Abstract
Targeted proteomics via selected reaction monitoring is a powerful mass spectrometric technique affording higher dynamic range, increased specificity and lower limits of detection than other shotgun mass spectrometry methods when applied to proteome analyses. However, it involves selective measurement of predetermined analytes, which requires more preparation in the form of selecting appropriate signatures for the proteins and peptides that are to be targeted. There is a growing number of software programs and resources for selecting optimal transitions and the instrument settings used for the detection and quantification of the targeted peptides, but the exchange of this information is hindered by a lack of a standard format. We have developed a new standardized format, called TraML, for encoding transition lists and associated metadata. In addition to introducing the TraML format, we demonstrate several implementations across the community, and provide semantic validators, extensive documentation, and multiple example instances to demonstrate correctly written documents. Widespread use of TraML will facilitate the exchange of transitions, reduce time spent handling incompatible list formats, increase the reusability of previously optimized transitions, and thus accelerate the widespread adoption of targeted proteomics via selected reaction monitoring.
Targeted proteomics using selected reaction monitoring (SRM)1 (also referred to as multiple reaction monitoring (MRM)) is a powerful technique that is widely used to quantify small molecules in complex matrices. More recently introduced in proteomics, it supports the identification and quantification of predetermined sets of peptides in complex samples, with a low limit of detection, wide dynamic range, high reproducibility and minimal redundancy (1, 2). For this technique, a specific mass spectrometric assay has to be developed once for each protein. Such assays are typically characterized by the identity of the analyte (i.e. peptide amino acid sequence), the parent ion m/z value, the approximate expected retention time of the targeted peptides, and the m/z and relative signal intensity of product ions that are specifically associated with each precursor ion. These measures, if detected, uniquely identify the targeted peptide in a complex sample. The assays are generally optimized with respect to their fragmentation pattern with the background matrix of the sample origin (i.e. plasma or cellular lysate). SRM assays can also be conducted using either native protein digests to detect targeted proteotypic peptides or can be incorporated in affinity capture routines such as N-glycocapture (3) or immunoaffinity isolation (4), to decrease complex digest solutions and increase both specificity and sensitivity to levels well within the pg/ml range (5). Because these assays need to be generated only once per peptide and are increasingly publicly accessible in publications and databases, a generally accepted and transparent format for communicating SRM assays is a significant advance for this powerful targeted proteomics technology.
At present, a wide array of software tools are available to predict, select, validate and optimize transitions, such as TIQAM (6), Skyline (7), ATAQS (8), as well as commercial offerings such as MRMPilot, Pinpoint, MassHunter, and VerifyE, from AB SCIEX, Thermo Scientific, Agilent, and Waters, respectively. These tools use a variety of different, mostly tabular formats. Furthermore, emerging resources and tools for the generation and databasing of transitions such as PeptideAtlas (9, 10), SRMAtlas (11, 12), MRMaid (13), MRMaid-DB (14), GPMDB (15), PASSEL (16), and QuAD (http://proteome.moffitt.org/QUAD) also support different formats.
The Human Proteome Organization (HUPO) Proteomics Standards Initiative (PSI; (17)) has been instrumental in developing and supporting several standards for mass spectrometry data, including mzML (18, 19) for mass spectrometer output files and mzIdentML (20) for the results of proteomics data processing. Each of the PSI formats is developed with similar concepts, such as controlled vocabularies and semantic validators. They follow a rigorous approval process that ensures that PSI formats are well tested and broadly applicable.
Toward unifying the fragmented state of SRM transition list formats, and facilitating communication between resources, tools, and instruments, the HUPO PSI Mass Spectrometry Standards Working Group has developed a new standardized format, TraML, that can be used to archive, share, and manage transition lists. In the following sections we describe the basic structure of the format, several use cases, and existing software implementations.
TraML Overview
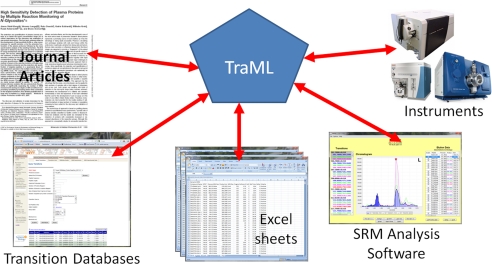
As summarized in Fig. 1, TraML is intended as a standardized format that can serve as an interchange between several components: published journal articles that include transition lists as part of their methods; transition databases such as MRMaid, MRMaid-DB, SRMAtlas, PASSEL, and QuAD that provide recommended transitions based on user input; the many existing transitions lists that are already in common use; SRM experiment design and analysis software such as ATAQS, TIQAM, Skyline, and others; and the instruments themselves via their control software. If all or most of these tools can exchange annotated transition lists in a common format, the hassle of transforming one format to another is severely reduced if not altogether eliminated.
Fig. 1.
TraML is a common, standard exchange format between published transition lists, transitions available in public or private databases, SRM design and analysis software, and instrument control software, as well as a bridge to legacy transition lists primarily stored in tab-delimited files or Excel worksheets.
TraML builds on the same design concepts that were used for mzML and mzIdentML. Like these formats previously developed for different data types, TraML is based on Extensible Markup Language (XML) and can be parsed and validated for structural correctness with many industry-standard tools. As with the other PSI formats, most of the metadata in the TraML file are encoded with the use of controlled vocabulary (CV) terms. These terms are all included in the PSI-MS CV, also used by mzML, mzIdentML, and mzQuantML and actively maintained by the PSI Mass Spectrometry Standards Working Group.
The proper use of CV terms can be validated with the PSI semantic validator (21), which uses the TraML mapping file to ensure that certain terms are used where required and that other terms are not used in semantically invalid locations in the document. An implementation of this semantic validator framework parses a TraML document to ensure well-formed XML that adheres to the XML schema definition (XSD) and also applies the rules encoded in the TraML mapping file, along with the latest (online) version of the CV, to ensure that all CV terms are properly used. There are currently two implementations of a semantic validator as described below in the “implementations” section. Links to all the auxiliary files that define the format are available at the official public TraML web page (http://www.psidev.info/traml) at the PSI web site.
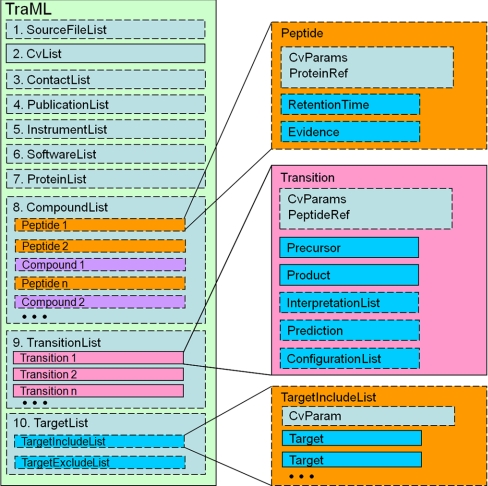
The TraML schema is organized into ten major top-level sets of information (Fig. 2), each of which can contain several levels of dependent information. The sets are numbered 1 through 10 in Fig. 2, and are described in more detail here. Element 1, <SourceFileList>, contains CV terms that allow the listing of one or more data files from which the transitions contained in the current file are derived. Element 2, <CvList>, is a required element containing a listing of the CVs referenced in the file. Note that, although the PSI-MS CV must always be listed here because every valid TraML will contain terms from this CV, additional CVs may be used to annotate the transition information in ways that are not yet supported by the PSI MS CV. Element 3, <ContactList>, provides a container to list one or more people involved in the generation, validation, and/or optimization of the transitions contained in the current file. Element 4, <PublicationList>, is a container for one or more publications from which the transitions are derived. An entire file may be the complete set of transitions from a single publication, or a merged transition set distilled from several publications into a single file with reference to the source of the individual transitions.
Fig. 2.
Schematic overview of the TraML schema. Each rectangle represents an XML element with the displayed name. Optional elements are depicted with a dashed outline. Elements can contain other dependent elements. Some of these elements are partly expanded on the right side of the diagram. The top of a TraML document contains general information about the contents of the document, and then the lists of compounds, transitions, and targets follow.
Element 5, <InstrumentList>, provides a container for specifying one or more instruments that can be referenced in the context of specifying validation and optimization information for the transitions. Element 6, <SoftwareList>, provides a container for describing software programs that were used to predict, validate, and/or optimize the transitions contained in the current file. Apart from CvList, all of these elements are optional, thus making it possible to encode very simple lists in TraML, while still allowing the option of adding rich metadata.
Following these initial 6 metadata containers is element 7, <ProteinList>, an optional list of protein identifiers that may be referenced by peptide entries. The protein entries may have accession numbers, full names, or even full sequences. Following this is element 8, <CompoundList>, which may contain any number of peptide or compound entries. A “compound” is used here to represent a biomolecule that is more generic than a peptide, allowing, for instance, the inclusion of chemical compounds and metabolites. These peptide or compound elements are then referenced in the subsequent transition or target lists.
Indeed, element 9, the <TransitionList> is encountered next. Unsurprisingly, this list forms the heart of the document. Each transition must at minimum contain the barest of information about the precursor and product m/z value, but may furthermore contain rich information about interpretations, predictions, as well as instrument configurations on which the transition has been tested or optimized. The transitions will typically reference the previously listed peptides or compounds.
Finally the optional element 10 is a general <TargetList> container, which may contain an inclusion list and/or an exclusion list. Each of these lists contains individual targets with at minimum a precursor m/z, but optionally also retention times and other attributes.
Although the format is primarily intended for the exchange of SRM transition lists, this final component was added to manage and exchange of ordinary inclusion or exclusion precursor m/z lists in product ion scans. It is expected that this is a relatively minor use case, however it is envisaged in future iterations of mass spectrometers this will become a major feature as whole proteome measurements will become more routine. There is no other suitable format for encoding such information, so it was suggested late in development that the format support this data type as well. It was considered to simply make <Transition> a more generic element that could also contain inclusion targets, but the working group decided that trying to force inclusion targets (with only a precursor m/z) into a <Transition> element would only lead to validation difficulties, and that this minor use case was therefore best left as a separate, optional component in the schema.
Use Cases
We expect TraML to be used in three primary ways: as an archival format, as an exchange format, and as a working format. For example, it can be used as an archival format to display supplemental material of journal articles. Currently, transition lists are stored in tables of varying formats, sometimes even as PDF files, from which it can be difficult to extract relevant data. If transitions are stored in an approved TraML format, any TraML-supporting software will immediately be able to read such a file, encouraging its reuse. Another important use will be for the general exchange of transition lists between labs or lab members. When one wants to share a list, it is now commonplace to send an Excel sheet of transitions, which must then often be adjusted to fit the workflow of the destination. With the emergence of public repositories of experimentally validated transitions for large numbers of proteins (10, 12, 14), we expect the need for efficient transition file exchange to increase dramatically. The exchange of transition lists is particularly important in the case of targeted proteomics as a set of transitions, once optimized, can be used perpetually. The final intended use is as a working format. Transition lists often need to undergo bulk modifications such as recalculation of retention times for the local instrumental setup, or optimization for the local instrument or specific instrument conditions, and we envision that the software tools that perform these recalculations or enhancement can use TraML as a document that undergoes active revision. It may be that individual software packages will continue to support their native formats, but the reuse of lists will be greatly enhanced by the common use of a standard format.
As the development of the format has occurred under the PSI, the primary intended use has been for proteomics experiments. However, the needs of metabolomics research, where SRM techniques have been used for far longer, have been incorporated into the format. The metadata associated with metabolomics experiments tends to be less complex than that for proteomics because the whole complexity of peptide-protein mapping can be excluded. This schema can also support similar targeted mass spectrometry for the SRM application to lipidomics given that the application of this technique provides for a similar common denominator of parent and transition mass tables. Instead of peptide sequences and protein mappings, basic compositional information and database accessions may be associated with targeted molecular compounds instead.
As noted above, the primary use case for this format is for targeted mass spectrometry SRM assays, for which transitions requiring both precursor m/z and product m/z are the key components. In addition, ordinary inclusion and exclusion lists are also supported for current and future developments in whole proteome approaches. Such lists are often employed to follow up on features detected in MS1 scans that have not yet been identified or confirmed with MS2 scans. TraML supports both inclusion lists, specifying which features to identify with fragmentation events, and exclusion lists, that specify features not to select for fragmentation in a future run. Broad sharing of inclusion or exclusion lists seems rare, but whole-proteome quantification via an inclusion list containing the top proteotypic peptides for each protein has been shown to be feasible (22) and may become a popular approach. In any case, the format can be used as a working format where inclusion lists are iteratively developed and optimized during an experimental workflow.
As a mechanism for supporting iterative workflows, various levels of confidence for a transition can be encoded in TraML, with appropriate references to the history of increasing confidence. Transitions can be marked as predicted based on some algorithm or as selected from a real MS/MS spectrum, although perhaps from a different kind of instrument. Transitions can be called “optimized” for a specific instrument model if they are based on selection from an MS/MS spectrum or chromatogram acquired with that instrument model, and “CE optimized” if the optimum collision energy is determined. Finally, a transition can be called “verified” if chromatograms have been acquired and minimal confusion with contaminating peaks is verified in the target sample. The history of such an optimization workflow can be encoded in TraML, thereby giving researchers who use the transitions the ability to assess the past history of the transitions.
An example of such an iterative workflow might occur as follows: a series of shotgun experiments are analyzed to select detectable peptides for a list of relevant proteins to create a list of candidate peptide and transition targets, and the resulting transitions written in TraML with an annotation that the transitions are selected from ion trap data. Synthetic peptides are acquired and the resulting peptides are measured via the candidate transitions on an Agilent QQQ instrument; the resulting mzML files analyzed by automated software and unsuitable transitions are discarded and a new, updated TraML file is written with verification results added via <ValidationStatus> elements. Then a collision energy optimization procedure is run to determine optimum energies for the remaining transitions, and the results are written to an updated TraML file. Finally, the selected transitions are monitored in a plasma sample to determine which transitions show unacceptable interferences with other ions in this type of sample, and the TraML file is again updated with new information in <ValidationStatus>. The final TraML file represents an optimized set of transitions and the history of their development, and it can be used to generate methods for the experiment assays and be submitted as supplemental material with a manuscript as the final transitions used in the experiment, eventually to be archived in public transition databases.
TraML is not intended to represent the results of an SRM experiment, but rather for use as the input for an SRM experiment. The direct results of an SRM experiment in the form of chromatograms can be stored in the mzML format. The quantitative measurements and subsequent statistical aggregation can be stored in a format currently being developed by the PSI, namely mzQuantML (http://www.psidev.info/mzquantml).
Implementations and Examples
TraML has already become quite mature; it has gone through several rounds of revision and refinement based on feedback from many experienced researchers from different institutions. Furthermore, instrument and software vendors actively participate in PSI and have been a part of the development of TraML. Most of the significant SRM-related software tools either already support the format, or their authors participated in the development of TraML with the intent to support the format soon.
There are already several existing software implementations of the TraML schema. This is important for several reasons. First, it means that potential users do not need to write their own software to begin using the format. Second, it is the act of implementing the format, and reading and writing real data, that provides a real-world test of the format (18). Finally, the existence of several software implementations prior to official completion of TraML indicates the need for and interest in a standard format.
The ProteoWizard project (23) aims to provide an extensive reusable C++ library as well as software applications for the analysis and manipulation of mass spectrometry data. ,bx-ProteoWizard is the reference implementation for mzML, and is distributed under a very permissive Apache 2.0 license, which allows it to be incorporated in any other software without constraints on the license of the final product. ProteoWizard now provides a set of classes for the TraML elements as well as code to read and write TraML files into memory.
The TraML schema has been implemented in an open-source Java library by the jTraML toolkit (24), also under the Apache 2.0 license. It provides a complete API for all TraML elements, along with syntactic and semantic validation support, and demonstrates the use of these classes with an on-line converter that can transform a variety of existing tab-separated-value formats to and from TraML, available at http://iomics.ugent.be/jtraml.
The OpenMS project (25) also provides a reusable set of C++ libraries for the processing and analysis of mass spectrometry data made available under the GNU Lesser Public License (LGPL). TraML support is built into the library, and tools are included for manipulating transition lists. An on-line TraML semantic validator is hosted by an OpenMS server (http://open-ms.sourceforge.net), and allows anyone to upload a TraML file and verify the validity of the file.
Skyline (7) is a C# client application for Windows that enables very flexible and user-friendly manipulation and maintenance of transition lists as well as chromatogram analysis. It is built on top of the ProteoWizard libraries and could readily derive its TraML support through ProteoWizard itself, although this has not yet been implemented.
The Automated Targeted And Quantitative System (8) (ATAQS) is a web-based collaborative system for managing an entire targeted proteomics workflow from beginning (protein and transition selection) to end (quantitative analysis of the chromatograms). Transition lists may be imported, stored, manipulated, and exported using ATAQS. Several formats, including TraML, are supported.
The SRMAtlas component (12) of the PeptideAtlas project (9, 10) provides a publicly accessible compendium of proteotypic peptides and transitions collated from several sources and specific species builds. This includes both the PeptideAtlas Transitions Resource (PATR), which stores curated lists of transitions collected from published articles, as well as community submissions. These are available for download in the native format and soon in the TraML format. Queries to the SRMAtlas compendium can be returned in several formats, soon in TraML as well.
The MRMaid-DB resource does not yet support TraML, but the MRMaid-DB journal article (14) indicates that TraML support will be forthcoming as soon as TraML is declared stable. The authors compared the MRMaid-DB database data model to the schema of an earlier development version of TraML and showed that TraML supports nearly all of the fields and concepts in their database. The only exception was the storage of coefficient of variance measures, which TraML now supports via a new controlled vocabulary term.
The Anubis software (http://www.quantitativeproteomics.org/anubis) provides a system for automated peptide quantification using SRM data. By its support for transition lists in TraML format, as well as raw MS data in mzML format, it is an example of software that can analyze data from all major instrument vendors through implementation of standards support.
Example TraML documents are available at the official TraML web page, including a hand-crafted “ToyExample” document that demonstrates the use of most elements, attributes, and CV terms. There are also examples of a real transition list for an SRM yeast experiment generated by ATAQS and a yeast inclusion list generated by the Proteios system (26, 27), which supports TraML for inclusion/exclusion lists, as well as a transition list converted by the jTraML toolkit.
CONCLUSION
We have developed the open TraML standard format for storage and exchange of SRM transitions. Along with the format, we demonstrate several initial implementations across the community, provide semantic validators and extensive documentation to ensure proper implementation, and furnish multiple example files to demonstrate correct implementations. Widespread use of TraML will facilitate the exchange of transitions, reduce time spent handling incompatible list formats, increase the reusability of previously optimized transitions, and thus accelerate the field of targeted proteomics via SRM. The format provides for rich annotation of transition lists with an extensive set of optional components. However, because these annotations are optional, very simple transition lists may also be encoded in TraML.
The PSI is currently developing a module for the Minimum Information About a Proteomics Experiment (MIAPE) (28) specification, called MIAPE-Quant. It specifies a set of minimum information that should be provided when publishing a quantitative proteomics experiment, including an SRM experiment. TraML will serve as the data format that can encode the minimum information concepts in MIAPE-Quant related to the input for a SRM experiment. All materials related to the TraML format are available at the PSI web page for this format at http://www.psidev.info/traml.
Acknowledgments
We thank the steering committee and the editors of the Proteomics Standards Initiative for the provision of the document process and feedback on the specification documents, as well as the numerous participants of the PSI Mass Spectrometry Standards Working Group. We acknowledge the contributions of our colleague Andreas Bertsch, who lost his life unexpectedly and far too early.
Footnotes
* This work has been funded in part by NIH with NHLBI under contract N01-HV-28179, NIGMS grant GM087221, NHGRI grant HG005805, EU FP7 grant ‘ProteomeXchange’ (grant number 260558), and the Systems Biology Initiative of the State of Luxembourg.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- SRM
- selected reaction monitoring
- MRM
- multiple reaction monitoring
- PSI
- Proteomics Standards Initiative
- CV
- controlled vocabulary.
REFERENCES
- 1. Domon B., Aebersold R. (2006) Mass spectrometry and protein analysis. Science 312, 212–217 [DOI] [PubMed] [Google Scholar]
- 2. Kuster B., Schirle M., Mallick P., Aebersold R. (2005) Scoring proteomes with proteotypic peptide probes. Nat. Rev. Mol. Cell Biol. 6, 577–583 [DOI] [PubMed] [Google Scholar]
- 3. Zhang H., Yi E. C., Li X. J., Mallick P., Kelly-Spratt K. S., Masselon C. D., Camp D. G., 2nd, Smith R. D., Kemp C. J., Aebersold R. (2005) High Throughput Quantitative Analysis of Serum Proteins Using Glycopeptide Capture and Liquid Chromatography Mass Spectrometry. Mol. Cell. Proteomics 4, 144–155 [DOI] [PubMed] [Google Scholar]
- 4. Anderson N. L., Anderson N. G., Haines L. R., Hardie D. B., Olafson R. W., Pearson T. W. (2004) Mass spectrometric quantitation of peptides and proteins using Stable Isotope Standards and Capture by Anti-Peptide Antibodies (SISCAPA). J Proteome Res 3, 235–244 [DOI] [PubMed] [Google Scholar]
- 5. Whiteaker J. R., Zhao L., Abbatiello S. E., Burgess M., Kuhn E., Lin C., Pope M. E., Razavi M., Anderson N. L., Pearson T. W., Carr S. A., Paulovich A. G. (2011) Evaluation of large scale quantitative proteomic assay development using peptide affinity-based mass spectrometry. Mol Cell Proteomics. M110.005645, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lange V., Malmström J. A., Didion J., King N. L., Johansson B. P., Schäfer J., Rameseder J., Wong C. H., Deutsch E. W., Brusniak M. Y., Bühlmann P., Björck L., Domon B., Aebersold R. (2008) Targeted quantitative analysis of Streptococcus pyogenes virulence factors by multiple reaction monitoring. Mol Cell Proteomics 7, 1489–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. MacLean B., Tomazela D. M., Shulman N., Chambers M., Finney G. L., Frewen B., Kern R., Tabb D. L., Liebler D. C., MacCoss M. J. (2010) Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brusniak M. Y., Kwok S. T., Christiansen M., Campbell D., Reiter L., Picotti P., Kusebauch U., Ramos H., Deutsch E. W., Chen J., Moritz R. L., Aebersold R. (2011) ATAQS: A computational software tool for high throughput transition optimization and validation for selected reaction monitoring mass spectrometry. BMC Bioinformatics 12, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Desiere F., Deutsch E. W., Nesvizhskii A. I., Mallick P., King N. L., Eng J. K., Aderem A., Boyle R., Brunner E., Donohoe S., Fausto N., Hafen E., Hood L., Katze M. G., Kennedy K. A., Kregenow F., Lee H., Lin B., Martin D., Ranish J. A., Rawlings D. J., Samelson L. E., Shiio Y., Watts J. D., Wollscheid B., Wright M. E., Yan W., Yang L., Yi E. C., Zhang H., Aebersold R. (2005) Integration with the human genome of peptide sequences obtained by high-throughput mass spectrometry. Genome Biol 6, R9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Deutsch E. W., Lam H., Aebersold R. (2008) PeptideAtlas: a resource for target selection for emerging targeted proteomics workflows. EMBO Rep 9, 429–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Picotti P., Lam H., Campbell D., Deutsch E. W., Mirzaei H., Ranish J., Domon B., Aebersold R. (2008) A database of mass spectrometric assays for the yeast proteome. Nat Methods 5, 913–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Deutsch E. W., Campbell D. S., Mendoza L., Sun Z., Farrah T., Kusebauch U., Chu C., Stevens J., Slagel J., Picotti P., Brusniak M.-Y., Lam H., Bletz J., Wang G., He W.-w., Hood L., Aebersold R., Moritz R. L. (in preparation) SRMAtlas: Generating targeted proteomics transition atlases for complete proteomes. [Google Scholar]
- 13. Mead J. A., Bianco L., Ottone V., Barton C., Kay R. G., Lilley K. S., Bond N. J., Bessant C. (2009) MRMaid, the web-based tool for designing multiple reaction monitoring (MRM) transitions. Mol Cell Proteomics 8, 696–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cham J. A., Bianco L., Barton C., Bessant C. (2010) MRMaid-DB: a repository of published SRM transitions. J Proteome Res 9, 620–625 [DOI] [PubMed] [Google Scholar]
- 15. Craig R., Cortens J. P., Beavis R. C. (2004) Open source system for analyzing, validating, and storing protein identification data. J Proteome Res 3, 1234–1242 [DOI] [PubMed] [Google Scholar]
- 16. Farrah T., Deutsch E. W., Kreisberg R., Sun Z., Campbell D. S., Mendoza L., Kusebauch U., Brusniak M.-Y., Hüttenhain R., Schiess R., Selevsek N., Aebersold R., Moritz R. L. PASSEL: The PeptideAtlas SRM Experiment Library. submitted [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Taylor C. F., Hermjakob H., Julian R. K., Jr., Garavelli J. S., Aebersold R., Apweiler R. (2006) This work of the Human Proteome Organisation's Proteomics Standards Initiative (HUPO PSI). OMICS 10, 145–151 [DOI] [PubMed] [Google Scholar]
- 18. Deutsch E. (2008) mzML: a single, unifying data format for mass spectrometer output. Proteomics 8, 2776–2777 [DOI] [PubMed] [Google Scholar]
- 19. Martens L., Chambers M., Sturm M., Kessner D., Levander F., Shofstahl J., Tang W. H., Römpp A., Neumann S., Pizarro A. D., Montecchi-Palazzi L., Tasman N., Coleman M., Reisinger F., Souda P., Hermjakob H., Binz P. A., Deutsch E. W. (2011) mzML–a community standard for mass spectrometry data. Mol Cell Proteomics 10, R110.000133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jones A. R., Eisenacher M., Mayer G., Kohlbacher O., Siepen J., Hubbard S., Selley J., Searle B., Shofstahl J., Seymour S., Julian R., Binz P. A., Deutsch E. W., Hermjakob H., Reisinger F., Griss J., Vizcaíno J. A., Chambers M., Pizarro A., Creasy D. (2012) The mzIdentML data standard for mass spectrometry-based proteomics results. Mol. Cell Proteomics. 11, M111.014381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Montecchi-Palazzi L., Kerrien S., Reisinger F., Aranda B., Jones A. R., Martens L., Hermjakob H. (2009) The PSI semantic validator: a framework to check MIAPE compliance of proteomics data. Proteomics 9, 5112–5119 [DOI] [PubMed] [Google Scholar]
- 22. Malmström J., Beck M., Schmidt A., Lange V., Deutsch E. W., Aebersold R. (2009) Proteome-wide cellular protein concentrations of the human pathogen Leptospira interrogans. Nature 460, 762–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kessner D., Chambers M., Burke R., Agus D., Mallick P. (2008) ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics 24, 2534–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Helsens K., Brusniak M. Y., Deutsch E., Moritz R. L., Martens L. (2011) jTraML: An open source java API for TraML, the PSI standard for sharing SRM transitions. J Proteome Res Epub 2011/10/05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sturm M., Bertsch A., Gröpl C., Hildebrandt A., Hussong R., Lange E., Pfeifer N., Schulz-Trieglaff O., Zerck A., Reinert K., Kohlbacher O. (2008) OpenMS - an open-source software framework for mass spectrometry. BMC Bioinformatics 9, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gärdän P., Alm R., Hakkinen J. (2005) PROTEIOS: an open source proteomics initiative. Bioinformatics 21, 2085–208715691852 [Google Scholar]
- 27. Häkkinen J., Vincic G., Månsson O., Warell K., Levander F. (2009) The proteios software environment: an extensible multiuser platform for management and analysis of proteomics data. J Proteome Res 8, 3037–3043 [DOI] [PubMed] [Google Scholar]
- 28. Taylor C. F., Paton N. W., Lilley K. S., Binz P. A., Julian R. K., Jr., Jones A. R., Zhu W., Apweiler R., Aebersold R., Deutsch E. W., Dunn M. J., Heck A. J., Leitner A., Macht M., Mann M., Martens L., Neubert T. A., Patterson S. D., Ping P., Seymour S. L., Souda P., Tsugita A., Vandekerckhove J., Vondriska T. M., Whitelegge J. P., Wilkins M. R., Xenarios I., Yates J. R., 3rd, Hermjakob H. (2007) The minimum information about a proteomics experiment (MIAPE). Nat Biotechnol 25, 887–893 [DOI] [PubMed] [Google Scholar]