol. 155, 663–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. López-Lázaro M. (2007) Dual role of hydrogen peroxide in cancer: possible relevance to cancer chemoprevention and therapy. Cancer Lett. 252, 1–8 [DOI] [PubMed] [Google Scholar]
- 41. Meza J. E., Scott G. K., Benz C. C., Baldwin M. A. (2003) Essential cysteine-alkylation strategies to monitor structurally altered estrogen receptor as found in oxidant-stressed breast cancers. Anal. Biochem. 320, 21–31 [DOI] [PubMed] [Google Scholar]
- 42. Schilling B., Yoo C. B., Collins C. J., Gibson B. W. (2004) Determining cysteine oxidation status using differential alkylation. Int. J. Mass Spectrom. 236, 117–127 [Google Scholar]
- 43. Zander T., Phadke N. D., Bardwell J. C. (1998) Disulfide bond catalysts in Escherichia coli. Methods Enzymol. 290, 59–74 [DOI] [PubMed] [Google Scholar]
- 44. Delaunay A., Isnard A. D., Toledano M. B. (2000) H2O2 sensing through oxidation of the Yap1 transcription factor. EMBO J. 19, 5157–5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rogers L. K., Leinweber B. L., Smith C. V. (2006) Detection of reversible protein thiol modifications in tissues. Anal. Biochem. 358, 171–184 [DOI] [PubMed] [Google Scholar]
- 46. Barradas R. G., Fletcher S., Porter J. D. (1976) The hydrolysis of maleimide in alkaline solution. Can. J. Chem. 54, 1400 [Google Scholar]
- 47. Wang X., Kettenhofen N. J., Shiva S., Hogg N., Gladwin M. T. (2008) Copper dependence of the biotin switch assay: modified assay for measuring cellular and blood nitrosated proteins. Free Radic. Biol. Med. 44, 1362–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang H., Xian M. (2011) Chemical methods to detect S-nitrosation. Curr. Opin. Chem. Biol. 15, 32–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bechtold E., Reisz J. A., Klomsiri C., Tsang A. W., Wright M. W., Poole L. B., Furdui C. M., King S. B. (2010) Water-soluble triarylphosphines as biomarkers for protein S-nitrosation. ACS Chem. Biol. 5, 405–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Saurin A. T., Neubert H., Brennan J. P., Eaton P. (2004) Widespread sulfenic acid formation in tissues in response to hydrogen peroxide. Proc. Natl. Acad. Sci. U.S.A. 101, 17982–17987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lind C., Gerdes R., Hamnell Y., Schuppe-Koistinen I., von Löwenhielm H. B., Holmgren A., Cotgreave I. A. (2002) Identification of S-glutathionylated cellular proteins during oxidative stress and constitutive metabolism by affinity purification and proteomic analysis. Arch. Biochem. Biophys. 406, 229–240 [DOI] [PubMed] [Google Scholar]
- 52. Reynaert N. L., Ckless K., Guala A. S., Wouters E. F., van der Vliet A., Janssen-Heininger Y. M. (2006) In situ detection of S-glutathionylated proteins following glutaredoxin-1 catalyzed cysteine derivatization. Biochim. Biophys. Acta 1760, 380–387 [DOI] [PubMed] [Google Scholar]
- 53. Jaffrey S. R., Erdjument-Bromage H., Ferris C. D., Tempst P., Snyder S. H. (2001) Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat. Cell. Biol. 3, 193–197 [DOI] [PubMed] [Google Scholar]
- 54. Gladwin M. T., Wang X., Hogg N. (2006) Methodological vexation about thiol oxidation versus S-nitrosation – a commentary on “An ascorbate-dependent artifact that interferes with the interpretation of the biotin-switch assay”. Free Radic. Biol. Med. 41, 557–561 [DOI] [PubMed] [Google Scholar]
- 55. Camerini S., Polci M. L., Restuccia U., Usuelli V., Malgaroli A., Bachi A. (2007) A novel approach to identify proteins modified by nitric oxide: the HIS-TAG switch method. J. Proteome Res. 6, 3224–3231 [DOI] [PubMed] [Google Scholar]
- 56. Forrester M. T., Thompson J. W., Foster M. W., Nogueira L., Moseley M. A., Stamler J. S. (2009) Proteomic analysis of S-nitrosylation and denitrosylation by resin-assisted capture. Nat Biotechnol. 27, 557–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hurd T. R., Prime T. A., Harbour M. E., Lilley K. S., Murphy M. P. (2007) Detection of reactive oxygen species-sensitive thiol proteins by redox difference gel electrophoresis: implications for mitochondrial redox signaling. J. Biol. Chem. 282, 22040–22051 [DOI] [PubMed] [Google Scholar]
- 58. Riederer I. M., Riederer B. M. (2007) Differential protein labeling with thiol-reactive infrared DY-680 and DY-780 maleimides and analysis by two-dimensional gel electrophoresis. Proteomics 7, 1753–1756 [DOI] [PubMed] [Google Scholar]
- 59. Le Moan N., Tacnet F., Toledano M. B. (2008) Protein-thiol oxidation, from single proteins to proteome-wide analyses. Methods Mol. Biol. 476, 181–198 [PubMed] [Google Scholar]
- 60. Chouchani E. T., Hurd T. R., Nadtochiy S. M., Brookes P. S., Fearnley I. M., Lilley K. S., Smith R. A., Murphy M. P. (2010) Identification of S-nitrosated mitochondrial proteins by S-nitrosothiol difference in gel electrophoresis (SNO-DIGE): implications for the regulation of mitochondrial function by reversible S-nitrosation. Biochem. J. 430, 49–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Leichert L. I., Jakob U. (2004) Protein thiol modifications visualized in vivo. PLoS Biol. 2, e333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gygi S. P., Rist B., Gerber S. A., Turecek F., Gelb M. H., Aebersold R. (1999) Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 17, 994–999 [DOI] [PubMed] [Google Scholar]
- 63. Sethuraman M., McComb M. E., Huang H., Huang S., Heibeck T., Costello C. E., Cohen R. A. (2004) Isotope-coded affinity tag (ICAT) approach to redox proteomics: identification and quantitation of oxidant-sensitive cysteine thiols in complex protein mixtures. J. Proteome Res. 3, 1228–1233 [DOI] [PubMed] [Google Scholar]
- 64. Sethuraman M., McComb M. E., Heibeck T., Costello C. E., Cohen R. A. (2004) Isotope-coded affinity tag approach to identify and quantify oxidant-sensitive protein thiols. Mol. Cell. Proteomics 3, 273–278 [DOI] [PubMed] [Google Scholar]
- 65. Fu C., Hu J., Liu T., Ago T., Sadoshima J., Li H. (2008) Quantitative analysis of redox-sensitive proteome with DIGE and ICAT. J. Proteome Res. 7, 3789–3802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Leichert L. I., Gehrke F., Gudiseva H. V., Blackwell T., Ilbert M., Walker A. K., Strahler J. R., Andrews P. C., Jakob U. (2008) Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc. Natl. Acad. Sci. U.S.A. 105, 8197–8202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kumsta C., Thamsen M., Jakob U. (2011) Effects of oxidative stress on behavior, physiology, and the redox thiol proteome of Caenorhabditis elegans. Antioxid. Redox. Signal 14, 1023–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kozarova A., Sliskovic I., Mutus B., Simon E. S., Andrews P. C., Vacratsis P. O. (2007) Identification of redox sensitive thiols of protein disulfide isomerase using isotope coded affinity technology and mass spectrometry. J. Am. Soc. Mass Spectrom. 18, 260–269 [DOI] [PubMed] [Google Scholar]
- 69. Sethuraman M., Clavreul N., Huang H., McComb M. E., Costello C. E., Cohen R. A. (2007) Quantification of oxidative posttranslational modifications of cysteine thiols of p21ras associated with redox modulation of activity using isotope-coded affinity tags and mass spectrometry. Free Radic. Biol. Med. 42, 823–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yi L., Jenkins P. M., Leichert L. I., Jakob U., Martens J. R., Ragsdale S. W. (2009) Heme regulatory motifs in heme oxygenase-2 form a thiol/disulfide redox switch that responds to the cellular redox state. J. Biol. Chem. 284, 20556–20561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Held J. M., Danielson S. R., Behring J. B., Atsriku C., Britton D. J., Puckett R. L., Schilling B., Campisi J., Benz C. C., Gibson B. W. (2010) Targeted quantitation of site-specific cysteine oxidation in endogenous proteins using a differential alkylation and multiple reaction monitoring mass spectrometry approach. Mol. Cell. Proteomics 9, 1400–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Danielson S. R., Held J. M., Oo M., Riley R., Gibson B. W., Andersen J. K. (2011) Quantitative mapping of reversible mitochondrial Complex I cysteine oxidation in a Parkinson disease mouse model. J. Biol. Chem. 286, 7601–7608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. MacLean B., Tomazela D. M., Shulman N., Chambers M., Finney G. L., Frewen B., Kern R., Tabb D. L., Liebler D. C., MacCoss M. J. (2010) Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Whiteaker J. R., Zhao L., Abbatiello S. E., Burgess M., Kuhn E., Lin C., Pope M. E., Razavi M., Anderson N. L., Pearson T. W., Carr S. A., Paulovich A. G. (2011) Evaluation of Large Scale Quantitative Proteomic Assay Development Using Peptide Affinity-based Mass Spectrometry. Mol. Cell. Proteomics 10, M110.005645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Anderson L., Hunter C. L. (2006) Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol. Cell. Proteomics 5, 573–588 [DOI] [PubMed] [Google Scholar]
- 76. Riley T., Sontag E., Chen P., Levine A. (2008) Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Biol. 9, 402–412 [DOI] [PubMed] [Google Scholar]
- 77. Hansen J. M., Go Y. M., Jones D. P. (2006) Nuclear and mitochondrial compartmentation of oxidative stress and redox signaling. Annu. Rev. Pharmacol. Toxicol. 46, 215–234 [DOI] [PubMed] [Google Scholar]
- 78. Chen Y. Y., Chu H. M., Pan K. T., Teng C. H., Wang D. L., Wang A. H., Khoo K. H., Meng T. C. (2008) Cysteine S-nitrosylation protects protein-tyrosine phosphatase 1B against oxidation-induced permanent inactivation. J. Biol. Chem. 283, 35265–35272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kim J. R., Yoon H. W., Kwon K. S., Lee S. R., Rhee S. G. (2000) Identification of proteins containing cysteine residues that are sensitive to oxidation by hydrogen peroxide at neutral pH. Anal. Biochem. 283, 214–221 [DOI] [PubMed] [Google Scholar]
- 80. Murphy M. P. (2008) Targeting lipophilic cations to mitochondria. Biochim. Biophys. Acta 1777, 1028–1031 [DOI] [PubMed] [Google Scholar]
- 81. Lin T. K., Hughes G., Muratovska A., Blaikie F. H., Brookes P. S., Darley-Usmar V., Smith R. A., Murphy M. P. (2002) Specific modification of mitochondrial protein thiols in response to oxidative stress: a proteomics approach. J. Biol. Chem. 277, 17048–17056 [DOI] [PubMed] [Google Scholar]
- 82. Reddie K. G., Carroll K. S. (2008) Expanding the functional diversity of proteins through cysteine oxidation. Curr. Opin. Chem. Biol. 12, 746–754 [DOI] [PubMed] [Google Scholar]
- 83. Charles R. L., Schröder E., May G., Free P., Gaffney P. R., Wait R., Begum S., Heads R. J., Eaton P. (2007) Protein sulfenation as a redox sensor: proteomics studies using a novel biotinylated dimedone analogue. Mol. Cell. Proteomics 6, 1473–1484 [DOI] [PubMed] [Google Scholar]
- 84. Poole L. B., Zeng B. B., Knaggs S. A., Yakubu M., King S. B. (2005) Synthesis of chemical probes to map sulfenic acid modifications on proteins. Bioconjug. Chem. 16, 1624–1628 [DOI] [PubMed] [Google Scholar]
- 85. Reddie K. G., Seo Y. H., Muse Iii W. B., Leonard S. E., Carroll K. S. (2008) A chemical approach for detecting sulfenic acid-modified proteins in living cells. Mol. Biosyst. 4, 521–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Leonard S. E., Reddie K. G., Carroll K. S. (2009) Mining the thiol proteome for sulfenic acid modifications reveals new targets for oxidation in cells. ACS Chem. Biol. 4, 783–799 [DOI] [PubMed] [Google Scholar]
- 87. Seo Y. H., Carroll K. S. (2011) Quantification of protein sulfenic acid modifications using isotope-coded dimedone and iododimedone. Angew Chem. Int. Ed. Engl. 50, 1342–1345 [DOI] [PubMed] [Google Scholar]
- 88. Shetty V., Spellman D. S., Neubert T. A. (2007) Characterization by tandem mass spectrometry of stable cysteine sulfenic acid in a cysteine switch peptide of matrix metalloproteinases. J. Am. Soc. Mass Spectrom. 18, 1544–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Seo Y. H., Carroll K. S. (2009) Profiling protein thiol oxidation in tumor cells using sulfenic acid-specific antibodies. Proc. Natl. Acad. Sci. U.S.A. 106, 16163–16168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Yusuf M. A., Chuang T., Bhat G. J., Srivenugopal K. S. (2010) Cys-141 glutathionylation of human p53: Studies using specific polyclonal antibodies in cancer samples and cell lines. Free Radic. Biol. Med. 49, 908–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Brennan J. P., Miller J. I., Fuller W., Wait R., Begum S., Dunn M. J., Eaton P. (2006) The utility of N,N-biotinyl glutathione disulfide in the study of protein S-glutathiolation. Mol. Cell. Proteomics 5, 215–225 [DOI] [PubMed] [Google Scholar]
- 92. Sullivan D. M., Wehr N. B., Fergusson M. M., Levine R. L., Finkel T. (2000) Identification of oxidant-sensitive proteins: TNF-alpha induces protein glutathiolation. Biochemistry 39, 11121–11128 [DOI] [PubMed] [Google Scholar]
- 93. Baillie T. A., Davis M. R. (1993) Mass spectrometry in the analysis of glutathione conjugates. Biol Mass Spectrom 22, 319–325 [DOI] [PubMed] [Google Scholar]
- 94. Wen B., Fitch W. L. (2009) Screening and characterization of reactive metabolites using glutathione ethyl ester in combination with Q-trap mass spectrometry. J. Mass Spectrom. 44, 90–100 [DOI] [PubMed] [Google Scholar]
- 95. Yan Z., Caldwell G. W. (2004) Stable-isotope trapping and high-throughput screenings of reactive metabolites using the isotope MS signature. Anal. Chem. 76, 6835–6847 [DOI] [PubMed] [Google Scholar]
- 96. Mutlib A., Lam W., Atherton J., Chen H., Galatsis P., Stolle W. (2005) Application of stable isotope labeled glutathione and rapid scanning mass spectrometers in detecting and characterizing reactive metabolites. Rapid Commun. Mass Spectrom. 19, 3482–3492 [DOI] [PubMed] [Google Scholar]
- 97. Sun J., Xin C., Eu J. P., Stamler J. S., Meissner G. (2001) Cysteine-3635 is responsible for skeletal muscle ryanodine receptor modulation by NO. Proc. Natl. Acad. Sci. U.S.A. 98, 11158–11162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Gow A. J., Chen Q., Hess D. T., Day B. J., Ischiropoulos H., Stamler J. S. (2002) Basal and stimulated protein S-nitrosylation in multiple cell types and tissues. J. Biol. Chem. 277, 9637–9640 [DOI] [PubMed] [Google Scholar]
- 99. Lane P., Hao G., Gross S. S. (2001) S-nitrosylation is emerging as a specific and fundamental posttranslational protein modification: head-to-head comparison with O-phosphorylation. Sci. STKE 2001, re1. [DOI] [PubMed] [Google Scholar]
- 100. Benhar M., Forrester M. T., Stamler J. S. (2009) Protein denitrosylation: enzymatic mechanisms and cellular functions. Nat. Rev. Mol. Cell Biol. 10, 721–732 [DOI] [PubMed] [Google Scholar]
- 101. Obin M., Shang F., Gong X., Handelman G., Blumberg J., Taylor A. (1998) Redox regulation of ubiquitin-conjugating enzymes: mechanistic insights using the thiol-specific oxidant diamide. FASEB J. 12, 561–569 [DOI] [PubMed] [Google Scholar]
- 102. Zhang D. D., Lo S. C., Cross J. V., Templeton D. J., Hannink M. (2004) Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol. Cell. Biol. 24, 10941–10953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Yao D., Gu Z., Nakamura T., Shi Z. Q., Ma Y., Gaston B., Palmer L. A., Rockenstein E. M., Zhang Z., Masliah E., Uehara T., Lipton S. A. (2004) Nitrosative stress linked to sporadic Parkinson's disease: S-nitrosylation of parkin regulates its E3 ubiquitin ligase activity. Proc. Natl. Acad. Sci. U.S.A. 101, 10810–10814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Bossis G., Melchior F. (2006) Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol. Cell 21, 349–357 [DOI] [PubMed] [Google Scholar]
- 105. Okumura F., Lenschow D. J., Zhang D. E. (2008) Nitrosylation of ISG15 prevents the disulfide bond-mediated dimerization of ISG15 and contributes to effective ISGylation. J. Biol. Chem. 283, 24484–24488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Xu Z., Lam L. S., Lam L. H., Chau S. F., Ng T. B., Au S. W. (2008) Molecular basis of the redox regulation of SUMO proteases: a protective mechanism of intermolecular disulfide linkage against irreversible sulfhydryl oxidation. FASEB J. 22, 127–137 [DOI] [PubMed] [Google Scholar]
- 107. Ago T., Liu T., Zhai P., Chen W., Li H., Molkentin J. D., Vatner S. F., Sadoshima J. (2008) A redox-dependent pathway for regulating class II HDACs and cardiac hypertrophy. Cell 133, 978–993 [DOI] [PubMed] [Google Scholar]
- 108. Diao Y., Liu W., Wong C. C., Wang X., Lee K., Cheung P. Y., Pan L., Xu T., Han J., Yates J. R., 3rd, Zhang M., Wu Z. (2010) Oxidation-induced intramolecular disulfide bond inactivates mitogen-activated protein kinase kinase 6 by inhibiting ATP binding. Proc. Natl. Acad. Sci. U.S.A. 107, 20974–20979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Kamata H., Honda S., Maeda S., Chang L., Hirata H., Karin M. (2005) Reactive oxygen species promote TNFalpha-induced death and sustained JNK activation by inhibiting MAP kinase phosphatases. Cell 120, 649–661 [DOI] [PubMed] [Google Scholar]
- 110. Lee S. R., Kwon K. S., Kim S. R., Rhee S. G. (1998) Reversible inactivation of protein-tyrosine phosphatase 1B in A431 cells stimulated with epidermal growth factor. J. Biol. Chem. 273, 15366–15372 [DOI] [PubMed] [Google Scholar]
- 111. Lee S. R., Yang K. S., Kwon J., Lee C., Jeong W., Rhee S. G. (2002) Reversible inactivation of the tumor suppressor PTEN by H2O2. J. Biol. Chem. 277, 20336–20342 [DOI] [PubMed] [Google Scholar]
- 112. Witze E. S., Old W. M., Resing K. A., Ahn N. G. (2007) Mapping protein post-translational modifications with mass spectrometry. Nat. Methods 4, 798–806 [DOI] [PubMed] [Google Scholar]
- 113. Wohlschlegel J. A. (2009) Identification of SUMO-conjugated proteins and their SUMO attachment sites using proteomic mass spectrometry. Methods Mol. Biol. 497, 33–49 [DOI] [PubMed] [Google Scholar]
- 114. Dansen T. B., Smits L. M., van Triest M. H., de Keizer P. L., van Leenen D., Koerkamp M. G., Szypowska A., Meppelink A., Brenkman A. B., Yodoi J., Holstege F. C., Burgering B. M. (2009) Redox-sensitive cysteines bridge p300/CBP-mediated acetylation and FoxO4 activity. Nat. Chem. Biol. 5, 664–672 [DOI] [PubMed] [Google Scholar]
- 115. Clark K. L., Oelke A., Johnson M. E., Eilert K. D., Simpson P. C., Todd S. C. (2004) CD81 associates with 14–3-3 in a redox-regulated palmitoylation-dependent manner. J. Biol. Chem. 279, 19401–19406 [DOI] [PubMed] [Google Scholar]
- 116. Kim S. O., Merchant K., Nudelman R., Beyer W. F., Jr., Keng T., DeAngelo J., Hausladen A., Stamler J. S. (2002) OxyR: a molecular code for redox-related signaling. Cell 109, 383–396 [DOI] [PubMed] [Google Scholar]
- 117. Andziak B., O'Connor T. P., Qi W., DeWaal E. M., Pierce A., Chaudhuri A. R., Van Remmen H., Buffenstein R. (2006) High oxidative damage levels in the longest-living rodent, the naked mole-rat. Aging Cell 5, 463–471 [DOI] [PubMed] [Google Scholar]
- 118. Labinskyy N., Csiszar A., Orosz Z., Smith K., Rivera A., Buffenstein R., Ungvari Z. (2006) Comparison of endothelial function, O2-* and H2O2 production, and vascular oxidative stress resistance between the longest-living rodent, the naked mole rat, and mice. Am. J. Physiol. Heart Circ Physiol 291, H2698–2704 [DOI] [PubMed] [Google Scholar]
- 119. Pérez V. I., Buffenstein R., Masamsetti V., Leonard S., Salmon A. B., Mele J., Andziak B., Yang T., Edrey Y., Friguet B., Ward W., Richardson A., Chaudhuri A. (2009) Protein stability and resistance to oxidative stress are determinants of longevity in the longest-living rodent, the naked mole-rat. Proc. Natl. Acad. Sci. U.S.A. 106, 3059–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]